Open Access Article
Open Access ArticleGlmU inhibitor from the roots of Euphorbia ebracteolata as an anti-tuberculosis agent†
Xiuyan Han‡
ab,
Changming Chen‡c,
Honglei Wangb,
Jian Kangb,
Qiulong Yanb,
Yufang Ma b,
Wenxin Wangb,
Shan Wub,
Chao Wang
b,
Wenxin Wangb,
Shan Wub,
Chao Wang *ab and
Xiaochi Ma
*ab and
Xiaochi Ma *a
*a
aSecond Affiliated Hospital, Institute of Integrative Medicine, Dalian Medical University, Dalian 116023, P.R. of China. E-mail: wach_edu@sina.com; maxc1978@163.com
bCollege of Pharmacy, Department of Biochemistry and Molecular Biology, Dalian Medical University, Dalian 116044, P.R. of China
cDepartment of Rheumatology and Immunology, The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang 550001, P.R. of China
First published on 22nd June 2022
Abstract
At present, the emerging drug-resistance of Mycobacterium tuberculosis (M. tb) against existing frontline drugs has prompted the development of novel anti-tuberculosis agents based on new targets. Activity of the bifunctional enzyme, glucosamine-1-phosphate acetyltransferase activity and N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) is essential for biosynthesis of the mycobacterium cell wall components and has been proposed as a potential drug target for therapeutic interventions. On the basis of the high-throughput screening of the GlmU AT inhibitor, an extract of Euphorbia ehracteolata displayed a significant inhibitory effect among 49 tested herbal medicines. Using the bioassay-guided separation, an aromatic diterpenoid ebractenoid F was identified as a GlmU AT inhibitor (IC50: 4.608 μg mL−1). Inhibition kinetics showed that ebractenoid F acted as a competitive inhibitor for substrate acetyl-CoA and an uncompetitive inhibitor for substrate GlcN-1-P. Ala434 was deduced to be the key active residue for the interaction between ebractenoid F and GlmU. Furthermore, ebractenoid F displayed an anti-mycobacterial effect against M. tb H37Ra with a minimal inhibitory concentration (MIC) of 12.5 μg mL−1 along with an inhibitory effect on the formation of biofilm and a synergistic effect with isoniazid against M. tb H37Ra. Above all, a GlmU inhibitor was identified from E. ehracteolata and is proposed to be a potential therapeutic anti-tumberculosis agent.
Introduction
Tuberculosis (TB) as the second greatest killer worldwide is caused by infection with bacterium Mycobacterium tuberculosis (M. tb) and has become a major global health risk. Although various anti-TB drugs, such as streptomycin, isoniazid, ethambutol, and rifampin, have been developed and are the currently available anti-tubercular drugs, they are ineffective for treating dormant M. tb. Meanwhile, drug resistance has become a new challenge in TB therapy. Therefore, the development of novel anti-tuberculosis drugs for treating TB is urgently needed. To our best knowledge, inhibition of specific pathogenic bacterial targets that are significantly different from those of the currently used antibiotics, is regarded as an efficient approach to discovering some new anti-tuberculosis agents for the effective treatment of TB.1The M. tb glucosamine-1-phosphate acetyltransferase activity and N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) protein is encoded by gene Rv1018c and is a bifunctional enzyme with both acetyltransferase and uridylyltransferase (pyrophosphorylase) activities, which are mainly responsible for the formation of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) from glucosamine-1-P (GlcN-1-P), UTP, and acetyl CoA (Ac-CoA). The C- and N-terminal domains of GlmU catalyze acetyltransferase and uridyltransferase activities, respectively.2 The final product, UDP-GlcNAc, is a critical precursor for two important biosynthetic pathways of the cell wall: (1) disaccharide linker (D-N-GlcNAc-L-rhamnose) and (2) peptidoglycan (PG).3,4 M. tb GlmU was reported to be essential for mycobacterium survival based on transposon site hybridization (TraSH) assay and knockout experiments.2,5,6 For therapeutic intervention, M. tb GlmU with the absence of a homolog in eukaryotes is the subject of study.7 Therefore, GlmU is regarded as a novel and vital target for the anti-tuberculosis substance development.
Natural products (NPs) include millions of compounds are important resources for bioactive substance discovery; some can even be considered as drugs.8,9 It is also recognized that anti-tuberculosis agents can be obtained from natural products, especially those considered phytomedicines.10–13 In China, several herbal preparations have been used for the treatment of tuberculosis, such as “Jieheling”.14,15 Therefore, in the present study, the inhibitory effects of various herbal extracts against the new target, GlmU, were evaluated using a high throughput screening technique, which indicated the presence of an active medicinal plant Euphorbia ehracteolata Hayata. Based on the bioassay, a GlmU inhibitor, which displayed significant inhibitory effect against M. tuberculosis was obtained from E. ehracteolata.
Results
Identification of ebractenoid F as an inhibitor of GlmU acetyltransferase
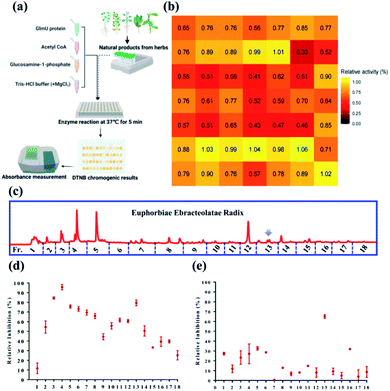
In this study, M. tb GlmU acetyltransferase activity can be assayed by a high throughput screening system using 5′5-dithio-bis (2-nitrobenzoic acid [DTNB]) colorimetric measurement. The ethanolic extracts of 49 herbal medicines were tested their inhibitory effects against M. tb GlmU acetyltransferase activity (Fig. 1a, Table S1†). Comparing the relative activity (%) of crude extracts from different herbaceous medicines, E. ebracteolata showed the strongest inhibitory effect on GlmU acetyltransferase with 33% relative activity at 100 μg mL−1 (Fig. 1b), which indicated the proposed anti-tuberculosis effect. Thus, the active constituents should be identified for E. ebracteolata with the combination of various chromatographic techniques and bioassay system. With the help of automatic purification system, 18 chromatographic fractions of E. ebracteolata (Fr.1–18) were prepared for inhibitory activity assay against GlmU acetyltransferase (Fig. 1c). As shown in Fig. 1d, three fractions (F3, F4 and F13) showed strong inhibition on GlmU at 100 μg mL−1. Furthermore, fractions 1–18 was subsequently tested for its efficacy against M. tb H37Ra using MABA. The antibacterial effect of Fraction F13 was much higher than that of other fractions (Fig. 1e). Therefore, fraction 13 was identified to be the active fraction, which prompted the further purification of bioactive compounds.After the further purification, seven compounds were obtained from active fraction 13. Analysis of nuclear magnetic resonance and high-resolution mass spectrometry (1H NMR, 13C NMR, and HRMS, respectively) indicated that the isolated compounds were aromatic rosane diterpenoids. Compared with previous studies,16–20 the chemical structure of the isolate was determined to be a series rosane type diterpenoid possessing an aromatic ring as shown in Fig. 2a. Using the DTNB colorimetric assay, ebractenoid F (3) displayed a significant inhibitory effect on GlmU acetyltransferase (IC50 4 ± 0.5 μg mL−1) as shown in Fig. 2b and c. Therefore, GlmU as a novel target for anti-tuberculosis agents provided a strategy for bioactive constituent investigation. Above all, using the high throughput screening technique together with bioassay results, the bioactive herbal medicines in addition to bioactive compounds can be efficiently identified.
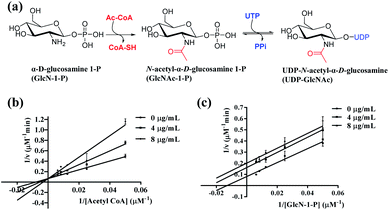
Inhibition modes of ebractenoid F on GlmU acetyltransferase
The GlmU protein is known as a bifunctional enzyme and is widely found in bacteria. GlcN-1-P is subsequently converted in two steps by M. tb GlmU (Fig. 3a): (1) acetyltransferase activity converts GlcN-1-P to GlcNAc-1-P in the first step and (2) uridyltransferase activity as converting GlcNAc-1-P to UDP-GlcNAc in the second step.21 As reported, the C-terminus is the key point of the acetyltransferase activity, and the N-terminus is related to the uridylyltransferase function. In this study, the ethanolic extract of E. ebracteolata could inhibit the acetyltransferase activity of M. tb GlmU. Thus, ebractenoid F was proposed to be the main active, inhibitory constituent.The inhibition kinetics of ebractenoid F against GlmU acetyltransferase were then investigated about two substrates acetyl CoA and GlcN-1-P, respectively. Lineweaver–Burk plots of GlmU acetyltransferase substrates acetyl CoA or GlcN-1-P were used to determine inhibition modes by measuring the inhibitory effects of ebractenoid F on enzymatic velocity. Increasing concentrations of ebractenoid F were set and either acetyl CoA or GlcN-1-P concentration was varied. The inhibition kinetics indicated that ebractenoid F was competitive with substrate acetyl CoA (Ki-com 18.592 μM), and uncompetitive with GlcN-1-P (Ki-uncom 26.771 μM) (Fig. 3b and c, and Table S2†). So, ebractenoid F could inhibit the acetyltransferase activity of GlmU with different inhibition mechanisms against either substrate, acetyl CoA or GlcN-1-P. Each point on the Lineweaver–Burk plot was assayed in triplicate, and the data represent the mean ± standard error of the three independent experiments.
Interaction between ebractenoid F and GlmU acetyltransferase
In consideration of the inhibitory effect of ebractenoid F against GlmU acetyltransferase, the interaction between ebractenoid F and GlmU was investigated in the present study. Based on a surface plasmon resonance (SPR) mechanism, the affinity of ebractenoid F for GlmU was measured. Concentration-dependent binding was observed between ebractenoid F and GlmU, and the KD value was determined to be 15.7 μM (Fig. 4a).To clarify the interaction sites between ebracternoid F and GlmU, in silico docking analysis was performed using the three-dimensional (3D) structure of GlmU obtained from the Protein Data Bank (PDB) database (PDB code: 4K6R). A schematic representation of inhibitor ebractenoid F–GlmU interactions was drawn and shown in Fig. 4b. Hydrogen-bonds (H-bond) formed between the Ala434 side chain of GlmU and ebracternoid F are represented with a purple arrowhead, which is a central point of the interaction site. In addition to the hydrogen-bonded interactions, ebractenoid F was involved in another interaction and further annotated, such as hydrophobic residues are coloured in light green and polar residues are coloured in light blue.
A site-directed mutagenesis method was used to verify the molecular docking results. Three amino acid residues (Ala434, Gly433, and Ala451) of GlmU were selected for mutagenesis investigation. The GlmU mutant proteins, Ala434Ser, Gly433Ala, and Ala451Ser were obtained. All mutant proteins were purified, identified, and quantified. Acetyltransferase activity was determined by method mentioned above.
Importantly, the Ala434Ser GlmU mutant protein lost 24% acetyltransferase activity (Fig. 4c), and ebractenoid F showed some reduction in inhibitory activity against Ala434Ser GlmU mutant protein (Fig. 4d). From the uniprot database UniProtKB-P9WMN3 (GLMU_MYCTU), it has been proven that Ala434 is the binding site of GlmU acetyltransferase with acetyl CoA.22 Therefore, it was confirmed that the inhibitor, ebractenoid F, formed strong hydrogen bonds with the Ala434 residue of GlmU acetyltransferase and prevented the substrate acetyl CoA from binding the Ala434 residue, resulting in GlmU loss of acetyltransferase activity. Finally, the interaction analysis between ebractenoid F and GlmU was consistent with the inhibition mode, which confirmed that ebractenoid F was competitive with substrate acetyl CoA.
Ebractenoid F is a potential anti-tuberculosis agent
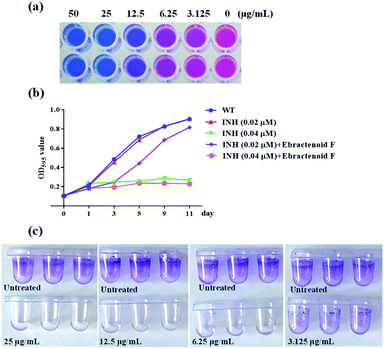
As mentioned above, ebractenoid F as an aromatic rosane diterpenoid from Euphorbia ebracteolata was identified to be a potential inhibitor of GlmU acetyltransferase. Using the AlamarBlue assay, ebractenoid F displayed significant inhibitory effect against M. tb H37Ra, and the minimal inhibitory concentration (MIC) was determined to be 12.5 μg mL−1 (Fig. 5a). Also, ebractenoid F demonstrated a synergistic effect with the anti-tuberculosis drug, isoniazid, on the growth curve M. tb H37Ra (Fig. 5b).Considering that biofilm is an important protective barrier for mycobacterium, it has been reported that GlmU is essential for biofilm formation in M. smegmatis and participated in the synthesis of the precursor required for biofilm production, suggesting the involvement of GlmU in the M. smegmatis defence mechanisms.23 The influence of ebractenoid F on biofilm formation in M. smegmatis was investigated. The results showed that ebractenoid F could inhibit biofilm formation at concentrations below the MIC value (Fig. 5c).
Destroyed integrity of M. tb H37Ra by ebractenoid F
UDP-GlcNAc is an essential precursor for peptidoglycan (PG) and disaccharide linker (D-N-GlcNAc-L-rhamnose) for M. tb cell wall biosynthesis.24 Inhibitor targeting the acetyltransferase activity of GlmU will block the UDP-GlcNAc synthesis pathway and destroy the integrity of bacterial cellular.With the help of scanning and transmission electron microscopy (SEM and TEM, respectively) the impact of ebractenoid F on bacterial morphology was observed. Compared with the untreated M. tb H37Ra cells, ebractenoid F (2 × MIC)-treated M. tb H37Ra cells indicated thinner cell walls (Fig. 6a), longer cell morphology, and more wrinkles on the cell surfaces (Fig. 6b).
Furthermore, propidium iodide (PI) dye as a fluorescence indicator was used to measure the permeability of M. tb H37Ra cells.25 The PI dye could not enter the cell with a fully integrated cell membrane but could pass through a disrupted membrane and bind with DNA, which is indicated by red fluorescence. After M. tb H37Ra was treated with ebractenoid F (2 × MIC), bacteria showed obvious red fluorescence, indicating lack of integrity of the cell wall and membrane, which allowed PI to penetrate the bacterial cells, whereas, untreated M. tb H37Ra showed no red fluorescence emission (Fig. 6c). Thus, the inhibitor ebractenoid F, targeting GlmU, could destroy the structure and integrity of the M. tb cell wall.
Discussion
UDP-GlcNAc is key constituent in the cell wall of many pathogens. When the biosynthesis of UDP-GlcNAc is blocked, the reproduction of pathogens is inhibited. Recently, the pathway for UDP-GlcNAc biosynthesis has been explored in the M. tuberculosis genome, and the results suggest that the GlmU is a key functional enzyme. Therefore, GlmU as an important functional enzyme for the biosynthesis of the cell wall is proposed to be a novel target for anti-tuberculosis.Natural products are important resources for new drug discovery. Therefore, in the present study, the bioactive herbal medicines targeting GlmU for anti-tuberculosis were investigated. As a result, the ethanolic extract of E. ebracteolata was screened as active inhibitors of GlmU. In China, E. ebracteolata was known as the traditional chinese medicine “Langdu” for the treatment of tuberculosis. Also, “Langdu” is the major pharmaceutical ingredient of clinical medicine “Jieheling” used for tuberculosis therapy. Therefore, the anti-tuberculosis effect of E. ebracteolata was unambiguous, and it was desirable to clarify the key active constituent together with the proposed target. Benefitting from the bioassay results, the key GlmU inhibitor was efficiently identified as ebractenoid F from E. ebracteolata, which displayed good affinity toward GlmU. When combined with in silico docking and protein amount technique, the interaction between ebractenoid F and GlmU protein was revealed. Furthermore, GlmU inhibitor ebracternoid F displayed a significant inhibitory effect against M. tb H37Ra by the pathway required for cell wall biosynthesis. Thus, this work also confirmed that the GlmU inhibitor could be used for further investigation involving anti-tuberculosis agents.
Conclusions
GlmU is an essential functional enzyme for the biosynthesis of cell wall of M. tuberculosis, which is recognized to be a novel target for the treatment of tuberculosis. Using the bioassay guidance of GlmU, E. ebracteolata was screened to be the active herb, and ebractenoid F was identified to be a key GlmU inhibitor (IC50 4 ± 0.5 μg mL−1). The interaction between ebractenoid F and GlmU was also revealed by KD value determination, in silico docking analysis, and protein mutant evaluations. Furthermore, ebractenoid F was shown to inhibit cell wall biosynthesis and the biofilm formation of M. tb H37Ra (MIC 12.5 μg mL−1) together with the synergistic effect with isoniazid against M. tb H37Ra. Therefore, E. ebracteolata was confirmed to be a traditional anti-tuberculosis herbal medicine, and ebractenoid F, acting as a GlmU inhibitor, displayed the potential as an anti-tuberculosis agent.Experimental section
Chemical materials
Methanol and acetonitrile for high-performance liquid chromatography (HPLC) were purchased from Sigma Aldrich. Ethanol, petroleum ether, ethyl acetate for the extracts preparation and purification of chemical constituents were productions of Tianjin Kermel (China).Apparatus
HPLC was performed using a Waters e2695 (Waters, USA). Preparative HPLC was manufactured by Agela technologies (Tianjin, China). NMR spectra were measured using Bruker-600 with tetramethylsilane (TMS) as the internal standard (Bruker, USA). High-resolution electrospray ionization mass spectrometry (HR-ESIMS) data were obtained using an Agilent 1290 infinity 6540 UHD accurate mass Q-TOF MS (Agilent, USA). A constant temperature incubator shaker (ZHWY-2012C) was obtained from Shanghai Zhicheng Analytical Instrument Co. Ltd (China).Plasmids, bacterial strains, and growth conditions
The M. tuberculosis H37Ra strain (ATCC 25177) as an attenuated strain was cultured in Difco Middlebrook 7H9 broth supplemented with 10% albumin–dextrose-catalase enrichment (ADC), 0.4% glycerol and 0.05% Tween 80 under shaking concentrations at 37 °C for drug susceptibility testing. Strains on solid medium were grown on Difco Middlebrook 7H10 agar supplemented with 10% ADC and 0.4% glycerol in a 37 °C incubator. M. smegmatis mc2 155 strain was grown in lysogeny broth (LB) broth containing 0.05% Tween 80.Escherichia coli NovaBlue and E. coli BL21(DE3) strains (Novagen) were grown in LB broth for gene cloning and protein expression analyses. Plasmid pJET1.2 blunt (Thermo) with ampicillin resistance was used for GlmU gene cloning. Expression vector pET16b (Novagen) with ampicillin resistance was used to express GlmU protein in E. coli BL21(DE3) strains.
Preparation of herbal extracts and purification of compounds
Forty-nine herbal materials used as traditional chinese medicines were obtained from the First Affiliated Hospital of Dalian Medical University (Table S1†) and were extracted by ultrasonic with 90% aqueous ethanol. After the evaporation of solvents, the residues were used for bioassay. The ethanolic extract of the air-dried roots of E. ebracteolata was separated by Waters e2695 preparative HPLC, and 18 fractions were collected. After solvent evaporation, the residues corresponding to 18 fractions were obtained for bioassays. Additionally, fraction 13 was further purified using pre-HPLC to give the compounds 1–7. The chemical structures were determined by the spectroscopic data analysis according to our previous work.16,17 Especially, compound 3 was determined to be ebractenoid F, an aromatic rosane type diterpenoid,18–20 based on 1H NMR (CDCl3, 600 MHz) δH 6.71 (1H, s), 5.86 (1H, dd, J = 17.4, 10.8 Hz), 4.96 (1H, dd, J = 17.4, 1.2 Hz), 4.89 (1H, dd, J = 10.8, 1.8 Hz), 2.64 (2H, m), 1.97 (1H, dt, J = 12.6, 3.0 Hz), 1.75 (1H, m), 1.66 (2H, m), 1.57 (3H, m), 1.44 (1H, t, J = 13.2 Hz), 1.39 (1H, m), 1.20 (1H, dt, J = 13.2, 2.4 Hz), 1.02 (3H, s), 1.01 (3H, s) (Fig. S1†), 13C NMR (CDCl3, 150 MHz) δC 151.1, 140.7, 140.4, 139.8, 127.1, 122.6, 108.9, 108.8, 39.6, 36.5, 36.4, 36.3, 34.1, 32.9, 26.9, 25.6, 22.8, 21.4, 11.4 (Fig. S2†), and HRMS: m/z 285.1856 [M–H]− (calcd C19H26O2, 285.1855) as shown in Fig. S3†.Expression and purification of M. tb GlmU protein
M. tb GlmU expression strain (E. coli BL21 (DE3)/pET16b-GlmUM. tb) was previously constructed in our lab and used to obtain M. tb GlmU as in our previous work.13Briefly, the E. coli BL21 (DE3)/pET16b-GlmUM. tb strain was cultured to an OD595 of 0.5 in LB broth containing 50 μg mL−1 ampicillin, adding 1 mM isopropyl β-1-D-1- thiogalactopyranoside (IPTG) to the bacterial culture and incubating at 37 °C for 6 h to induce GlmU gene expression. The bacterial culture was centrifuged (4 °C, 3000 × g, 10 min) after which cell pellets were resuspended using lysis buffer (20 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.5 M NaCl, 20% glycerol and 1 mM PMSF) followed by sonication. The supernatant was collected by centrifugation (4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 15 min), and loaded onto a Ni-NAT column (Qiagen) for purification of His-tagged GlmU protein. The concentration of purified GlmU protein was determined using the Bradford method. The purity and identification of GlmU protein were checked by sodium dodecyl polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
000 × g, 15 min), and loaded onto a Ni-NAT column (Qiagen) for purification of His-tagged GlmU protein. The concentration of purified GlmU protein was determined using the Bradford method. The purity and identification of GlmU protein were checked by sodium dodecyl polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
Inhibitor screening of GlmU acetyltransferase
A colorimetric assay coupled with 5,5′-dithio-bis-(2-nitrobenzoic acid [DTNB]) was performed to screen inhibitors of GlmU acetyltransferase.26 According to the method in our previous work, the enzymatic reaction was a 50 μL mixture performed in a 96-well microtiter plate containing 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 0.4 mM glucosamine-i-phosphate (GlcN-1-P), 0.4 mM acetyl CoA (Ac-CoA) and purified GlmU protein at 37 °C for 5 min. The reaction was then terminated by adding 50 μL 6 M guanidine hydrochloride and 50 μL Ellman's reagent (0.2 mM DTNB and 1 mM ethylenediaminetetraacetic acid [EDTA] in 50 mM Tris–HCl, pH 7.5). The absorbance value was measured at a wavelength of 405 nm by using Multiscan Fc (Thermo scientific). Separately, the “background group” containing the substrates (Ac-CoA and GlcN-1-P) and tested compound was used to correct the error. Percent inhibition: inhibition (%) = 100 (1 − [A405 − AMin]/[AMax − AMin]) for which A405 was the absorbance of the test reaction that contained compound, AMin was the absorbance of the background group, and AMax was the absorbance of the uninhibited reaction. GraphPad was used to calculate the concentration of 50% inhibition (IC50).Mode-of-inhibition studies
To investigate inhibition modes, the dual-substrate reactions of GlmU were performed in presence of ebractenoid F. The concentrations of both substrates, GlcN-1-P and acetyl CoA, were set as 0.0, 0.04, 0.08, 0.12, and 0.16 mM. The fixed substrate was set at 0.4 mM. The inhibitory constant (Ki) and inhibition modes were determined for the co-incubation of ebractenoid F at different concentrations.Interaction studies
First, the affinity between GlmU and ebractenoid F was measured using Biacore T200 (GE, USA) based on surface plasmon resonance (SPR).27,28 Briefly, Ni was captured by NTA chip and then used to capture GlmU protein by His tag. ebractenoid F was flowed to detect the affinity with GlmU protein with a concentration gradient (1.56–25 μg mL−1).Additionally, binding interactions of identified inhibitor ebractenoid F at M. tb GlmU acetyltransferase domain were analysed via molecular docking studies. Docking studies were performed to explore the binding conformation and potential interactions between ebractenoid F and GlmU protein. The three-dimensional (3D) structure of GlmU was downloaded from the RCSB Protein Data Bank as a protein databank file (PDB code: 4K6R). The 3D structures of ligands were sketched based on the correct atom type and chirality using Sybyl software (Tripos Inc., St.Louis, MO, USA). Both energy optimization and the calculation of charge (Gasteiger–Huckel) were needed for the preparation of ligands. The Surflex-Dock program was used for the docking calculations with default parameters. The 2D diagrams of protein–ligand interactions were analysed using the Web-based online tool Pose View (http://poseview.zbh.uni-hamburg.de/poseview).
A site-directed mutagenesis assay was used to probe the roles of interaction residues in GlmU acetyltransferase domain according to the molecular docking results. These amino acid residues of GlmU were substituted (Gly433Ala, Ala434Ser, and Ala451Ser) by designing specific primers, and a mutation was introduced into the cloned M. tb GlmU gene using the MutanBEST Kit (Table S3†, Fig. S4†). All GlmU mutant proteins were expressed, purified, quantified, and an acetyltransferase enzymatic activity was determined by the method described above (Fig. S5†).
MIC determination against M. tuberculosis
The Fr.1–18 fractions of E. ebracteolata were screened against M. tb H37Ra for the growth inhibitory activity by Microlate-based AlamarBlue assay (MABA).29 Briefly, log phase M. tb H37Ra cultures were diluted using 7H9 broth (supplemented with 10% ADC) and divided into 400 μL (1 × 105 cfu mL−1 bacteria) into each well of 48-well plates. The 100 μL 7H9 broth containing different fractions were added individually to each well. The total volume in each well was 500 μL in the 48-well plate, and the final concentrations of fractions were 10 μg mL−1. The negative group (only including bacteria) and positive group (including bacteria and kanamycin) were set as controls. Plates were sealed and carefully incubated at 37 °C for seven days after which 400 μL resazurin working solution was added to detect bacterial survival and growth. Also, the absorbance of the colour changes was captured at a wavelength of 595 nm by using Multiscan Fc spectrometer (Thermo Scientific). The relative inhibition ratio (%) was calculated and the absorbance value was interpreted after subtraction of the background absorbance of the negative controls. The MIC was defined as the lowest concentration of drug that prevented the colour change from blue to pink.In vitro combination studies with anti-tuberculosis drug
Briefly, log phase M. tb H37Ra cultures were diluted to an OD595 0.105 using 7H9 broth (supplemented with 10% ADC). Then the diluted cultures were divided 20 mL into each flask and cultured at 37 °C under shaking conditions. The experimental groups were specifically set: (1) 0.02 μM isoniazid (INH) treated group, (2) 0.04 μM INH treated group, (3) 0.02 μM INH and 12.5 μg mL−1 ebractenoid F treated group, (4) 0.04 μM INH and 12.5 μg mL−1 ebractenoid F treated group, and (5) untreated group. The 200 μL from each flask were used to measure OD595 on days 0, 1, 3, 5, 9, and 11 for constructing the time-depended growth curve.Inhibitory effects of ebracternoid F on biofilm formation
The M. smegmatis mc2 155 strain was used as a model to test biofilm formation.23,30 To produce a biofilm, log phase M. smegmatis cultures were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in M63 medium supplemented with 1 mM MgSO4, 0.7 mM CaCl2, and 0.5% casamino acids. The bacterial dilutions were transferred (150 μL) into each well of a sterile 96-well polypropylene microtiter plate. Ebractenoid F was added into each well at a gradient of final concentrations ranging from 50 to 0 μg mL−1. A kanamycin-containing well was set as a positive control. Plates were cultured at 37 °C incubator without disturbance for six days. Liquid was then removed from the wells of plate and washed three times with saline solution after which 150 μL of 0.1% crystal violet was added into each well for 15 min at room temperature to stain the biofilm. The wells were then washed three times with water, and 150 μL of 95% ethanol was added for biofilm quantitation at OD570.
1000 in M63 medium supplemented with 1 mM MgSO4, 0.7 mM CaCl2, and 0.5% casamino acids. The bacterial dilutions were transferred (150 μL) into each well of a sterile 96-well polypropylene microtiter plate. Ebractenoid F was added into each well at a gradient of final concentrations ranging from 50 to 0 μg mL−1. A kanamycin-containing well was set as a positive control. Plates were cultured at 37 °C incubator without disturbance for six days. Liquid was then removed from the wells of plate and washed three times with saline solution after which 150 μL of 0.1% crystal violet was added into each well for 15 min at room temperature to stain the biofilm. The wells were then washed three times with water, and 150 μL of 95% ethanol was added for biofilm quantitation at OD570.
Bacterial cellular morphology and integrity testing
The effects of GlmU acetyltransferase inhibitor on cellular integrity was assessed using the PI staining method.31 M. tb H37Ra (OD595 at 0.5) cultures were treated with ebractenoid F at MIC concentration for 24 h and untreated M. tb H37Ra cultures were set as control group. Bacterial cultures were centrifuged at 3000 × g for 10 min at 4 °C. Cell pellets were resuspended in 1 mL saline solution, and 3 μL PI solution (Sigma) was added for a 15 min incubation period in the dark. The PI solution was then removed by centrifugation and samples were washed by saline solution for three times. For the last time, cell pellets were resuspended in 100 μL saline solution and observed under fluorescence microscope with a 10 × lens.Morphology observation of untreated M. tb H37Ra and ebractenoid F-treated M. tb H37Ra was performed by scanning and transmission electron microscopy (SEM and TEM, respectively). The samples were prepared as described previously.2,13 Briefly, M. tb H37Ra treated with or without ebractenoid F were centrifuged at 3000 × g for 10 min at 4 °C. For SEM, the pellets were washed three times with 0.1 M phosphate buffer (pH 7.4) and fixed with 2.5% glutaraldehyde and 1% OsO4, followed by dehydration through a graded series of ethanol (20%, 40%, 60%, 70%, 80%, 90%, and 100%). Finally, the cell pellet was applied to a silicon wafer slide and then coated with gold to a thickness of 5 nm. SEM images were captured using JSM-6369-LV scanning electron microscope. For TEM, the pellets were fixed with 2.5% glutaraldehyde for 2 h and dehydrated in graded series of alcohol. Finally, samples embedded in epoxy resin and ultra-thin slices were prepared for TEM observation using a JEM-1400 PLUS transmission electron microscope.
Author contributions
Xiuyan Han: data curation, software, writing – review & editing. Changming Chen, Honglei Wang: data curation. Jian Kang and Qiulong Yan: validation. Yufang Ma: review & editing. Wenxin Wang and Shan Wu: data curation, validaiton. Chao Wang: writing – review & editing, supervision. Xiaochi Ma: writing – review & editing, supervision.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81930112 and 81872970), Distinguished Professor of Liaoning Province (XLYC2002008), Dalian Science and Technology Leading Talents Project (2019RD15 and 2020RJ09), and Liaoning Revitalization Talents Program (XLYC1907017).Notes and references
- J. C. Sacchettini, E. J. Rubin and J. S. Freundlich, Nat. Rev. Microbiol., 2008, 6, 41–52 CrossRef CAS PubMed.
- W. Zhang, V. C. Jones, M. S. Scherman, S. Mahapatra, D. Crick, S. Bhamidi, Y. Xin, M. R. McNeil and Y. Ma, Int. J. Biochem. Cell Biol., 2008, 40, 2560–2571 CrossRef CAS PubMed.
- M. R. McNeil and P. J. Brennan, Res. Microbiol., 1991, 142, 451–463 CrossRef CAS PubMed.
- P. J. Brennan and D. C. Crick, Curr. Top. Med. Chem., 2007, 7, 475–488 CrossRef CAS PubMed.
- C. M. Sassetti, D. H. Boyd and E. J. Rubin, Mol. Microbiol., 2003, 48, 77–84 CrossRef CAS PubMed.
- V. Soni, S. Upadhayay, P. Suryadevara, G. Samla, A. Singh, P. Yogeeswari, D. Sriram and V. K. Nandicoori, PLoS Pathog., 2015, 11, e1005235 CrossRef PubMed.
- M. Agarwal, V. Soni, S. Kumar, B. Singha and V. K. Nandicoori, Biochem. J., 2021, 478, 2081–2099 CrossRef CAS PubMed.
- G. Cazzaniga, M. Mori, L. R. Chiarelli, A. Gelain, F. Meneghetti and S. Villa, Eur. J. Med. Chem., 2021, 224, 113732 CrossRef CAS PubMed.
- H. Yuan, Q. Ma, L. Ye and G. Piao, Mol, 2016, 21 Search PubMed.
- K. W. Nam, W. S. Jang, M. A. Jyoti, S. Kim, B. E. Lee and H. Y. Song, Phytomedicine, 2016, 23, 578–582 CrossRef CAS PubMed.
- V. P. Baldin, R. B. L. Scodro, M. A. Lopes-Ortiz, A. L. de Almeida, Z. C. Gazim, L. Ferarrese, V. D. S. Faioes, E. C. Torres-Santos, C. T. A. Pires, K. R. Caleffi-Ferracioli, V. L. D. Siqueira, D. A. G. Cortez and R. F. Cardoso, Phytomedicine, 2018, 47, 34–39 CrossRef CAS PubMed.
- S. Kim, H. Seo, H. A. Mahmud, M. I. Islam, B. E. Lee, M. L. Cho and H. Y. Song, Phytomedicine, 2018, 46, 104–110 CrossRef CAS PubMed.
- C. Chen, X. Han, Q. Yan, C. Wang, L. Jia, A. Taj, L. Zhao and Y. Ma, Front. Cell. Infect. Microbiol., 2019, 9, 251 CrossRef CAS PubMed.
- J. M. Li, Q. W. Shi, Y. Jiang, J. L. Gao, M. J. Li and H. Y. Ma, Anal. Methods, 2016, 8, 5218–5227 RSC.
- S. S. Huang, P. Li, B. J. Zhang, S. Deng, H. L. Zhang, C. P. Sun, X. K. Huo, X. G. Tian, X. C. Ma and C. Wang, Phytochem. Lett., 2017, 19, 151–155 CrossRef CAS.
- J. Zhou, J. H. Wen, J. H. Huang, P. F. Chen, X. C. Ma, R. J. Sun and C. Wang, Rec. Nat. Prod., 2021, 15, 202–206 CrossRef CAS.
- L. Li, D. Li, C. Wang, L. Feng, Z. Yu, J. Ning, B. Zhang, H. Zhang, C. Wang and X. Ma, Bioorg. Chem., 2020, 94, 103360 CrossRef CAS PubMed.
- Z. G. Liu, Z. L. Li, J. Bai, D. L. Meng, N. Li, Y. H. Pei, F. Zhao and H. M. Hua, J. Nat. Prod., 2014, 77, 792–799 CrossRef CAS PubMed.
- S. Z. Mu, C. R. Jiang, T. Huang and X. J. Hao, Helv. Chim. Acta, 2013, 96, 2299–2303 CrossRef CAS.
- J. W. Lee, C. Lee, Q. Jin, H. Jang, D. Lee, H. J. Lee, J. W. Shin, S. B. Han, J. T. Hong, Y. Kim, M. K. Lee and B. Y. Hwang, J. Nat. Prod., 2016, 79, 126–131 CrossRef CAS PubMed.
- P. D. Craggs, S. Mouilleron, M. Rejzek, C. de Chiara, R. J. Young, R. A. Field, A. Argyrou and L. P. S. de Carvalho, Biochemistry, 2018, 57, 3387–3401 CrossRef CAS PubMed.
- P. K. Jagtap, V. Soni, N. Vithani, G. D. Jhingan, V. S. Bais, V. K. Nandicoori and B. Prakash, J. Biol. Chem., 2012, 287, 39524–39537 CrossRef PubMed.
- A. Di Somma, M. Caterino, V. Soni, M. Agarwal, P. di Pasquale, S. Zanetti, P. Molicotti, S. Cannas, V. K. Nandicoori and A. Duilio, Res. Microbiol., 2019, 170, 171–181 CrossRef CAS PubMed.
- C. Rani and I. A. Khan, Eur. J. Pharm. Sci., 2016, 83, 62–70 CrossRef CAS PubMed.
- Y. Yang, Y. Xiang and M. Xu, Sci. Rep., 2015, 5, 18583 CrossRef CAS PubMed.
- Y. Zhou, Y. Xin, S. Sha and Y. Ma, Arch. Microbiol., 2011, 193, 751–757 CrossRef CAS PubMed.
- G. D. Healey, A. Frostell, T. Fagge, D. Gonzalez and R. S. Conlan, Antibodies, 2019, 8, 7 CrossRef CAS PubMed.
- Y. Huang, S. H. Yu, W. X. Zhen, T. Cheng, D. Wang, J. B. Lin, Y. H. Wu, Y. F. Wang, Y. Chen, L. P. Shu, Y. Wang, X. J. Sun, Y. Zhou, F. Yang, C. H. Hsu and P. F. Xu, Theranostics, 2021, 11, 6891–6904 CrossRef CAS PubMed.
- X. Han, C. Chen, Q. Yan, L. Jia, A. Taj and Y. Ma, Front. Microbiol., 2019, 10, 1799 CrossRef PubMed.
- J. Kang, L. Xu, S. Yang, W. Yu, S. Liu, Y. Xin and Y. Ma, PLoS One, 2013, 8, e61589 CrossRef CAS PubMed.
- R. Sharma, C. Rani, R. Mehra, A. Nargotra, R. Chib, V. S. Rajput, S. Kumar, S. Singh, P. R. Sharma and I. A. Khan, Appl. Microbiol. Biotechnol., 2016, 100, 3071–3085 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02044k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |