Open Access Article
Open Access ArticleSingle-molecule study of the effects of temperature, pH, and RNA base on the stepwise enzyme kinetics of 10–23 deoxyribozyme†
Jiwon Jung,
Seon Yong Kim and
Seong Keun Kim *
*
Department of Chemistry, Seoul National University, Seoul 08826, Korea. E-mail: seongkim@snu.ac.kr; Fax: +82-2-889-5719; Tel: +82-2-880-6659
First published on 18th May 2022
Abstract
We investigated how the stepwise enzyme kinetics of 10–23 deoxyribozyme was affected by temperature, pH, and RNA residue of the substrate at the single-molecule level. A deoxyribozyme-substrate system was employed to temporally categorize a single-turnover reaction into four distinct steps: binding, cleavage, dissociation of one of the cleaved fragments, and dissociation of the other fragment. The dwell time of each step was measured as the temperature was varied from 26 to 34 °C, to which the transition state theory was applied to obtain the enthalpy and entropy of activation for individual steps. In addition, we found that only the cleavage step was significantly affected by pH, indicating that it involves deprotonation of a single proton. We also found that different RNA residues specifically affect the cleavage step and cause the dwell time to change by as much as 5 times.
Since the first discovery of the starch-breaking protein diastase by Anselme Payen in 1833,1 numerous types of protein enzymes have been discovered and found to play crucial roles in all life forms.2,3 Another type of biological catalysts was discovered in the 1980s in the intron of an RNA transcript,4 followed by a diverse range of kindred ribozymes discovered in nature,5–8 which have been studied at both the ensemble9 and single-molecule levels.10,11
Although DNA is structurally similar to RNA except for the 2′-hydroxyl group replaced by hydrogen, catalytic DNAs have not yet been discovered in nature. In the 1990s, Joyce and Breaker conducted in vitro selection assays and found that DNAs can function as enzymes, with these deoxyribozymes in vitro playing a role akin to ribozymes in vivo.12 Among various deoxyribozymes, the RNA-cleaving 10–23 deoxyribozyme has a high enzymatic activity13 and has been extensively studied for its chemical and biological properties in ensemble.14–19 The much higher chemical stability of DNAs over RNAs make deoxyribozymes a highly promising enzyme in their applications in medicine, sensing, and bio-engineering.20–26
Since their first optical detection at the single-molecule level,27 ribozymes and deoxyribozymes have been investigated for their single-molecule properties.10,11,28–30 In order to understand the working mechanism of ribozymes from changes in their structure, immobilization-based single-molecule FRET experiments were conducted.31 On the other hand, diffusion-based FRET assays have also been adopted to correlate enzymatic activity with enzyme structure by molecule-by-molecule sorting.28–30
Here, we present our new study on the uniquely single-molecule features of the kinetics of 10–23 deoxyribozymes that are hidden at the ensemble level. A single-turnover reaction is temporally subdivided into individual reaction steps that include substrate binding, catalytic cleavage, and sequential dissociations of cleaved substrates. We also examined how different physical and chemical conditions such as temperature and pH affect each reaction step of the enzymatic pathway. With the application of the transition state theory and using the pH-dependent data for kinetics, we were able to construct a detailed chemical model for the enzymatic cleavage step. Additionally, the RNA sequence of the substrate was varied to compare the enzymatic activity of 10–23 deoxyribozyme influenced by RNA bases.
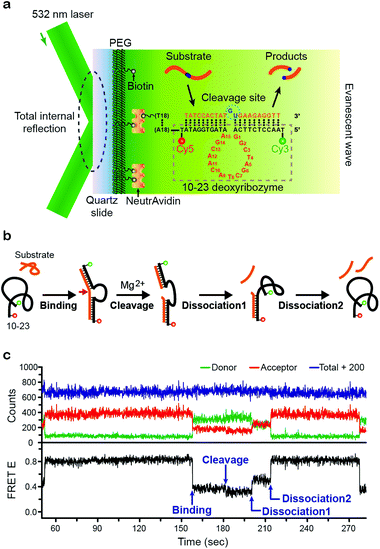
We used a homemade prism-type total internal reflection fluorescence (TIRF) microscope described in our previous work.32,33 To immobilize our enzyme, biotin-labeled 10–23 deoxyribozyme was attached to a PEG-coated quartz slide via the biotin-avidin interaction. For single-molecule FRET measurements, the Cy3–Cy5 FRET pair was labeled on each end of 10–23 deoxyribozyme, and a 532 nm laser diode was used to generate an evanescent wave. The substrate was designed to consist of an RNA-based cleavage site in the middle and two DNA-based arms that facilitated enzyme–substrate binding by complementary base pairing to form a dsDNA (Fig. 1a).
The scheme for our overall enzyme–substrate reaction is shown in Fig. 1b. Diffusing substrate in its natural coiled form binds to the immobilized 10–23 deoxyribozyme (also initially in coiled form) and forms a dsDNA structure with the two arms. In the presence of the Mg2+ cofactor in solution, 10–23 deoxyribozyme cleaves the RNA-based cleavage site of the DNA substrate. Once the substrate is cleaved, dissociation of the cleaved substrate pieces (“half-substrates”) from deoxyribozyme takes place in successive steps.
The corresponding time traces of our FRET data for the above scheme are shown in Fig. 1c. The free enzyme has a compact coiled-structure (thus with a high FRET efficiency E of ∼0.8) because of the reduced electrostatic repulsion between the negatively charged phosphate groups due to the cations in the buffer. Upon substrate binding, the complimentary base pairing results in a stretched dsDNA structure for the enzyme–substrate complex, leading to a low E value of ∼0.4. In a few tens of seconds, the cleavage event occurs that unlocks the two RNA bases in the middle of the substrate, giving more structural freedom that allows an even further stretched form of the deoxyribozyme with a smaller value of E (∼0.3). Finally, the two half-substrates dissociate from the enzyme in two steps: in the first step, only one of the half-substrates dissociates so that the enzyme is still in partially stretched form (E ∼ 0.5), but in the next step, the remaining half-substrates comes off so that it returns to its initial coiled structure (E ∼ 0.8).
As with most biological enzymes that exhibit their maximum activity at “physiological temperatures”, 10–23 deoxyribozyme shows a bell-shaped activity profile for temperature in the range of 30 to 40 °C.16 It is known that most enzymes also have their optimal range of pH that affects their chemical structure and catalytic activity.
We studied how each step of our overall kinetic scheme depends on reaction conditions such as temperature and pH at the single-molecule level. For temperature dependence, we found that the rates of all reaction steps were accelerated as the temperature was increased over the range from 26 to 34 °C under our experimental condition (pH 7.52). As for pH dependence, we varied pH from 7.3 to 7.8 at 30 °C and found that different steps were affected differently by a change in pH. Finally, we also changed the RNA base sequence of the substrate and found that the reaction rate is greatly affected by it.
In all our measurements, only the kinetic rates of the forward reactions were measured. For example, the reverse reaction of the substrate binding step was ignored since spontaneous dissociation of the full enzyme–substrate complex was considered not likely at the thermal energy of the system. Likewise, for the cleavage and dissociation steps, we assumed that the spontaneous ligation and re-binding of the fragmented substrates that diffuse away were deemed unlikely on entropic grounds. Measured duration times of each step were turned into histogram distribution, which was fitted with a single-exponential curve to yield the reaction time and hence the rate constant for each step.
We applied the standard Eyring–Polanyi equation of the transition state theory to obtain the enthalpy, entropy, and Gibbs energy of activation. The data fit well with the equation for all reaction steps (Fig. 2 and Table 1). The substrate-binding step of 10–23 deoxyribozyme involves sequential binding of its two arms to the substrate, with the binding of the first arm playing the rate-determining step. Table 1 shows that the activation enthalpy of binding is 17.5 kcal mol−1, which is the cost in enthalpy when a pair of fully hydrated enzyme and substrate partially sacrifice their respective hydration shell to form a hybridized enzyme–substrate complex that has a smaller degree of hydration overall in exchange for their newly formed enzyme–substrate bonds. The entropy of activation is −29.3 J mol−1 K−1, whose negative value indicates an increase of order associated with the transition from the random coil structures of the enzyme and substrate to a hybridized structure of their complex.
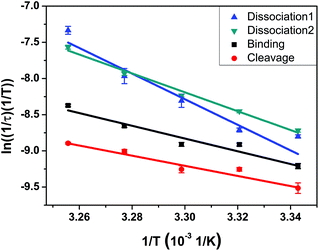 | ||
| Fig. 2 Arrhenius plot for each step of the overall reaction in the temperature range from 26 to 34 °C at pH 7.52. Optimized linear fitting parameters yield the values of enthalpy, entropy, and Gibbs free energy of activation given in Table 1. | ||
| ΔH‡ (kcal mol−1) | ΔS‡ (J mol−1 K−1) | ΔG‡ at 26 °C (kcal mol−1) | |
|---|---|---|---|
| Binding | 17.5 | −29.3 | 19.6 |
| Cleavage | 13.9 | −82.0 | 19.8 |
| Dissociation1 | 35.1 | 219.1 | 19.5 |
| Dissociation2 | 25.9 | 92.2 | 19.3 |
The cleavage step is the most important kinetic step in the 10–23 deoxyribozyme reaction. There have been many studies that investigated the RNA cleavage step by ensemble experiments, but we provide here kinetic constants from an individual cleavage step without ensemble averaging. It is notable that the entropy of activation for the cleavage step is negative (−82.0 J mol−1 K−1), indicating that it is a process of gaining order on its way to the transition state. We note that the structure of the enzyme–substrate complex has a higher coordination of bonds in the transition state, i.e., a 5-bond coordination rather than the 3- or 4-bond coordination seen at the phosphate backbone.34
Finally, for the dissociation step, the entropy of activation is a very large positive value (219.1 J mol−1 K−1) while the enthalpy of activation (35.1 kcal mol−1) is also much larger than for binding or cleavage, both of which indicate the large energy cost and the resulting increase of disorder associated with the dissociation of the enzyme–substrate complex.
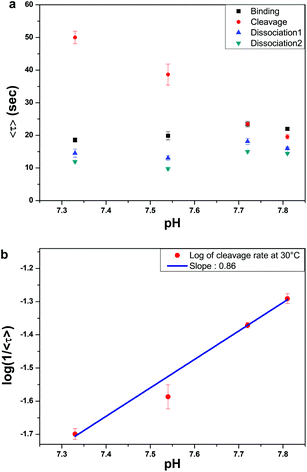
As with protein- and RNA-enzymes, pH is one of the most important governing factors for the enzymatic activity of DNA-enzymes.14 However, there has been no direct measurement of the pH-dependent reaction rates based on single-molecule measurement. In this study, we varied the pH from 7.3 to 7.8 to measure the kinetics of our 4-step enzyme reaction at the single-molecule level. Fig. 3a shows that undoing ensemble averaging allowed us to see that only the cleavage step is significantly affected by pH. The average dwell time for the cleavage step decreased drastically from 50 to 20 s as the pH increased from 7.3 to 7.8. The same data plotted in the log-linear scale (Fig. 3b) gives a slope of ∼0.86, suggesting that the rate-determining step of the cleavage reaction involves deprotonation of a single proton, which is the same conclusion drawn indirectly from earlier ensemble studies.14
In contrast to the cleavage step, the binding and the sequential dissociation steps exhibit no significant effect of pH (Fig. 3a). For renaturation and hybridization, there have been few reports on the effect of pH. In 1968, Wetmer and Davidson reported that they found no effect of pH in the range of pH 6 to 8,35 although their limited buffer conditions and poor data quality make this conclusion somewhat dubious.
Another observation of interest is that the slowest step is switched from the cleavage to the binding step at pH 7.8, as a result of the pH-independence of DNA hybridization and the pH-sensitive cleavage reaction of our enzyme reaction. In fact, at pH 7.8, all 4 dwell times lie within a factor of 2.
10–23 deoxyribozyme is a powerful enzyme that cleaves a variety of RNA substrates. Because it cleaves any type of RNA substrate that is composed of unpaired purines and paired pyrimidines, we investigated how the single RNA residue of the substrate affects the reaction rate of the four individual enzyme reaction steps. We prepared five types of the substrate with the same two arms but different RNA residues (AU, UU, GU, CU, and U) at the cleavage site and measured their average dwell time for each reaction step at 30 °C and pH 7.7. Since the RNA residue does not directly participate in the binding or dissociation reaction, we anticipated that only the cleavage step would be significantly affected. Our measurement of dwell times showed that the substrate with a GU residue is most readily cleaved. In fact, Fig. 4 show that the rate of cleavage is ∼5 times faster for the substrate with a GU residue than with an AU residue, leading to an overall turnover rate ∼2 times larger. We failed to obtain the entire reaction trace for the substrate with a CU, UU, and U residue under our experimental conditions, since their reaction rates were too slow for our limited detection time of ∼10 min imposed by the photobleaching of labeling dyes.
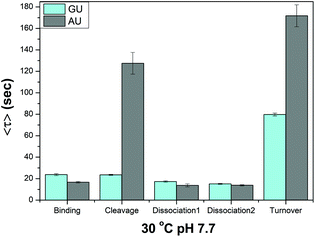 | ||
| Fig. 4 Effect of a mismatched residue in the substrate for each reaction step. Only the cleavage step is affected drastically while other steps are little affected. | ||
In conclusion, we investigated the effects of temperature, pH, and RNA base residue on the kinetics of the four distinct reaction steps of 10–23 deoxyribozyme by single-molecule detection using TIRF microscopy. With regard to the temperature dependence, both the substrate binding and cleavage steps have a negative entropy of activation, indicating that they are a process of increasing order on their way to the transition state. In contrast, for the dissociation step of the cleaved substrate fragments, the entropy of activation is a large positive number indicative of the increase in disorder accompanying the dissociation. The enthalpy of activation is likewise much larger for the substrate dissociation step than for the binding or cleavage step due to the high energy cost of the complex dissociation. With regard to the pH dependence, only the cleavage step was significantly affected by pH, whose linear power dependence suggests that the rate-determining step should involve deprotonation of a single proton. Finally, with regard to the effect of RNA residue, we found that the cleavage step is again greatly affected, with a difference in dwell time being as large as 5 times between the GU and AU residues.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea grants to S. K. K. (NRF-2018R1A2B2001422). We also acknowledge the BK21 Plus Program and SNU Brain Fusion Grant.Notes and references
- A. Payen and J. F. Persoz, Ann. Chim. Phys., 1833, 53, 73 Search PubMed.
- M. Abdelmonem, H. Durwald and H. Hoffmannberling, Eur. J. Biochem., 1976, 65, 441 CrossRef CAS PubMed.
- I. R. Lehman, M. J. Bessman, E. S. Simms and A. Kornberg, J. Biol. Chem., 1958, 233, 163 CrossRef CAS PubMed.
- K. Kruger, P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling and T. R. Cech, Cell, 1982, 31, 147 CrossRef CAS PubMed.
- B. J. Saville and R. A. Collins, Cell, 1990, 61, 685 CrossRef CAS PubMed.
- L. Sharmeen, M. Y. P. Kuo, G. Dintergottlieb and J. Taylor, J. Virol., 1988, 62, 2674 CrossRef CAS PubMed.
- M. Y. P. Kuo, L. Sharmeen, G. Dintergottlieb and J. Taylor, J. Virol., 1988, 62, 4439 CrossRef CAS PubMed.
- G. A. Prody, J. T. Bakos, J. M. Buzayan, I. R. Schneider and G. Bruening, Science, 1986, 231, 1577 CrossRef CAS PubMed.
- S. C. Dahm and O. C. Uhlenbeck, Biochemistry, 1991, 30, 9464 CrossRef CAS PubMed.
- X. W. Zhuang, L. E. Bartley, H. P. Babcock, R. Russell, T. J. Ha, D. Herschlag and S. Chu, Science, 2000, 288, 2048 CrossRef CAS PubMed.
- B. Okumus, T. J. Wilson, D. M. J. Lilley and T. Ha, Biophys. J., 2004, 87, 2798 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U.S.A., 1997, 94, 4262 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Biochemistry, 1998, 37, 13330 CrossRef CAS PubMed.
- Z. Zaborowska, J. P. Furste, V. A. Erdmann and J. Kurreck, J. Biol. Chem., 2002, 277, 40617 CrossRef CAS PubMed.
- S. Schubert, D. C. Gul, H. P. Grunert, H. Zeichhardt, V. A. Erdmann and J. Kurreck, Nucleic Acids Res., 2003, 31, 5982 CrossRef CAS PubMed.
- M. J. Cairns, A. King and L. Q. Sun, Nucleic Acids Res., 2003, 31, 2883 CrossRef CAS PubMed.
- B. Nawrot, K. Widera, M. Wojcik, B. Rebowska, G. Nowak and W. J. Stec, FEBS J., 2007, 274, 1062 CrossRef CAS PubMed.
- Y. Liu, Z. W. Li, G. F. Liu, Q. Wang, W. Chen, D. Zhang, M. S. Cheng, Z. B. Zheng, K. L. Liu and J. L. He, Chem. Commun., 2013, 49, 5037 RSC.
- T. G. Cha, J. Pan, H. R. Chen, J. Salgado, X. Li, C. D. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39 CrossRef CAS PubMed.
- M. A. Hallett, B. Teng, H. Hasegawa, L. P. Schwab, T. N. Seagroves and T. Pourmotabbed, Breast Cancer Res., 2013, 15, R12 CrossRef CAS PubMed.
- J. C. Achenbach, R. Nutiu and Y. F. Li, Anal. Chim. Acta, 2005, 534, 41 CrossRef CAS.
- Y. Tian, Y. He, Y. Chen, P. Yin and C. D. Mao, Angew. Chem., Int. Ed., 2005, 44, 4355 CrossRef CAS PubMed.
- B. K. Ruble, J. L. Richards, J. C. Cheung-Lau and I. J. Dmochowski, Inorg. Chim. Acta, 2012, 380, 386 CrossRef CAS PubMed.
- S. Deborggraeve, J. Y. Dai, Y. Xiao and H. T. Soh, Chem. Commun., 2013, 49, 397 RSC.
- S. A. McManus and Y. F. Li, J. Am. Chem. Soc., 2013, 135, 7181 CrossRef CAS PubMed.
- W. E. Moerner and L. Kador, Phys. Rev. Lett., 1989, 62, 2535 CrossRef CAS PubMed.
- J. Jung, K. Y. Han, H. R. Koh, J. Lee, Y. M. Choi, C. Kim and S. K. Kim, J. Phys. Chem. B, 2012, 116, 3007 CrossRef CAS PubMed.
- N. K. Lee, H. R. Koh, K. Y. Han and S. K. Kim, J. Am. Chem. Soc., 2007, 129, 15526 CrossRef CAS PubMed.
- N. K. Lee, H. R. Koh, K. Y. Han, J. Lee and S. K. Kim, Chem. Commun., 2010, 46, 4683 RSC.
- M. Steiner, K. S. Karunatilaka, R. K. O. Sigel and D. Rueda, Proc. Natl. Acad. Sci. U.S.A., 2008, 105, 13853 CrossRef CAS PubMed.
- Y. B. Lim, S. Y. Bak, S. H. Lee and S. K. Kim, J. Phys. Chem. Lett., 2020, 11, 8048 CrossRef CAS PubMed.
- Y. B. Lim, S. Y. Bak, K. Sung, E. Jeong, S. H. Lee, J. S. Kim, S. Bae and S. K. Kim, Nat. Commun., 2016, 7, 13350 CrossRef CAS PubMed.
- K. Ichikawa, M. Tarnai, M. K. Uddin, K. Nakata and S. Sato, J. Inorg. Biochem., 2002, 91, 437 CrossRef CAS PubMed.
- J. G. Wetmur and N. Davidson, J. Mol. Biol., 1968, 31, 349 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02131e |
| This journal is © The Royal Society of Chemistry 2022 |