Open Access Article
Open Access ArticleIntracellular mechanism of antimicrobial peptide HJH-3 against Salmonella pullorum
Qing Wang,
Yanzhao Xu* and
Jianhe Hu
College of Animal Science and Veterinary Medicine, Henan Institute of Science and Technology, Xinxiang 453003, China. E-mail: xuyanzhao@hist.edu.cn
First published on 13th May 2022
Abstract
To explore the potential intracellular mechanism of the antimicrobial peptide HJH-3 in killing Salmonella, a DNA blocking test and scanning electron microscopy (SEM) were used to determine the ability of the peptide to bind bacterial DNA in vitro. Laser confocal analysis and electron microscopy were used to observe the binding of antimicrobial peptide HJH-3 and Salmonella DNA, and flow cytometry was used to analyze the effect of antimicrobial peptides on cell division in vivo. The results showed that HJH-3 can bind to DNA to block the diffusion and migration of DNA in agarose gel. Laser confocal microscopy revealed that antimicrobial peptide HJH-3 penetrated the bacterial cell membrane and bound with bacterial DNA. Transmission electron microscopy showed that antimicrobial peptide HJH-3 aggregated in the nucleoid of Salmonella cells, and through a channel in the membrane destroyed by the antimicrobial peptide, DNA and other intracellular contents were excreted, and polymerized DNA was fragmented. The results of the flow cytometry analysis confirmed that the death rate of Salmonella increased significantly after exposure to antimicrobial peptide HJH-3 and increased with increasing antimicrobial peptide concentration. These results suggest that AMP HJH-3 may be a candidate antimicrobial agent to treat infectious diseases caused by Salmonella pullorum.
1. Introduction
Antimicrobial peptides (AMPs) are components of the innate immune system.1,2 To date, more than 3000 natural AMPs have been discovered.3 AMPs have become important alternatives to antibiotics because of their unique mechanisms of action against a variety of bacteria and viruses, including multidrug-resistant bacteria4,5 and human immunodeficiency viruses.6,7Compared with that of traditional antibiotics, the mechanisms of action of AMPs are not sufficiently thorough.8 The mechanisms of action of AMPs are debated, and among the many opinions, no theory explains the mechanisms of all AMPs.9–11 However, it is clear that the interaction between the positive charge and amphiphilic structure of the cationic peptide and the negative charge of cell membrane lipids plays an important role in the biological functions of these peptides.12,13 The diversity of the AMP structure determines the diversity of its modes of action and function. Because of their different structures and functions, AMPs belong to different families, and the action modes and mechanisms of AMPs in different families differ. The main bactericidal mechanisms of AMPs are extracellular bactericidal mechanisms and intracellular bactericidal mechanisms.12,14 Many studies on the mechanisms of AMPs have shown that even the same AMP can kill bacteria by simultaneously invoking different bactericidal mechanisms;15–17 in contrast, different kinds of AMPs can evoke the same bactericidal mechanism.12,18
Most of the AMPs can kill bacteria by breaking the structure of the bacterial cell membrane, and the “blanket” model, the “barrel plate” model, the “ring pore” model and the “coacervation” model are all based on the membrane action mechanism of AMPs.19 In recent years, scientists have found that some AMPs can successfully kill bacteria without destroying the integrity of cell membrane. So, it follows that AMPs have other antibacterial mechanisms in addition to the membrane mechanism of action. Intracellular action targeting doctrine of AMPs is a typical representative of the intracellular action mechanism of AMPs.20 The intracellular bactericidal mechanism of AMPs refers to that under the action of static electricity, some AMPs attract and bind to the bacterial cell membrane without damaging the cell membrane, but they accumulate in the cell after penetrating the cell plasma membrane and specifically bind with the intracellular target, thus interfering with the normal metabolism of cells,21 Investigation of AMPs using laser confocal microscopy techniques can indirectly verify the existence of non-membrane destruction mechanism of AMPs.
Therefore, only through in-depth exploration of the bactericidal mechanism of AMPs can we further modify the structure of AMPs to improve their bactericidal activity and reduce their cytotoxicity and thereby lay a theoretical foundation for promoting the clinical application of AMPs.
In our previous study, AMP P3 was isolated from the bovine erythrocyte haemoglobin alpha subunit.8,9,22
To further enhance the antimicrobial activity of P3 and reduce its cytotoxicity, in this study, we modify P3 according to the principle of AMP modification to obtain an HJH-3 analogue with higher antimicrobial activity than P3 and study its intracellular mechanism of action. The findings lay a theoretical foundation for the development and application of AMP HJH-3.
2. Materials and methods
2.1 AMPs and bacteria strains
The AMP HJH-3 was synthesized by solid-phase synthesis. The standard strain of Salmonella pullorum (S. pullorum, CVCC533, bought from China Veterinary Culture Collection Center) was used.2.2 Physicochemical properties of AMP HJH-3
In our previous study, the sequence of the natural bovine erythrocyte-derived AMP P3 was modified, and the AMP analogue HJH-3 was identified as having better antimicrobial activity than AMP P3. The ProtParam online tool was used to analyse the isoelectric point (pI), lipid solubility index (aliphatic index), average hydrophobic index (total average of hydropathicity) and instability index (instability index) of AMP HJH-3 in different systems. The hydrophobicity of AMP HJH-3 was predicted by the ProtScale online tool. The structure of HJH-3 was predicted by the method of I-TASSER (Iterative Threading ASSEmbly Refinement). I-TASSER is a hierarchical approach to protein structure prediction and structure-based function annotation. The website of this method is https://zhanggroup.org/I-TASSER/. The predicted parameters are set according to the system default settings.2.3 Synthesis, purification, and identification of AMP HJH-3
The AMP HJH-3 was synthesized by chemical synthesis using an automated solid-phase peptide synthesizer with reference to the peptide synthesis procedure provided by Protein Technologies, Inc. (PTI). Before synthesis, biological software was used to predict the difficulty of coupling corresponding amino acids in advance. The antibacterial peptide HJH-3 was synthesized through the processes of resin swelling, deprotection of amino acids, gradual coupling and extension of amino acids, peptide cleavage and peptide precipitation. Among synthesis procedures, the coupling of amino acids was the key to the successful synthesis. Different amino acids had different degrees of difficulty in coupling. The standard synthesis time of each amino acid was 5 minutes. According to the prediction of the software, every time the difficulty coefficient increased by 0.1, the synthesis time increased by 1 minute.The crude extract of the synthetic AMP HJH-3 was purified by semipreparative reverse phase high-performance liquid chromatography (RP-HPLC). The crude AMP was purified with a C18 column, and acetonitrile plus 0.01% TFA was used as the mobile phase. The detector of HPLC (LC-3000, LIANZHONG) is UV3000 UV/Vis Spectroscopic Detector.
Purified AMP HJH-3 was obtained by rotating evaporation that removed acetonitrile and was dried. According to the operation requirements of liquid-mass spectrometry (LC-MS), HJH-3 products were dissolved and identified by mass spectrometry. Mass spectrometry measurement conditions were as follows: positive ionization mode; the capillary voltage, 3.00 kV; the capillary outlet voltage, 50 V; the fragmentation voltage, 5 V; the drying gas flow rate, 1.5 L min−1; the drying gas temperature, 350 °C; and the scanning range, 400–1900 m/z. The MIC value of AMPs HJH-3 was detected with standard methods.
2.4 Gel retardation test of DNA-HJH-3
By determining the migration rate of genomic DNA of S. pullorum with agarose gel electrophoresis, the degree of antibacterial peptide HJH-3 binding to bacterial DNA was determined. Then, the destructive effect of AMPs on DNA of S. pullorum was assessed.The extraction method of S. pullorum genomic DNA was carried out according to the instructions of the bacterial genomic DNA extraction kit.
AMP HJH-3 was dissolved and diluted with DNA-bound buffer (with the following formulation, composition, and proportion: 5% glycerol (v/v); 10 mM Tris–HCl, pH 8.0; 1 mM EDTA; 1 mM DTT; 20 mM KCl; and 50 μg mL−1 BSA). A 5 μL-aliquot of genomic DNA of S. pullorum in solution was poured into a centrifugal tube. The total mass of DNA added to each centrifuge tube was 500 ng. Then, the AMP solution was diluted with a 5 μL gradient and then added to the DNA and incubated at room temperature for 10 minutes. The negative control was a mixture of genomic DNA and PBS buffer.
The DNA loading buffer was mixed with each sample in proportion, and then the products were analysed by agarose gel electrophoresis at 0.7%. After electrophoresis, a UV gel imaging system was used to observe and take photographs. The test was repeated three times.
2.5 Scanning electron microscopic observation of AMP HJH-3 activity with the genomic DNA of S. pullorum
First, genomic DNA of 100 μg mL−1 S. pullorum was incubated with 5 × MIC AMP HJH-3 at room temperature for 20 minutes. Then, the sample was dripped onto the silicon wafer and dried by a freeze-drying apparatus. Third, the dried silicon wafer was adhered to the conductive adhesive of the sample table, sprayed with gold by ion sputtering instrument and observed under scanning electron microscope. Genomic DNA without peptides was used as a blank control. The experiments were performed three times. The magnification focus for SEM was 3000 times.2.6 TEM observation of AMP HJH-3 activity in the cytoplasm of S. pullorum
S. pullorum at a logarithmic growth stage in 10 mL was centrifuged at 2420 g for 15 minutes. The bacteria were washed three times with sterilized PBS and suspended again in 100 μL of PBS (the concentration of bacteria was approximately 1 × 108 CFU mL−1), and 100 μL of HJH-3 was added to a final concentration of 2 × MIC. The bacteria were cultured in a constant temperature incubator at 37 °C for 1 hour. Bacteria without AMPs were used as blank controls. Samples were collected and centrifuged at 2420g for 15 minutes. The bacteria were immobilized with 2.5% glutaraldehyde (prepared in 0.1 M PBS buffer) overnight and rinsed 3 times with 0.1 M PBS for 15 minutes each time. Then, the bacteria were re-immobilized for 2–3 hours with 1% osmium acid stationary solution and rinsed 3 times with 0.1 M PBS for 15 minutes each time. The test was repeated three times. After fixation, dehydration, and embedding, the specimen sections were stained with uranyl acetate and alkaline lead citrate for 15 minutes and observed under a transmission electron microscope.2.7 Laser confocal microscopic observation of AMP HJH-3 effects on the genomic DNA of S. pullorum
S. pullorum were cultured to logarithmic growth phase and centrifuged at 2420 g for 15 minutes. After washed with sterilized PBS for three times, the S. pullorum were suspended again with 100 μL PBS (approximately 1 × 108 CFU mL−1). Then the harvested bacteria were treated with AMP HJH-3 on final concentration of 2 × MIC at 37 °C for 1 hour. Centrifuged and washed by PBS, the bacteria were incubated with PI and Hoechst 33528 for 10 minutes at 37 °C. Finally, the treated samples were dripped onto slides, covered with cover slip. Fluorescence images were taken with Zeiss LSM 780 confocal laser microscope with laser excitation wavelength 535 nm and emission wavelength 615 nm (PI) and excitation wavelength 352 nm and emission wavelength 461 nm (Hoechst 33528). Bacteria without AMPs were used as blank controls. All the experiments were performed three times.2.8 Flow cytometric analysis of AMP HJH-3 effects on the genomic DNA of S. pullorum
The prepared bacteria were added to AMP HJH-3 to final peptide concentrations of 5 × MIC and 10 × MIC, were fully mixed, and incubated for 0.5 hour at 1 g in a shaker at 37 °C. The bacteria were collected in 1 mL aliquots and centrifuged at 1685 g for 3 minutes, and the supernatant was discarded. Then, the cells were washed with sterilized PBS (pH 7.0) 3 times, PI solution was added to assess the cell cycle, and the cells were incubated in the dark at 4 °C for 30 minutes. The experiment was repeated three times. The samples were detected by flow cytometer.3. Results
3.1 Physicochemical properties of AMP HJH-3
A sequence analysis showed that the half-life of HJH-3 was more than 100 hours in mammals and reticular cells (in vitro), 20 hours in yeast (in vivo) and more than 10 hours in Escherichia coli (in vivo). The instability index of HJH-3 was 18.42, and the aliphatic index was 162.22. It was concluded that HJH-3 is a stable and heat-resistant AMP that may contribute to the development of its antimicrobial activity. The amino acid sequence and physicochemical properties of AMP HJH-3 are shown in Table 1.3.2 Synthesis, purification and identification of AMP HJH-3
According to the design of the experiment, 1.04 g of AMP product was obtained theoretically, and 0.75 g of crude AMP product was obtained via this synthetic method. Therefore, the yield of the AMP synthesized by this polypeptide synthesizer was 0.75÷1.04 ≈ 72.16%. The crude extract of the dried polypeptide was a yellow-white powder (data not shown).The crude extract of HJH-3 was purified by semipreparative RP-HPLC. The results showed that the baseline of the chromatogram of the crude product was very stable, and the main peak showed adequate symmetry. The chromatographic purification process of crude AMP products was shown in Fig. 1A. The synthesized HJH-3 product yield was 0.69 g by chromatographic purification, and the purified yield was 0.69÷0.75 = 92.00%.
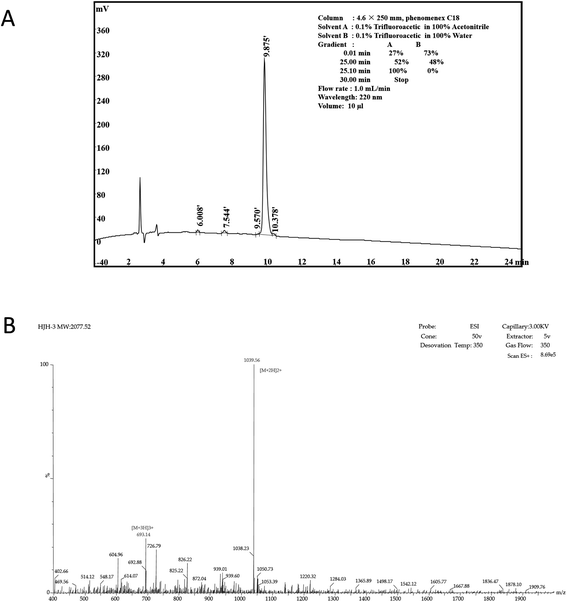 | ||
| Fig. 1 Characterization of AMP HJH-1 (A) purification of AMP HJH-3; (B) electrospray ionization (ESI) mass spectrometer of AMP HJH-3. | ||
According to the operation protocol of liquid-mass spectrometry (LC-MS), HJH-3 products were dissolved and identified by mass spectrometry. The test results were shown in Fig. 1B. AMP products were ionized into different fragments and protons via mass spectrometry. After the analysis, peaks were evident at [M + 2H] and [M + 3H], as expected. Hence, the substance detected by mass spectrometry was the synthetic AMP HJH-3.
The MIC against S. pullorum was 1.5625 μg mL−1.
3.3 DNA-HJH-3 binding gel retardation test
The binding of the AMP HJH-3 to the genomic DNA of S. pullorum increased the molecular weight of the genomic DNA, resulting in slow movement and hysteresis of the DNA during agarose gel electrophoresis. The interaction between HJH-3 and the genomic DNA of S. pullorum was detected in the agarose electrophoresis gel, as shown in Fig. 2. At a concentration of 20 μg mL−1, antibacterial peptide HJH-3 had an effect on DNA that was visible to the naked eye. With increasing peptide concentration, the blocking effect was more obvious. When the concentration reached 400 μg mL−1, the peptide blocked the entire genomic DNA in the sample well, resulting in the failure of the DNA to move in the gel.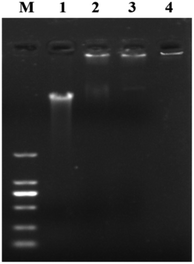 | ||
| Fig. 2 Gel blocking effect of AMP on genomic DNA of S. pullorum M. DNA marker DL 2000; (1) genomic DNA of S. pullorum without AMP; (2–4) S. pullorum treated with 20, 200, 400 μg mL−1 HJH-3. | ||
3.4 SEM observation of AMP HJH-3 activity with the genomic DNA of S. pullorum
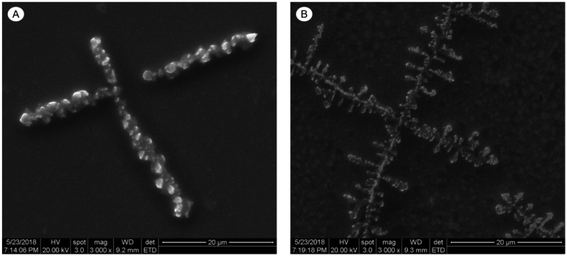
The morphology of the genomic DNA of S. pullorum treated with HJH-3 was observed by scanning electron microscopy (SEM). The degree of AMP HJH-3 binding to DNA of S. pullorum was determined. It was indirectly confirmed that AMPs can destroy bacterial DNA. The results were shown in Fig. 3. The DNA molecules that were not bound to AMPs were dispersed, uniform in structure, and arranged in a “rope-like” arrangement (Fig. 3A). After binding with AMPs HJH-3, DNA molecules were destroyed into small segments, aggregated with each other and appeared as “ice crystals-like” (Fig. 3B). These results indicated that AMP HJH-3 could destroy DNA molecules and bound with DNA molecules to change their structure. It was speculated that the peptide can cleave DNA molecules (but whether this cleavage is specific or random remains to be further studied) and thus rearrange DNA molecules.3.5 TEM observation of AMP HJH-3 acting on nucleoplasm of S. pullorum
By observing the changes in the internal ultrastructure of S. pullorum treated with HJH-3, the damage of AMPs to bacterial cells was reanalysed. The results were shown in Fig. 4. The intracellular nucleoplasm was relatively regular before the action of AMPs, the nucleoid and cytoplasm of the cells were relatively homogeneous, and the bacterial stroma was relatively clean. S. pullorum were observed in the division stage (Fig. 4A). After the action of AMPs, the nucleoid region of the bacterial cells was concentrated or lost, and a lot of loosened, damaged nucleoid components were observed between bacterial stromata (Fig. 4B).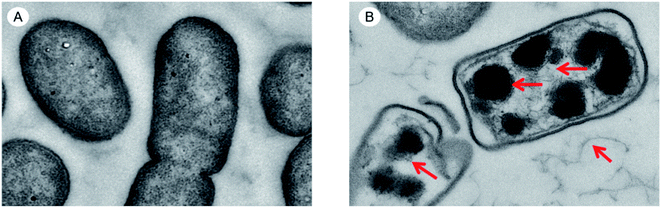 | ||
| Fig. 4 TEM results of genomic DNA acted by AMPs HJH-3 (A) TEM results of Sclerotinia of normal S. pullorum; (B) TEM results of Sclerotinia of S. pullorum treated with AMP HJH-3. | ||
3.6 Laser confocal microscopic observation of AMP HJH-3 activity on the genomic DNA of S. pullorum
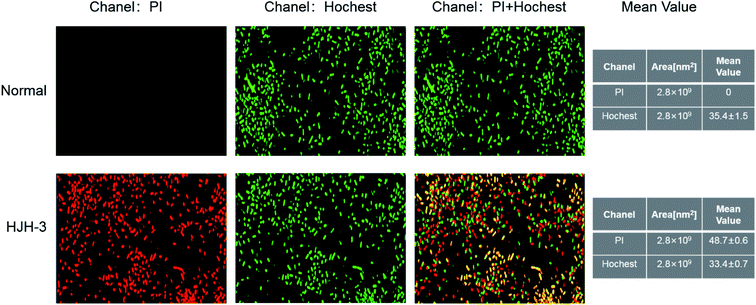
DNA staining in bacteria treated with HJH-3 was detected by PI and Hoechst 33528 double fluorescence labelling. The results were shown in Fig. 5. As shown in Fig. 5A, the bacteria that did not interact with AMP had complete cell membranes and no PI entered the bacteria; in these cases, only one type of fluorescence was emitted. Fig. 5B showed, after interaction with AMP HJH-3, S. pullorum entered the bacterium with the fluorescent dye PI due to the destruction of the cell membrane and then bound to DNA. The two fluorescent dyes emitted two kinds of fluorescence.3.7 Flow cytometric analysis of AMP HJH-3 activity with the genomic DNA of S. pullorum
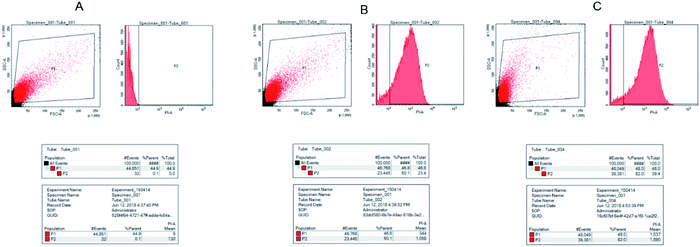
Flow cytometry was used to observe the binding of S. pullorum to PI, a dye used to detect the cell cycle phase, after AMP HJH-3. The results were shown in Fig. 6. After the action of 5 × MIC AMP HJH-3 on S. pullorum, the dead cells accounted for 50.1% and living cells for 49.9% of the total cells. After the action of 10 × MIC AMP HJH-3 on S. pullorum, the dead cells accounted for 82.0% and living cells for 18.0% of the total cells. Therefore, with the increase of AMP concentration, the number of dead cells showed an increasing trend, indicating that AMP HJH-3 had strong antibacterial activity, and its bactericidal activity was enhanced to some extent with increasing concentrations.4. Discussion
The drug resistance in bacteria poses a serious threat to livestock production and human health. The intracellular parasitic bacteria represented by Salmonella exacerbate bacterial drug resistance due to the limited amount of antibiotic ingested by the cells or the difficulty in reaching the parasitic site of the bacteria.It was found that most of the AMPs can permeate through the bacterial cell membrane to exert bactericidal effects, even some AMPs exerted their antibacterial effects through multiple mechanisms. This study found that in addition to the traditional bactericidal mechanism, the AMP HJH-3 can also bind with bacterial DNA and inhibit bacterial reproduction (the target acting with DNA have not been found). Therefore, HJH-3 had great advantages in preventing and controlling infection of intracellular cells.
In recent years, increasing attention has been focused on the intracellular mechanisms of action of AMPs. It has been found that AMPs such as Buforin II and Indolicidin can enter bacterial cells and play antimicrobial roles by interfering with certain important metabolic processes of the cells.11,23–27 The previous experimental results of this study have confirmed that the AMP HJH-3 can effectively kill Escherichia coli, Staphylococcus aureus and Salmonella. Moreover, the AMP HJH-3 and its analogues can play an antibacterial role by interacting with bacterial cell membrane.8,9,22
The research results of Huo et al.28 found that the fusion antibacterial peptide TAT-KR-12 can kill intracellular parasitic Staphylococcus aureus. Its mechanism is that Tat acts as a “Trojan horse” responsible for transmitting AMPs to cells, and AMP KR-12 is responsible for interacting with DNA in cells. To further explore whether the AMP HJH-3 plays an antibacterial role by acting on intracellular organelles, this study was carried out in vitro with DNA-peptide gel retardation tests, scanning electron microscopy, laser confocal microscopy and flow cytometry. The results showed that AMP HJH-3 can bind to Salmonella DNA.
The results showed that the AMP HJH-3 can bind to the DNA of many other bacteria. However, AMP HJH-3 had no obvious cytotoxicity on mammalian cells. It is speculated that the action mode of AMP HJH-3 on prokaryotic and eukaryotic cells was inconsistent. Whether the binding mode between AMP HJH-3 and prokaryotic DNA is specific or stochastic requires further validation.
AMPs are potential substitutes for environment-friendly antibiotics because of their small molecular weight, extensive antimicrobial effects, easy degradation and no pollution. Therefore, functional research on AMPs needs direct evidence. It is believed that with the progress of technology, the mechanisms of action of AMPs will be gradually revealed.3,13,29
5. Conclusions
The results of this study showed that the self-synthesized AMP HJH-3 can directly enter the cell through the cell membrane and play an antimicrobial role. It can bind to intracellular DNA and then specifically block cell division, thus achieving an antimicrobial effect. This study further enriches the study on the intracellular bacteriostasis mechanism of AMPs against bacteria and lays a theoretical foundation for the development of HJH-3.Author contributions
Designed research and data curation, Q. W. and Y. X. Funding acquisition, J. H.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This research was funded by the National Key R&D Program of China (2021YFD1301200), Leading Talents of Scientific and Technological Innovation in the Central Plains (224200510024), National Natural Science Foundation of China (31402208), Key Project of Science and Technology Research of Henan Provincial Education Department (19A230001). The authors would like to thank Shi-Jun Chen for assistance with operation of TEM.Notes and references
- K. Radek and R. Gallo, Semin. Immunopathol., 2007, 29, 27–43 CrossRef CAS PubMed.
- M. Pasupuleti, A. Schmidtchen and M. Malmsten, Crit. Rev. Biotechnol., 2012, 32, 143–171 CrossRef CAS PubMed.
- C. H. Chen and T. K. Lu, Antibiotics, 2020, 9, 24 CrossRef CAS PubMed.
- S. J. Lam, N. M. O'Brien-Simpson, N. Pantarat, A. Sulistio, E. H. Wong, Y. Y. Chen, J. C. Lenzo, J. A. Holden, A. Blencowe, E. C. Reynolds and G. G. Qiao, Nat. Microbiol., 2016, 1, 16162 CrossRef CAS PubMed.
- H. R. Lee, D. G. You, H. K. Kim, J. W. Sohn, M. J. Kim, J. K. Park, G. Y. Lee and Y. D. Yoo, mBio, 2020, 11 CAS.
- L. V. Kovalchuk, L. V. Gankovskaya, O. A. Gankovskaya and V. F. Lavrov, Adv. Exp. Med. Biol., 2007, 601, 369–376 CrossRef PubMed.
- Y. Yu, C. L. Cooper, G. Wang, M. J. Morwitzer, K. Kota, J. P. Tran, S. B. Bradfute, Y. Liu, J. Shao, A. K. Zhang, L. G. Luo, S. P. Reid, S. H. Hinrichs and K. Su, iScience, 2020, 23, 100999 CrossRef CAS PubMed.
- Q. Wang, Y. Xu, M. Dong, B. Hang, Y. Sun, L. Wang, Y. Wang, J. Hu and W. Zhang, Molecules, 2018, 23, 2026 CrossRef PubMed.
- Q. Zhang, Y. Xu, Q. Wang, B. Hang, Y. Sun, X. Wei and J. Hu, Antimicrob. Agents Chemother., 2015, 59, 2835–2841 CrossRef CAS PubMed.
- T. Ganz, Nat. Rev. Immunol., 2003, 3, 710–720 CrossRef CAS PubMed.
- T. D. Vo, C. Spahn, M. Heilemann and H. B. Bode, ACS Chem. Biol., 2021, 16, 447–451 CrossRef CAS PubMed.
- C. F. Le, C. M. Fang and S. D. Sekaran, Antimicrob. Agents Chemother., 2017, 61 Search PubMed.
- J. K. Boparai and P. K. Sharma, Protein Pept. Lett., 2020, 27, 4–16 CrossRef CAS PubMed.
- X. Luo, Y. Liu, Z. Qin, Z. Jin, L. Xu, L. Cao, F. He, X. Gu and X. Ouyang, J. Biomed. Nanotechnol., 2018, 14, 601–608 CrossRef CAS PubMed.
- T. V. Vineeth Kumar and G. Sanil, Curr. Protein Pept. Sci., 2017, 18, 1263–1272 CAS.
- G. Rispoli, Methods Mol. Biol., 2017, 1548, 255–269 CrossRef CAS PubMed.
- N. B. Last and A. D. Miranker, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6382–6387 CrossRef CAS PubMed.
- N. P. Chongsiriwatana, J. S. Lin, R. Kapoor, M. Wetzler, J. A. C. Rea, M. K. Didwania, C. H. Contag and A. E. Barron, Sci. Rep., 2017, 7, 16718 CrossRef PubMed.
- B. R. da Silva, V. A. de Freitas, L. G. Nascimento-Neto, V. A. Carneiro, F. V. Arruda, A. S. de Aguiar, B. S. Cavada and E. H. Teixeira, Peptides, 2012, 36, 315–321 CrossRef CAS PubMed.
- S. Peng, M. Yang, R. N. Sun, Y. Liu, W. Wang, Q. Xi, H. Gong and C. Chen, Protein Cell, 2018, 9, 890–895 CrossRef CAS PubMed.
- G. B. Taveira, E. O. Mello, A. O. Carvalho, M. Regente, M. Pinedo, L. de La Canal, R. Rodrigues and V. M. Gomes, Biopolymers, 2017, 108, e23008 CrossRef PubMed.
- L. Wang, X. Q. Zhao, X. J. Xia, C. L. Zhu, W. H. Qin, Y. Z. Xu, B. L. Hang, Y. W. Sun, S. J. Chen, H. H. Zhang, J. Q. Jiang, J. H. Hu, H. N. Fotina and G. P. Zhang, Probiotics Antimicrob. Proteins, 2019, 11, 1379–1390 CrossRef CAS PubMed.
- M. P. Smirnova, N. I. Kolodkin, A. A. Kolobov, V. G. Afonin, I. V. Afonina, L. I. Stefanenko, V. M. Shpen and O. V. Shamova, Peptides, 2020, 132, 170356 CrossRef CAS PubMed.
- I. V. Kutepov, Y. D. Lyashev, E. B. Artyushkova, A. V. Solin, V. S. Serikov, A. Y. Lyashev and A. R. Chahine, Bull. Exp. Biol. Med., 2019, 167, 47–49 CrossRef CAS PubMed.
- Y. Xie, E. Fleming, J. L. Chen and D. E. Elmore, Peptides, 2011, 32, 677–682 CrossRef CAS PubMed.
- C. Munoz-Camargo, V. A. Salazar, L. Barrero-Guevara, S. Camargo, A. Mosquera, H. Groot and E. Boix, Int. J. Mol. Sci., 2018, 19, 2170 CrossRef PubMed.
- D. E. Elmore, Peptides, 2012, 38, 357–362 CrossRef CAS PubMed.
- S. Huo, C. Chen, Z. Lyu, S. Zhang, Y. Wang, B. Nie and B. Yue, ACS Infect. Dis., 2020, 6, 3147–3162 CrossRef CAS PubMed.
- E. J. H. Bartels, D. Dekker and M. Amiche, Front. Pharmacol., 2019, 10, 1421 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |