Open Access Article
Open Access ArticleA mechanistic study on electro-Fenton system cooperating with phangerochate chrysosporium to degrade lignin
Yingjian Qin†
a,
Na Wang†a,
Zhongmin Maa,
Jinsheng Lib,
Yaozong Wangb and
Lihua Zang *a
*a
aCollege of Environmental Science and Engineering, Qilu University of Technology (Shandong Academy of Science), Jinan, 250353, Shandong, P.R. China. E-mail: Zlh@qlu.edu.cn; Tel: +86 13325127799
bWeifang Ensign Industry Co., Ltd, Changle, 262499, Shandong, P.R. China
First published on 10th June 2022
Abstract
The combined catalytic system of Electro-Fenton (E-Fenton) and Phanerochaete chrysosporium (P. chrysosporium) was constructed in liquid medium with additional potential to overcome the limitations of lignin degradation by white rot fungi alone. To further understand the mechanism of synergistic catalysis, we optimized the optimum potential for lignin catalysis by P. chrysosporium and built synergistic versus separate catalyses. After 48 h of incubation, the optimum growth environment and the highest lignin degradation rate (43.8%) of P. chrysosporium were achieved when 4 V was applied. After 96 h, the lignin degradation rate of the cocatalytic system was 62% (E-Fenton catalysis alone 22% and P. chrysosporium catalysis alone 19%), the pH of the growth maintenance system of P. chrysosporium was approximately 3.5, and the lignin peroxidase (LiP) and manganese-dependent peroxidase (MnP) enzyme activities, were significantly better than those of the control. The qPCR results indicated that the expression of both MnP and LiP genes was higher in the cocatalytic system. Meanwhile, FTIR and 2D-HSQC NMR confirmed that the synergistic catalysis was effective in breaking the aromatic functional groups and the side chains of the aliphatic region of lignin. This study showed that the synergistic catalytic process of electro-Fenton and P. chrysosporium was highly efficient in the degradation of lignin. In addition, the synergetic system is simple to operate, economical and green, and has good prospects for industrial application.
1. Introduction
Lignocellulose, which is rich in renewable carbon resources, is considered one of the most promising energy alternatives to reduce the overdependence on fossil fuels.1,2 Unfortunately, the resistance of lignin to filling the cellulose framework to support the cell wall hinders the efficient use of lignocellulose.3,4 Similarly, black liquor produced in the pulp industry has limited recycling and utilization due to the presence of lignin.5 Therefore, the degradation of lignin is pivotal for maximizing resource utilization and reducing the environmental pollution burden.6The chemical recalcitrance of lignin stems from its structural complexity.7 Lignin is an aromatic polymer formed by cross-coupling phenolic units through a variety of ether bonds and carbon–carbon bonds.8 Among the different methods of lignin degradation, the microbial method has many advantages, such as the conditions are mild and it is simple to operate, with no expensive or designed catalysts, and its secretion of enzyme catalytic activity and the complex metabolic action degradation of lignin has become an effective natural pathway.9 Filamentous fungi, especially white rot fungi,10 have received widespread attention due to their significant effects on wood degradation,11 papermaking wastewater treatment, and lignocellulose pretreatment.12 Phanerochaete chrysosporium (P. chrysosporium) is a typical white rot fungus that penetrates wood cells to produce lignin peroxidase (LiP) and Mn-dependent peroxidase (MnP) to degrade lignin.13 Between these peroxidases, LiP undergoes a series of oxidations and generates free radicals under the start of hydrogen peroxide (H2O2).14 In most cases, LiP can degrade nonphenolic lignin units by electron transfer via an invariant tryptophan residue (trp171), such as Cα–Cβ, which breaks the propyl side chain.7
In the degradation of lignin, the two peroxidases secreted by P. chrysosporium can effectively break the bonds between lignin units and increase the subsequent enzymatic hydrolysis of sugar.15,16 Unfortunately, LiP and MnP mainly attack the end groups or simple substrates of polymers and form short free radicals, which is due to their limited electron transfer.7 Therefore, their oxidation of lignin lacks the ability to penetrate the entire lignin structure and provide oxidants over a long time. In effect, ferrous ions in the active centre of peroxidase catalyse H2O2 to produce the reactive oxygen species (ROSs, mainly including hydroxyl radicals and superoxide radicals), which can be regarded as the Fenton system.17 A previous study showed that the natural degradation of lignin by white rot fungi simulated by the Fenton system in vitro could accelerate the depolymerization of lignin, and the mechanism of lignin degradation by the Fenton system was preliminarily revealed.18 It was found that the demethoxylation (Lars Hildén et al., 2000) of lignin was carried out, after which the electron transfer to the side chain results in the rupture of the β-O-4 ether bond.18 In addition, a previous study showed that the Fenton process combined with bioremediation technology had high efficiency in the degradation of organic polymer compounds in wastewater.19 However, previous studies, only focused on the application of P. chrysosporium or the Fenton process for lignin degradation, and refined in-depth studies on the combination of the two were lacking. Meanwhile, the fact that the addition of H2O2 brings economic restrictions to the industrialization of technology cannot be ignored. Subsequently, we learned that the continuous in situ generation of H2O2 by electro-Fenton (E-Fenton) technology requires only an applied electrical potential under acidic conditions to improve the industrial cost and safety risk limits inherent to the use of additional agents.20 The stimulation of the applied potential and generation of hydrogen peroxide in situ activated LiP and MnP and triggered the enzyme cycle, thereby expediting the degradation of lignin.21 The above shows that electro-Fenton is superior to conventional Fenton technology in terms of economic costs and practical applications. On the other hand, the acidic conditions of the E-Fenton reaction are provided by the production of small organic acids and fatty acids during the culture and metabolism of P. chrysosporium.22 Therefore, the interesting combined synergistic effect of the E-Fenton process and enzymatic hydrolysis of white rot fungi provides a novel idea for lignin degradation.
The combination of E-Fenton and P. chrysosporium has the advantages of simple operation, low cost, mildness and greenness, therefore, this paper attempts to construct a composite system of the two to degrade lignin and studies the synergistic mechanism. High purity graphite felt was used as the cathode material for the in situ generation of H2O2 due to its advantages of high conductivity, large specific surface area, corrosion resistance and good biological resistance. The successful construction of the synergistic system is closely dependent on the adjustment of pH in the E-Fenton system by P. chrysosporium and the rapid start of LiP and MnP enzymatic hydrolysis with the addition of trace Fe2+ and 4 V potential. Thus, the degradation effect due to the synergy of the two systems was studied. Moreover, related characterization methods, such as enzyme gene expression, FTIR and 2D-HSQC NMR, were systematically carried out to analyse the structural changes of lignin and study the codegradation mechanism.
2. Materials and methods
2.1 Strain, medium and inoculation
P. chrysosporium BKMF-1767 (ATCC 20696) was procured from the Global Bioresource Center (ATCC). The strain was stored on potato dextrose agar (PDA) plates at 4 °C.23Test liquid medium: a 250 mL conical flask was filled with 150 mL of culture solution consisting of lignin (dealkaline) (as a carbon source, 0.5 g L−1), NaCl (0.1 g L−1), MgSO4 (0.1 g L−1), KH2PO4 (1 g L−1), CaCl2·6H2O (0.1 g L−1), ammonium tartrate (0.1 g L−1), and trace element solution (3 mL L−1). The trace element solutions included FeSO4·7H2O (1.5 g L−1), CoSO4·7H2O (1 g L−1), ZnSO4·7H2O (1 g L−1), CuSO4·5H2O (20 mg L−1), MnSO4 (0.6 g L−1), and Vitamin B1 (0.6 g L−1). The above reagents were supplied by Sinopharm Chemical Reagent Co., Ltd (Beijing, China).
The PDA slant liquid medium previously stored in the refrigerator at 4 °C was removed, and the mycelium was inoculated into the liquid medium (using glucose as the carbon source) with an inoculation loop, incubated in a shaker at 38 °C for 96 hours and then prepared for use. Mycelium was obtained by filtration using filter paper, washed, weighed and divided equally into subsequent experiments.
2.2 Optimization of catalytic systems
The lignin degradation reactions were carried out in a 250 mL conical flask. A highly stable platinum electrode was used as the anode and a high purity graphite carbon felt made of polyacrylonitrile-based activated carbon fibre was used as the cathode electrode material, maintaining a 10 cm spacing between them.Determination of optimum potential: The lignin liquid medium inoculated with P. chrysosporium was applied with 3 V, 3.5 V, 4 V, 4.5 V, 5 V, and 5.5 V and incubated for 48 h at 60 rpm and 38 °C. Afterwards, the pH, lignin content, and enzymatic activity of the two enzymes were measured by sampling.
2.3 Construction and comparison of the catalytic degradation system of lignin
E-Fenton/enzyme catalysis (E-F/P): P. chrysosporium was inoculated into the test liquid medium and then a constant 4 V current was applied. Afterwards, the reaction vessels were placed in a constant temperature shaker with a sterile film seal and incubated for 96 h at 60 rpm and 38 °C. The pH, lignin content, and enzymatic activity of LiP and MnP in the catalytic system were measured every 12 h. Before the experiment, high-temperature autoclave treatment was performed, and the culture unit was assembled on a sterile table.E-Fenton catalysis (E-F): In this degradation system, no bacteria were inoculated, only the applied potential was provided, and other conditions and assays were consistent with E-F/P.
Enzyme catalysis (P): in this system, only the bacteria were inoculated, no power was applied, and all other conditions and assays were kept consistent with E-F/P.
In addition, a one-way analysis of variance (ANOVA) was performed to examine for significant variations with the statistical function of origin.
2.4 Calculation of lignin degradation rate
The principle was to quantify the change in lignin content by acetylating the phenolic hydroxyl group in lignin with the test reagent and then measuring its characteristic peak at 280 nm with an enzyme labeller (Tecan Infinite 200Pro).The lignin degradation ratio was calculated using the following formula:
| R(%) = 100 × (m0 − m)/m0 |
2.5 Measurement of enzyme activity
LiP activity was measured based on an ultraviolet spectrophotometry that monitored the oxidation process of veratryl aldehyde at 310 nm. The reaction was initiated by the addition of H2O2 to the standard reaction mixture. The standard reaction mixture consisted of 1.8 mL of sodium tartrate buffer (250 mM, pH 3.0), 0.6 mL of veratryl alcohol (10 mM), 0.03 mL of hydrogen peroxide solution (40 mM), and 0.6 mL of aqueous enzyme extract.24The MnP activity was determined by ultraviolet spectrophotometer at 290 nm to determine the rate of formation of Mn3+ (obtained by oxidation of Mn2+). The reaction was initiated by adding 60 mL of H2O2 (10 mM) to the reaction mixture. The reaction mixture contained 2.4 mL of sodium succinate (50 mM, pH 4.5) and 0.6 mL of enzyme sample.25
2.6 Determination of the relative expression of the Lip and MnP genes
The total RNA of P. chrysosporium was extracted from E-Fenton and lignin-induced and lignin-only induced P. chrysosporium in a glucose liquid medium by the liquid nitrogen grinding method. Then, the reaction solution was put into the PCR instrument, and the reaction was carried out according to the following reaction procedure: 37 °C for 15 min; 85 °C for 5 s; and 4 °C for holding. After finishing, it was removed and stored at −20 °C. Finally, qRT–PCR was performed on the three sets of samples with a fluorescent quantitative PCR instrument (Roche, LightCycler 480) to determine the mRNA levels of the LiP and MnP genes produced by the three sets of P. chrysosporium. The real-time PCR primer pairs for MnP and LiP of P. chrysosporium were: F: 5′-ACTAGTATGGCCTTCAAGTCCCTCA-3′, R: 5′-CCATGGTTATGCAGGGCCGTTGAAC-3′; F: 5′-ACTAGTATGGCCTTCAAGCAGCTCT-3′, R: 5′-GCGGCCGCAAGCACCCGGAGGCGG-3′.2.7 Lignin structural analysis
3. Results and discussion
3.1 Effect of external potential on the catalytic degradation of bacteria
In this study, potential gradients of 3 V, 3.5 V, 4 V, 4.5 V and 5 V were set to determine the optimum potential for P. chrysosporium catalysis by electric fields. The pre-experiments showed that the data of the E-Fenton combined with P. chrysosporium catalytic system at different potentials were basically stable and that the comparison trend was obvious after 48 h, so 48 h was chosen as the observation time point. Fig. 1 shows that the catalytic effect of P. chrysosporium was not proportional to the potential intensity of the electric field with a tendency of promoting and then inhibiting. This was attributed to the activating effect of H2O2, which was provided by the E-Fenton system constructed by the applied electric field, on MnP and LiP. However, larger potentials (>4 V) produce more H2O2, causing the high concentration of ROSs to exceed the antioxidant capacity of P. chrysosporium, resulting in oxidative stress.26,27 Further, cell membrane rupture or damage to DNA and proteins in P. chrysosporium leads to cell death and impaired catalytic lignin breakdown. In agreement with the results of Fig. 1c and d, the inhibition of Lip and Mnp activity occurred at >4 V.28,29 According to Fig. 1, when the externally applied potential was 4 V, the lignin degradation rate reached 43.8% after 48 h, and the activities of MnP and LiP reached maximum values of 355.1 (U L−1) and 50.15 (U L−1), respectively. This was attributed to the fact that ROSs, which has a high oxidation capacity, was produced by the catalytic decomposition of H2O2 generated by E-Fenton in the presence of ferrous ions, which promoted the oxidation of lignin.30 In addition, within the voltage range tolerated by the bacterium, approximately 4 V optimally increased the permeability of the bacterium's cell membrane, facilitating the transport of nutrients31 and increasing the stimulation and release of enzymes, thus improving the degradation of lignin. In addition, 4 V promoted the rapid growth of P. chrysosporium, causing it to metabolize lignin fragments to secrete organic acids, promoting a rapid decrease in environmental pH and providing the optimum acidic conditions for E-Fenton.32,33 These results indicated that 4 V was the optimum potential for P. chrysosporium to catalyse lignin degradation within a certain potential range. | ||
| Fig. 1 Degradation rate of lignin (a), pH value (b) and MnP (c) and LiP (d) activity of P. chrysosporium cultured for 48 h under different applied potentials. | ||
3.2 Synergistic catalysis for lignin degradation
Based on the above results, the external application of E-Fenton combined with P. chrysosporium-catalysed lignin degradation (E-F/P) was set as the experimental group, only potentiometric formation of the E-Fenton system to catalyse lignin degradation (E-F) was set as control Group 1, and only P. chrysosporium-catalysed lignin degradation (P) was set as control Group 2. After 96 h of incubation, the catalytic degradation rates of the three groups for lignin are shown in Fig. 2(a): E-F/P 62%, E-F 22%, and P 19%. By tracking the lignin degradation effect of each group, it was found that the degradation rate of lignin by E-F/P was greater than that of the control group in all time periods, which indicated that the combined effect of E-Fenton and P. chrysosporium greatly promoted the degradation of lignin (p < 0.05., indicating that there was a significant difference between the experimental group and the control group). This was related to the strong oxidation of ROSs provided by electro-Fenton to break the lignin functional group. | ||
| Fig. 2 Degradation rate of lignin (a), pH value (b) and MnP (c) and LiP (d) activity of E-Fenton/enzyme catalysis (E-F/P), E-Fenton catalysis (E-F) and Enzyme catalysis (P) systems. | ||
The pH values of different culture stages are shown in Fig. 2(b). As the incubation progressed, the pH of E-F/P dropped more rapidly and eventually stabilized at a lower value than that in the control group. The pH of E-F/P dropped rapidly from an initial value of 4.7 to approximately 3, and then the pH began to rise slowly and eventually stabilized at approximately 3.5, as a result of the synergistic effect of E-Fenton and P. chrysosporium. This was because when the fungal cells were placed in an applied potential of a suitable intensity, the permeability of the cell membrane was improved, which was beneficial to the introduction of various exogenous substances into living cells34 and accelerated the metabolic activity of P. chrysosporium.28,35–37 Microorganisms convert polysaccharides to organic acids during lignin degradation, which leads to a faster pH drop in E-F/P. Therefore, the acidic environment created by P. chrysosporium became necessary to constitute an E-Fenton. No additional acid was required to adjust the pH making the whole process greener and more economical. In addition, the microorganisms made further use of organic acids, promoting a rise in pH at the end of the culture.
As shown in Fig. 2c and d, E-F/P and P showed similar trends in the enzyme activity tests for LiP or MnP, but the enzyme activities of both enzymes were higher in the E-F/P synergistic system than in the control. The experimental data showed that the changes in the two enzymes could be roughly divided into three stages. With increasing culture time, the changes first decreased, then increased, and gradually began to decline after reaching a certain level. This may be attributed to the period of adaptation shown by P. chrysosporium to lignin as a carbon source, a period of growth that occurs with the use of lignin or electrical potential stimulation, and the period of weakening as the degradation and conversion of the carbon source leads to a gradual decrease in P. chrysosporium metabolism. The figure demonstrates the ability of E-Fenton to promote the catalytic reaction of enzymes in P. chrysosporium, which is consistent with the qPCR results. This was attributed to H2O2, which was cathodically generated in E-Fenton (4 V) cocatalysis with P. chrysosporium and rapidly initiated the hydrolysis of LiP and MnP to accelerate the process of lignin biodegradation.
3.3 Gene expression analysis of LiP and MnP
As shown in Fig. 3, the mRNA levels of P. chrysosporium were compared after E-Fenton and lignin induction, and lignin only induction was compared for 48 h. Among them, the utilisation of LiP and MnP played a vital role in the catalytic degradation of lignin. MnP is a peroxidase containing haemoglobin and requires H2O2 to initiate the reaction. MnP uses Mn2+ as an electron donor, peroxidizing to produce Mn3+ and then chelating with dicarboxylic acids to form a stable chelated Mn3+, which acts as a mediator in redox reactions.38 LiP was also initiated by H2O2, which degraded the alkyl side chains of lignin by generating aryl cation radicals for a variety of catalytic oxidation reactions.39–41 During enzymatic transcription, the gene expression of both MnP and LiP was higher in the E-F/P system than in the P system, which may be induced by the stress response mechanism42 generated by E-Fenton. In addition, based on the changes in gene expression within the two enzymes in the three groups of samples, it was known that the expression of the MnP gene was higher than that of LiP under potential induction. This indicates that MnP is the main enzyme gene that catalyses the degradation of lignin under this potential condition. Meanwhile, a genetic perspective confirmed that the E-Fenton combined with P. chrysosporium catalytic system favours the degradation of lignin. | ||
| Fig. 3 Gene expression changes in P. chrysosporium, where A was the strain coinduced with E-Fenton and lignin, and B was the strain induced with lignin only. | ||
The above results indicated that there was indeed some synergy of action between E-Fenton and the bacteria to promote lignin degradation. The reasons for the synergistic effect can be summarized as follows: (1) at the appropriate electrical potential, H2O2 was rapidly produced by the cathodic oxygen reduction reaction (ORR) and slowly secreted by P. chrysosporium, initiating the enzymatic digestion of LiP and MnP.43 (2) Simultaneously, the appropriate electrical potential increased P. chrysosporium cell membrane permeability, accelerating material transport and promoting lignin degradation.31,34 (3) As P. chrysosporium grew at 4 V electrical potential, its secreted organic acids lowered the pH of the environment, and together with Fe2+ in the system, the generated H2O2 constituted an electro-Fenton reaction, generating ROSs to act indiscriminately on the lignin functional group.44 (4) Low molecular weight compounds with reducing ability secreted by P. chrysosporium, such as organic acids, fatty acids, Fe3+ chelators and catechol derivatives, can reduce Fe3+ to Fe2+ and carry out a cyclic synergistic oxidation of lignin.45,46 The mechanism is shown in Fig. 4. This synergistic catalytic system provided H2O2 and an acidic environment to effectively reduce the cost, while the enzyme and ROSs cocatalysis enhanced the degradation of lignin, making it an effective strategy for lignin degradation.
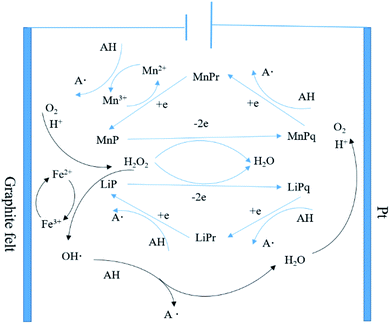 | ||
| Fig. 4 Proposed mechanism of the synergistic degradation of lignin by the E-Fenton reaction and P. chrysosporium. | ||
3.4 Structural analysis of lignin
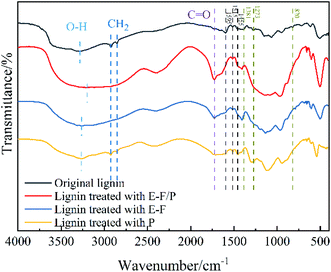
The alteration of the structure of untreated lignin and three groups of treated lignin was investigated by FTIR. As shown in Fig. 5, the O–H stretching originating from both aromatic and aliphatic groups can be clearly observed at approximately 3270 cm−1. However, the centres of their absorption peaks were clearly different, and all three treatment groups showed varying degrees of redshift here relative to the original lignin sample (3284.56 cm−1). It was possible that the hydrogen bond formed during the catalysis process weakened the O–H bond stretching vibration, and at the same time, there may also be a conjugated system formed after the reaction that caused the peak to shift to a low wavenumber.47 Among them, the strongest redshift (3199.13 cm−1) was observed after the E-F/P synergistic treatment. The characteristic signals at 2925 and 2854 cm−1 were attributed to methylene groups in the lignin side chain structure,48 and for the three sets of experiments, only the lignin retained the side chain after enzyme treatment. This indicated that P. chrysosporium treatment alone was very mild, while the externally applied potential helped to improve the catalytic effect. More importantly, the characteristic signal of the carbonyl group was significantly enhanced by the E-F/P treatment compared to the P and E-F treatments (approximately 1725 cm−1).The aromatic skeleton of lignin corresponds to approximately 1595 cm−1, 1515 cm−1 and 1455 cm−1. After P treatment alone, the characteristic signal did not change significantly, but the characteristic signal overlapped or disappeared when the potential was given, especially when the enzyme was combined with E-Fenton, indicating that electricity plays an important role in the induction of catalysis of the enzyme. The characteristic signals at approximately 1381 cm−1, 1273 cm−1 and 820 cm−1 corresponded to guaiacyl (G), syringyl (S) and p-hydroxyphenyl (H) in the lignin structure, respectively, which clearly showed that the three catalytic groups had certain degradation effects on the lignin monomer.49 In summary, the FTIR results showed that the magnitude of the effect of the three catalytic systems on the lignin structure occurs in the following order: E-F/P > F > P, and that the combined treatment of E-Fenton and enzyme has a synergistic catalytic effect. The structure and composition of untreated-lignin and three groups of treated lignin were resolved by 2D-HSQC NMR. The 2D-HSQC spectra of lignin are shown in Fig. 6 and attributed according to the published literature,49–51 and its corresponding signal attribution can be found in Table 1. The overall analysis of Fig. 6 shows that after the three catalysts, the signal intensity of the side chain structure in the aliphatic region (δC/δH 50–90/2.5–5.8) was correspondingly weakened and the signal of the functional group in the lignin aromatic region (δC/δH 90–150/5.5–8.5) almost disappeared. Fig. 6 shows that the intensity of the C–H signal of the methoxyl group, one of the characteristic functional groups of lignin, was as follows in the spectrum of the three groups of catalysed lignin: Origin > P > E-F > E-F/P. Therefore, E-Fenton and P. chrysosporium played a synergistic catalytic role in the degradation of methoxyl. Bγ was consistent with the methoxyl change in catalysis. In addition, β-O-4 was the main bonding signal of the lignin side chain structure,52 and Aγ was more favourably degraded in the E-F/P treatment, but the Aα(S) intensity was enhanced with the degradation of lignin. Moreover, the G2′, PCA3.5, G5 and H2,6 found in the initial lignin spectra all disappeared almost completely in the E-F and E-F/P treatments, indicating that the electrocatalysis seen here was more complete than that of pure bacteria. In conclusion, the synergistic degradation effect on lignin was exerted by the catalytic system of E-Fenton combined with P. chrysosporium, which was consistent with the FTIR results of lignin before and after the three sets of catalytic treatments.
4. Conclusions
The catalytic system of E-Fenton combined with P. chrysosporium synergistically promoted the degradation of lignin. Among them, 4 V became the optimum potential to promote the growth and codegradation of lignin by P. chrysosporium. In a control experiment with 96 h of lignin degradation, the lignin degradation rate (62%) and the enzymatic activities of MnP and LiP were significantly higher in E-F/P than in E-F and P, which had the lowest acidity. There was an increase in the gene expression of both MnP and LiP after the application of a suitable electrical potential. In the synergistic system, aliphatic side chains such as aromatic framework β-O-4 and the corresponding functional groups of lignin were completely broken. In summary, E-Fenton and P. chrysosporium were mutually reinforcing, were closely mutually beneficial and had significant advantages in the catalytic degradation of lignin.Author contributions
Yingjian Qin: visualization, writing – original draft, data curation. Na Wang: visualization, writing – original draft, data curation. Zhongmin Ma: data curation, investigation. Jinsheng Li: investigation. Yaozong Wang: investigation. Lihua Zang: conceptualization, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Integration of Science, Education and Industry Innovation Pilot Project of Qilu University of Technology (Shandong Academy of Sciences) (2020KJC-ZD12), and the Shandong Province Key R & D Program (Major Science and technology innovation project) (2021CXGC010802).References
- D. M. Kato, N. Elia, M. Flythe and B. C. Lynn, Bioresour. Technol., 2014, 162, 273–278 CrossRef CAS PubMed.
- R. Zhou, R. Zhou, S. Wang, U. G. Mihiri Ekanayake, Z. Fang, P. J. Cullen, K. Bazaka and K. K. Ostrikov, Bioresour. Technol., 2020, 318, 123917 CrossRef CAS PubMed.
- C. Xu, J. Zhang, Y. Zhang, Y. Guo, H. Xu, C. Liang, Z. Wang and J. Xu, Int. J. Biol. Macromol., 2019, 141, 484–492 CrossRef CAS PubMed.
- W. Zhang, W. Wang, J. Wang, G. Shen, Y. Yuan, L. Yan, H. Tang and W. Wang, Appl. Environ. Microbiol., 2021, 87, e01321–e01355 Search PubMed.
- S. Zhang, J. Xiao, G. Wang and G. Chen, Bioresour. Technol., 2020, 304, 122975 CrossRef CAS PubMed.
- E. Subbotina, T. Rukkijakan, M. D. Marquez-Medina, X. Yu, M. Johnsson and J. S. M. Samec, Nat. Chem., 2021, 13, 1118–1125 CrossRef CAS PubMed.
- K. E. Hammel and D. Cullen, Curr. Opin. Plant Biol., 2008, 11, 349–355 CrossRef CAS PubMed.
- X. Shen, Y. Xin, H. Liu and B. Han, ChemSusChem, 2020, 13, 4367–4381 CrossRef CAS PubMed.
- C. Cai, Z. Xu, H. Zhou, S. Chen and M. Jin, Sci. Adv., 2021, 7, eabg4585 CrossRef CAS PubMed.
- M. Asgher, A. Wahab, M. Bilal and H. M. Nasir Iqbal, Biocatal. Agric. Biotechnol., 2016, 6, 195–201 CrossRef.
- E. Bari, K. Ohno, N. Yilgor, A. P. Singh, J. J. Morrell, A. Pizzi, M. A. Tajick Ghanbary and J. Ribera, Microorganisms, 2021, 9, 247 CrossRef CAS PubMed.
- X. Qin, X. Su, H. Luo, R. Ma, B. Yao and F. Ma, Biotechnol. Biofuels, 2018, 11, 58 CrossRef PubMed.
- L. Jin, G. Zeng, H. Chen, L. Wang, H. Ji, S. Lin, R. Peng and D. Sun, BioResources, 2021, 16, 5494–5507 CAS.
- L. Thanh Mai Pham, M. H. Eom and Y. H. Kim, Enzyme Microb. Technol., 2014, 61–62, 48–54 CrossRef CAS PubMed.
- R. Potumarthi, R. R. Baadhe, P. Nayak and A. Jetty, Bioresour. Technol., 2013, 128, 113–117 CrossRef CAS PubMed.
- F. Q. Wang, H. Xie, W. Chen, E. T. Wang, F. G. Du and A. D. Song, Bioresour. Technol., 2013, 144, 572–578 CrossRef CAS PubMed.
- R. D. Villa, A. G. Trovo and R. F. Nogueira, Chemosphere, 2008, 71, 43–50 CrossRef CAS PubMed.
- J. Zeng, C. G. Yoo, F. Wang, X. Pan, W. Vermerris and Z. Tong, ChemSusChem, 2015, 8, 861–871 CrossRef CAS PubMed.
- C. Hu, D. Huang, G. Zeng, M. Cheng, X. Gong, R. Wang, W. Xue, Z. Hu and Y. Liu, Chem. Eng. J., 2018, 338, 432–439 CrossRef CAS.
- C.-H. Feng, F.-B. Li, H.-J. Mai and X.-Z. Li, Environ. Sci. Technol., 2010, 44, 1875–1880 CrossRef CAS PubMed.
- C. Yang, S. Maldonado and C. R. J. Stephenson, ACS Catal., 2021, 11, 10104–10114 CrossRef CAS.
- V. Arantes, A. M. Milagres, T. R. Filley and B. Goodell, J. Ind. Microbiol. Biotechnol., 2011, 38, 541–555 CrossRef CAS PubMed.
- F. C. Michel, E. A. Grulke and C. A. Reddy, J. Ind. Microbiol., 1990, 5, 103–112 CrossRef CAS.
- H. Tanaka, K. Koike, S. Itakura and A. Enoki, Enzyme Microb. Technol., 2009, 45, 384–390 CrossRef CAS.
- X. Peng, X.-z. Yuan, G.-m. Zeng, H.-j. Huang, H. Wang, H. Liu, S. Bao, Y.-j. Ma, K.-l. Cui, L.-j. Leng and Z.-h. Xiao, Sep. Purif. Technol., 2014, 123, 164–170 CrossRef CAS.
- M. Jaszek, J. Żuchowski, E. Dajczak, K. Cimek, M. Gra̢z and K. Grzywnowicz, Int. Biodeterior. Biodegrad., 2006, 58, 168–175 CrossRef CAS.
- D. J. Jamieson, Redox Rep., 1995, 1, 89–95 CrossRef CAS PubMed.
- L. Loghavi, S. K. Sastry and A. E. Yousef, Biotechnol. Bioeng., 2007, 98, 872–881 CrossRef CAS PubMed.
- B. Ezraty, A. Gennaris, F. Barras and J. F. Collet, Nat. Rev. Microbiol., 2017, 15, 385–396 CrossRef CAS PubMed.
- M. A. Fernandez de Dios, A. G. del Campo, F. J. Fernandez, M. Rodrigo, M. Pazos and M. A. Sanroman, Bioresour. Technol., 2013, 148, 39–46 CrossRef CAS PubMed.
- L. Hou, D. Ji, W. Dong, L. Yuan, F. Zhang, Y. Li and L. Zang, Front. Bioeng. Biotechnol., 2020, 8, 99 CrossRef PubMed.
- J. Jellison, J. Connolly, B. Goodell, B. Doyle, B. Illman, F. Fekete and A. Ostrofsky, Int. Biodeterior. Biodegrad., 1997, 39, 165–179 CrossRef CAS.
- M. Humar, M. Petrič and F. Pohleven, Eur. J. Wood Wood Prod., 2001, 59, 288–293 CrossRef CAS.
- A. Garg, I. M. Mishra and S. Chand, Clean: Soil, Air, Water, 2010, 38, 27–34 CAS.
- J. Cameron Thrash and J. D. Coates, Environ. Sci. Technol., 2008, 42, 3921–3931 CrossRef PubMed.
- A. Schievano, T. Pepe Sciarria, K. Vanbroekhoven, H. De Wever, S. Puig, S. J. Andersen, K. Rabaey and D. Pant, Trends Biotechnol., 2016, 34, 866–878 CrossRef CAS PubMed.
- N. Matsumoto, S. Nakasono, N. Ohmura and H. Saiki, Biotechnol. Bioeng., 1999, 64, 716–721 CrossRef CAS PubMed.
- G.-M. Zeng, M.-H. Zhao, D.-L. Huang, C. Lai, C. Huang, Z. Wei, P. Xu, N.-J. Li, C. Zhang, F.-L. Li and M. Cheng, Int. Biodeterior. Biodegrad., 2013, 85, 166–172 CrossRef CAS.
- O. D. V. Biko, M. Viljoen-Bloom and W. H. van Zyl, Enzyme Microb. Technol., 2020, 141, 109669 CrossRef CAS PubMed.
- G. Janusz, K. H. Kucharzyk, A. Pawlik, M. Staszczak and A. J. Paszczynski, Enzyme Microb. Technol., 2013, 52, 1–12 CrossRef CAS PubMed.
- D. W. Wong, Appl. Biochem. Biotechnol., 2009, 157, 174–209 CrossRef CAS PubMed.
- N. Garcia-Ortiz, F. J. Figueroa-Martinez, U. Carrasco-Navarro, E. Favela-Torres and O. Loera, Fungal Biol., 2018, 122, 487–496 CrossRef CAS PubMed.
- J. A. Mir-Tutusaus, R. Baccar, G. Caminal and M. Sarra, Water Res., 2018, 138, 137–151 CrossRef CAS PubMed.
- F. Li, H. Zhao, R. Shao, X. Zhang and H. Yu, J. Agric. Food Chem., 2021, 69, 7104–7114 CrossRef CAS PubMed.
- V. Arantes and A. M. F. Milagres, Process Biochem., 2006, 41, 887–891 CrossRef CAS.
- A. Gutierrez, J. C. del Rio, M. J. Martinez-Inigo, M. J. Martinez and A. T. Martinez, Appl. Environ. Microbiol., 2002, 68, 1344–1350 CrossRef CAS PubMed.
- M. Lei, S. Wu, C. Liu, J. Liang and R. Xiao, Fuel Process. Technol., 2021, 217, 106812 CrossRef CAS.
- A. Tejado, C. Pena, J. Labidi, J. M. Echeverria and I. Mondragon, Bioresour. Technol., 2007, 98, 1655–1663 CrossRef CAS PubMed.
- Y.-h. Ci, F. Yu, C.-x. Zhou, H.-e. Mo, Z.-y. Li, Y.-q. Ma and L.-h. Zang, Green Chem., 2020, 22, 8713–8720 RSC.
- J. C. del Rio, J. Rencoret, P. Prinsen, A. T. Martinez, J. Ralph and A. Gutierrez, J. Agric. Food Chem., 2012, 60, 5922–5935 CrossRef CAS PubMed.
- H. Kim, J. Ralph and T. Akiyama, BioEnergy Res., 2008, 1, 56–66 CrossRef.
- B. Wang, X.-J. Shen, J.-L. Wen and R.-C. Sun, RSC Adv., 2016, 6, 57986–57995 RSC.
Footnote |
| † Equal authors contribution. |
| This journal is © The Royal Society of Chemistry 2022 |