Open Access Article
Open Access ArticleQuinolizinium-based fluorescent probes for formaldehyde detection in aqueous solution, serum, and test strip via 2-aza-Cope rearrangement†
Ajcharapan Tantipanjaporn‡
b,
Karen Ka-Yan Kung‡b,
Hoi-Yi Sitab and
Man-Kin Wong *ab
*ab
aThe Hong Kong Polytechnic University Shenzhen Research Institute, Shenzhen, 518057, China. E-mail: mankin.wong@polyu.edu.hk
bState Key Laboratory of Chemical Biology and Drug Discovery, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hum, Hong Kong, China
First published on 13th April 2022
Abstract
Formaldehyde is an abundant contaminant in food and environments causing various diseases. Thus, the development of fast, simple, and selective formaldehyde detection is of great interest. Herein, novel quinolizinium-based fluorescent probes were designed based on a 2-aza-Cope rearrangement reaction and showed high selectivity to formaldehyde by fluorescence emission shift. We successfully reduced the detection time by increasing the bulkiness of the homoallylic moiety. The probes were applied to detect formaldehyde in aqueous solution, serum, and paper format.
Formaldehyde (FA) is one of the most abundantly reactive carbonyl species (RCS) which can cause contamination in environments such as wood, furniture, textiles, construction, carpeting, disinfectants and preservatives. FA can be found naturally in a variety of fruits, vegetables, dried mushrooms, meat, alcoholic beverages, milk products, crustaceans, and fish in which the level is up to a range of 300 to 400 mg kg−1.1 Moreover, FA can be endogenously generated in living systems during diverse biological processes including one-carbon metabolism, and demethylation of DNA, RNA, and histones.2–4 Although, FA is also a ubiquitous and essential metabolite in biological systems, accumulation of excess FA can cause various acute and chronic adverse effects, such as genotoxicity, carcinogenicity, and teratogenicity.5,6 The toxicities of FA arise from reaction of FA mostly with guanine and adenine in DNA and produce various types of DNA lesions including DNA adducts, DNA intrastrand crosslinks, DNA inter-strand crosslinks and DNA-protein crosslinks.7 The normal concentration of endogenous FA in human blood is about 2–3 mg L−1 or 67–100 μM but abnormal level of FA can be up to 800 μM indicating an association with many types of diseases including diabetes, neurodegenerative diseases, chronic liver diseases, atherosclerosis, and heart disorders.8–11 Thus, the development of facile and selective strategies for FA detection is vital for environmental and medicinal assessments.
Nowadays, the research developments for selective detection of FA have been focused on two reaction types involving Schiff base (amine and hydrazine) and 2-aza-Cope rearrangement reactions (Scheme 1).12–15 These two main approaches are highly selective and sensitive to FA but the 2-aza-Cope reaction based-approach showed much longer detection time than the Schiff base reaction (Table S1, ESI†).12 Thus, it is still challenging to develop fluorescent probes based on 2-aza-Cope rearrangement with fast detection time. In 2017, Brewer et al. have incorporated a geminal dimethyl substituent to the homoallylic amine (R = Me) resulting in increasing reactivity (detection time was 2 h).16 This result could indicate that the bulky homoallylic amine could accelerate the reaction rate. However, few 2-aza-Cope based-fluorescent probes have utilized the geminal dimethyl substituted homoallylic amine as the reaction site (Table S2, ESI†).17–19
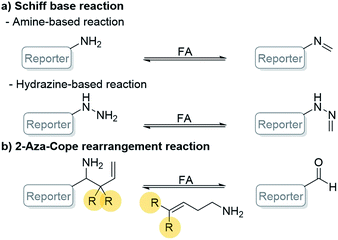 | ||
| Scheme 1 General reaction-based FA detection involving (a) Schiff base reaction and (b) 2-aza-Cope rearrangement reaction. | ||
Herein, we report the design and synthesis of quinolizinium-based fluorescent probes to detect FA by 2-aza-Cope rearrangement reaction with different homoallylic amine moieties (Scheme 2).17 We found that the incorporation of the geminal dimethyl substituted homoallylic amine to quinolizinium improved the detection time as low as 15 min. To our knowledge, QA2 showed the shortest detection time to detect FA comparing with other 2-aza-Cope reaction-based probes. Moreover, using quinolizinium as a core reporter for sensor development has various advantages due to its good quantum yield, high water solubility due to cationic aromatic heterocycles, diversity of structural modification, and excellent photostability.20
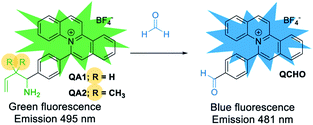 | ||
| Scheme 2 Our strategy to detect FA by quinolizinium-based fluorescent probes QA1–2 through a 2-aza-Cope rearrangement reaction. | ||
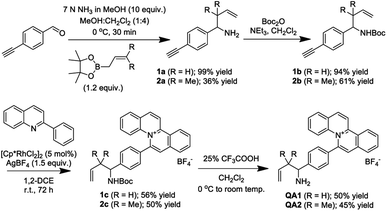
The synthesis of QA1–2 probes was outlined in Scheme 3. Moreover, we synthesized QCHO for the photophysical property studies to compare with the photophysical properties of the 2-aza-Cope reaction adducts from the reactions between QA1–2 and FA. The synthetic procedures and structural characterization of all compounds (confirmed by 1H NMR, 13C NMR and HRMS) were reported in ESI.† The synthesis was briefly described by taking QA1 probe as an example, 4-ethynylbenzaldehyde was firstly reacted with ammonia and then allylboronic acid pinacol ester to obtain 1a (99% yield). Secondly, the amino group was protected using di-tert-butyl decarbonate to form 1b (94% yield). Then, a [Cp*RhCl2]2-catalyzed C–H activation and annulation reaction of terminal alkyne 1b and 2-phenylquinoline to yield N-Boc protected quinolizinium product 1c (56% yield). Finally, the deprotection was carried out to give QA1 probe in 50% yield (Scheme 3).
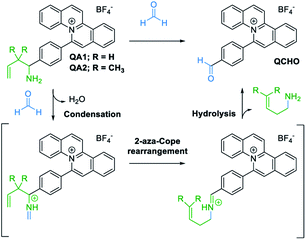
In the presence of FA, the probes (QA1–2) underwent condensation with FA followed by a 2-aza-Cope rearrangement reaction and subsequent hydrolysis to obtain an electron withdrawing aldehyde moiety in QCHO product which was confirmed by HRMS (QCHO C24H16NO+: calcd 334.1226; found, 334.1235 (for QA1) and 334.1232 (for QA2)) (Scheme 4 and Fig. S1, ESI†). Thus, QA1–2 could be employed for the FA detection.
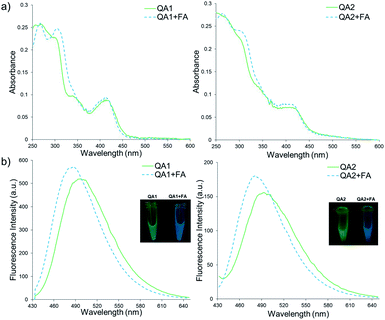
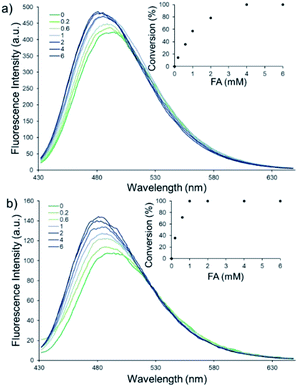
Firstly, the photophysical properties of QA1–2 were studied. Upon the addition of FA, the absorption peaks of probes QA1 and QA2 slightly shifted from 415 to 413 nm (Fig. 1a) and the emission peaks of both probes also shifted from 495 nm to a new blue emissive peak at 481 nm (Fig. 1b) when measured in CH3CN-10 mM PBS buffer pH 7.4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) at 37 °C. The blue shift of both absorption and emission spectra indicated the formation of QCHO via 2-aza-Cope rearrangement in which the absorption and emission spectra of QA1–2 in the presence of FA were similar with the spectra of the synthesized QCHO (Fig. S2, ESI†). QCHO showed the emissive blue shift comparing with QA1 and QA2 because of more electron deactivation effect of the resulting aldehyde group than alkyl amine which correlate with DFT/TD-DFT calculations of quinolizinium in our previous work.20 Interestingly, the fluorescence colour change was clearly observed through 365 nm UV light irradiation from intense green to blue colour upon addition of FA, correlating with their emission wavelengths (Fig. 1b; inset images). Fluorescence quantum yields of QA1 and QA2 in CH3CN-10 mM PBS buffer pH 7.4 (1
9, v/v) at 37 °C. The blue shift of both absorption and emission spectra indicated the formation of QCHO via 2-aza-Cope rearrangement in which the absorption and emission spectra of QA1–2 in the presence of FA were similar with the spectra of the synthesized QCHO (Fig. S2, ESI†). QCHO showed the emissive blue shift comparing with QA1 and QA2 because of more electron deactivation effect of the resulting aldehyde group than alkyl amine which correlate with DFT/TD-DFT calculations of quinolizinium in our previous work.20 Interestingly, the fluorescence colour change was clearly observed through 365 nm UV light irradiation from intense green to blue colour upon addition of FA, correlating with their emission wavelengths (Fig. 1b; inset images). Fluorescence quantum yields of QA1 and QA2 in CH3CN-10 mM PBS buffer pH 7.4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) was 0.14 and 0.12, respectively. The incubation of FA with QA1–2 displayed similar quantum yield as 0.14 which was the same as the quantum yield of QCHO (Table S3, ESI†).
9, v/v) was 0.14 and 0.12, respectively. The incubation of FA with QA1–2 displayed similar quantum yield as 0.14 which was the same as the quantum yield of QCHO (Table S3, ESI†).
The time-dependent fluorescence responses of QA1–2 measured in CH3CN-10 mM PBS buffer pH 7.4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) at 37 °C were then performed (Fig. S3, ESI†). To our delight, the fluorescence response of QA2 to FA reached an equilibrium in 15 min which was much faster than QA1 (60 min) suggesting that the geminal dimethyl substituted homoallylic amine in QA2 could improve the reactivity and shorten the reaction time as reported by Brewer et al.16 Subsequently, the fluorescence responses of QA1–2 at different pH values (2–9) with and without FA were studied. From acidic to slightly basic conditions (pH 2–8), QA1–2 showed maximum emission around 495 nm and the conditions with FA showed the blue shift emissive peak around 481 nm but at pH 9 showed less blue shift (Fig. S4, ESI†). These results showed that QA1–2 were insensitive towards the pH changes between pH 2–8 allowing the probes to detect FA at various pH values.
9, v/v) at 37 °C were then performed (Fig. S3, ESI†). To our delight, the fluorescence response of QA2 to FA reached an equilibrium in 15 min which was much faster than QA1 (60 min) suggesting that the geminal dimethyl substituted homoallylic amine in QA2 could improve the reactivity and shorten the reaction time as reported by Brewer et al.16 Subsequently, the fluorescence responses of QA1–2 at different pH values (2–9) with and without FA were studied. From acidic to slightly basic conditions (pH 2–8), QA1–2 showed maximum emission around 495 nm and the conditions with FA showed the blue shift emissive peak around 481 nm but at pH 9 showed less blue shift (Fig. S4, ESI†). These results showed that QA1–2 were insensitive towards the pH changes between pH 2–8 allowing the probes to detect FA at various pH values.
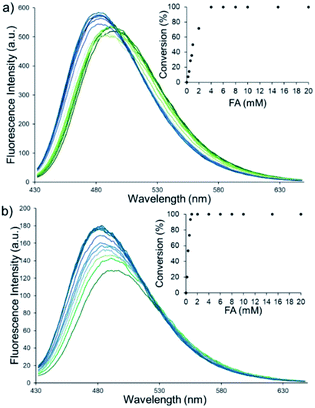
The fluorescence response of QA1–2 towards different concentrations of FA (0–20 mM) was then investigated (Fig. 2). Similar linear response of QA1 and QA2 to the FA concentrations were found in a range of 0–1 mM. The detection limits of QA1 and QA2 were calculated to be 5 and 1 μM, respectively (Fig. S5, ESI†).
To evaluate the selectivity of QA1–2 towards FA, a variety of interfering compounds including acetaldehyde, propionaldehyde, butyraldehyde, 2-furaldehyde, glyoxal, acetone, NaCl, KCl, CaCl2, FeSO4·7H2O, CuSO4·5H2O, ZnCl2, L-GSH, L-cysteine, H2O2, KNO3, and NaNO2 were tested. Each interfering compound (5 mM) was added into QA1–2 (10 μM) in CH3CN-10 mM PBS buffer pH 7.4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) conditions. All interfering compounds did not cause the fluorescence shift to 481 nm showing that QA1–2 possessed a high selectivity to FA (Fig. S6a, ESI†). Next, competition experiments of QA1–2 were investigated by adding the interfering compounds (5 mM) into QA1–2 (10 μM) in the presence of FA (5 mM). The results showed that the interfering compounds did not alter the fluorescent shift to 481 nm suggesting that QA1–2 can effectively detect FA even in the presence of the interfering compounds (Fig. S6b, ESI†). Thus, it is suggested that QA1–2 could be applied for FA detection in complex reaction mixtures.
9, v/v) conditions. All interfering compounds did not cause the fluorescence shift to 481 nm showing that QA1–2 possessed a high selectivity to FA (Fig. S6a, ESI†). Next, competition experiments of QA1–2 were investigated by adding the interfering compounds (5 mM) into QA1–2 (10 μM) in the presence of FA (5 mM). The results showed that the interfering compounds did not alter the fluorescent shift to 481 nm suggesting that QA1–2 can effectively detect FA even in the presence of the interfering compounds (Fig. S6b, ESI†). Thus, it is suggested that QA1–2 could be applied for FA detection in complex reaction mixtures.
We then examined the feasibility of QA1–2 for practical applications through the FA detection in mouse serum (Fig. 3) with literature ref. 21–23. The mouse serum was prepared as mentioned in the ESI.† In the CH3CN-10 mM PBS buffer pH 7.4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) conditions containing 0.1% mouse serum, QA1–2 (10 μM) exhibited an intense emission band centered at 495 nm. Upon addition of FA with various concentrations at 37 °C, the maximum emission wavelength at 495 nm gradually shifted to 481 nm (blue shift) with increasing concentrations of FA (0–6 mM). The incubation time of QA1 and QA2 were 60 and 15 min, respectively. We found that the linear range of QA1 and QA2 to FA was 0–1 mM. This result indicated that QA1–2 were able to detect FA in the mouse serum. We also studied the effect of mouse serum concentrations (0.1, 1, and 5%) on the FA detection (Fig. S7, ESI†). We found that QA1 and QA2 showed the blue shift upon the treatment with FA (10 mM) in all percentages of the mouse serum used. Thus, the presence of mouse serum did not interfere the detection of FA by our probes.
9, v/v) conditions containing 0.1% mouse serum, QA1–2 (10 μM) exhibited an intense emission band centered at 495 nm. Upon addition of FA with various concentrations at 37 °C, the maximum emission wavelength at 495 nm gradually shifted to 481 nm (blue shift) with increasing concentrations of FA (0–6 mM). The incubation time of QA1 and QA2 were 60 and 15 min, respectively. We found that the linear range of QA1 and QA2 to FA was 0–1 mM. This result indicated that QA1–2 were able to detect FA in the mouse serum. We also studied the effect of mouse serum concentrations (0.1, 1, and 5%) on the FA detection (Fig. S7, ESI†). We found that QA1 and QA2 showed the blue shift upon the treatment with FA (10 mM) in all percentages of the mouse serum used. Thus, the presence of mouse serum did not interfere the detection of FA by our probes.
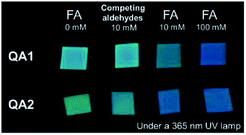
We further demonstrated another application of QA1–2 as the test strips for FA detection (Fig. 4). Filter paper was loaded with QA1–2 (10 mM in CH3CN) and let the strips dried. To detect FA, FA solution was dropped on the strip. After 10 min, the test strips were irradiated under the 365 nm UV lamp. We found that QA1 and QA2 test strips showed green fluorescence emission. In the presence of FA, the emission colour changed from green to blue moderately observed at 10 mM and clearly observed at 100 mM. Moreover, the strips of QA1 and QA2 were also tested with 10 mM of each acetaldehyde, propionaldehyde, butyraldehyde, 2-furaldehyde, glyoxal, and acetone. The results showed that the emission colour did not change to blue suggesting that the test strips tolerated with other competing aldehydes demonstrating the potential of our probes to detect FA qualitatively. These results implied that QA1–2 test strips could be applicable with FA detection in contaminated food samples as formaldehyde has been utilized as food preservative and bleaching agent illegally with extreme cases of over 4250 mg kg−1.24
In summary, we have achieved the design, synthesis, and applications of probes QA1–2 for FA detection. The QA1–2 probes utilized a quinolizinium as the fluorophore moiety appended with homoallylamine as the FA-responsive group based on the 2-aza-Cope rearrangement. In the presence of FA, QA1–2 showed the blue shift of fluorescence emission from 495 to 481 nm in which the fluorescence colour changed from green to blue as observed by naked eyes. Remarkably, we have demonstrated that the incorporation of geminal dimethyl substituents to the homoallylic amine moiety of QA2 reduced the FA detection time to 15 min which showed extremely fast response to FA comparing with other 2-aza-Cope-based fluorescent probes. The probes showed high selectivity with FA and were able to function at various pH (pH 2–8) as well as in the presence of the competing agents. The linear response of QA1 and QA2 showed in the wide range of 0–1 mM FA. The detection limits of QA1 and QA2 were evaluated to be 5 μM and 1 μM, respectively. Furthermore, we have carried out applications of the QA1–2 probes to detect FA in the mouse serum and utilized as the FA test strips.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the financial support by the Shenzhen Science and Technology Innovation Commission (JCYJ20170818104257975), Hong Kong Research Grants Council (15300117), the State Key Laboratory of Chemical Biology and Drug Discovery, The Hong Kong Polytechnic University (G-ZVVG and G-UACN), and Innovation and Technology Commission (PRP/075/19FX), and the supply of the mouse serum by Dr Ben Chi-Bun Ko and Dr Wing-Cheung Chan.Notes and references
- J. Chiou, A. H. H. Leung, H. W. Lee and W.-T. Wong, J. Integr. Agric., 2015, 14, 2243–2264 CrossRef CAS.
- C. Umansky, A. E. Morellato and L. B. Pontel, Nat. Commun., 2021, 12, 1–3 CrossRef PubMed.
- H. Reingruber and L. B. Pontel, Curr. Opin. Toxicol., 2018, 9, 28–34 CrossRef.
- T. Li, Y. Wei, M. Qu, L. Mou, J. Miao, M. Xi, Y. Liu and R. He, Genes, 2021, 12, 913 CrossRef CAS PubMed.
- J. A. Swenberg, B. C. Moeller, K. Lu, J. E. Rager, R. C. Fry and T. B. Starr, Toxicol. Pathol., 2013, 41, 181–189 CrossRef PubMed.
- R. Baan, Y. Grosse, K. Straif, B. Secretan, F. El Ghissassi, V. Bouvard, L. Benbrahim-Tallaa, N. Guha, C. Freeman and L. Galichet, Lancet Oncol., 2009, 10, 1143–1144 CrossRef PubMed.
- M. Kawanishi, T. Matsuda and T. Yagi, Front. Environ. Sci. Eng., 2014, 2, 1–8 Search PubMed.
- Z. Tong, C. Han, W. Luo, X. Wang, H. Li, H. Luo, J. Zhou, J. Qi and R. He, Age, 2013, 35, 583–596 CrossRef PubMed.
- T. Szarvas, E. Szatlóczky, J. Volford, L. Trézl, E. Tyihák and I. Rusznak, J. Radioanal. Nucl. Chem., 1986, 106, 357–367 CrossRef CAS.
- D. Takeshita, C. Nakajima-Takenaka, J. Shimizu, H. Hattori, T. Nakashima, A. Kikuta, H. Matsuyoshi and M. Takaki, Basic Clin. Pharmacol. Toxicol., 2009, 105, 271–280 CrossRef CAS PubMed.
- Z. Tong, W. Wang, W. Luo, J. Lv, H. Li, H. Luo, J. Jia and R. He, J. Alzheimer's Dis., 2017, 55, 1031–1038 CAS.
- S. K. Manna, T. K. Achar and S. Mondal, Anal. Methods, 2021, 13, 1084–1105 RSC.
- K. J. Bruemmer, T. F. Brewer and C. J. Chang, Curr. Opin. Chem. Biol., 2017, 39, 17–23 CrossRef CAS PubMed.
- K. Wechakorn, S. Supalang and S. Suanpai, Tetrahedron, 2020, 76, 131411 CrossRef CAS.
- S. Huang, Z. Li, M. Liu, M. Zhou, J. Weng, Y. He, Y. Jiang, H. Zhang and H. Sun, Chem. Commun., 2022, 58, 1442–1453 RSC.
- T. F. Brewer, G. Burgos-Barragan, N. Wit, K. J. Patel and C. J. Chang, Chem. Sci., 2017, 8, 4073–4081 RSC.
- Z. Xie, B. Yin, J. Shen, D. Hong, L. Zhu, J. Ge and Q. Zhu, Org. Biomol. Chem., 2018, 16, 4628–4632 RSC.
- K. J. Bruemmer, O. Green, T. A. Su, D. Shabat and C. J. Chang, Angew. Chem., Int. Ed., 2018, 130, 7630–7634 CrossRef.
- Y. Du, Y. Zhang, M. Huang, S. Wang, J. Wang, K. Liao, X. Wu, Q. Zhou, X. Zhang and Y.-D. Wu, Chem. Sci., 2021, 12, 13857–13869 RSC.
- W.-M. Yip, Q. Yu, A. Tantipanjaporn, W.-C. Chan, J.-R. Deng, B. C.-B. Ko and M.-K. Wong, Org. Biomol. Chem., 2021, 19, 8507–8515 RSC.
- W.-Y. O, W.-C. Chan, C. Xu, J.-R. Deng, B. C.-B. Ko and M.-K. Wong, RSC Adv., 2021, 11, 33294–33299 RSC.
- Y. Zhou, J. Yan, N. Zhang, D. Li, S. Xiao and K. Zheng, Sens. Actuators, B, 2018, 258, 156–162 CrossRef CAS.
- H. Yang, F. Wang, J. Zheng, H. Lin, B. Liu, Y. D. Tang and C. J. Zhang, Chem.–Asian J., 2018, 13, 1432–1437 CrossRef CAS PubMed.
- K.-F. Wong, J.-R. Deng, X.-Q. Wei, S.-P. Shao, D.-P. Xiang and M.-K. Wong, Org. Biomol. Chem., 2015, 13, 7408–7411 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra01397e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |