Open Access Article
Open Access ArticleAptamer fluorescence anisotropy assays for detection of aflatoxin B1 and adenosine triphosphate using antibody to amplify signal change†
Yapiao Liab,
Hao Yuab and
Qiang Zhao *abc
*abc
aState Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China. E-mail: qiangzhao@rcees.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cSchool of Environment, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310000, China
First published on 7th March 2022
Abstract
Fluorescence polarization/anisotropy (FP/FA) is an attractive technology for determining small molecules in homogeneous solution based on rotation changes of a fluorescent reporter. Binding induced conformation change is a specific property of aptamers. This property has been integrated into aptamer based FA assays for small molecules. In this work, we reported aptamer FA assays for aflatoxin B1 (AFB1) and adenosine triphosphate (ATP) by using antibody conjugated complementary DNA at the 3′ end and a fluorescein (FAM)-labeled aptamer at the 5′ end. The hybridization of aptamer and cDNA induced a FAM label close to the large-sized antibody, which restricts the local rotation of FAM and gives high FA signal. With the addition of target, the aptamer probe binds with the target, and the aptamer–cDNA duplex is inhibited, causing FA signal decreases. This method achieved detection of 25 pM AFB1 and 1 μM ATP, respectively. The assay is promising for application.
1. Introduction
Fluorescence polarization/anisotropy (FP/FA) assays for small molecules detection have wide applications in drug discovery, clinical diagnosis, food safety, environmental sensing, and molecular interaction study.1–4 FA analysis only requires mixing a probe and target with other reagents in homogeneous solution. As a ratiometric technique, FP/FA technique is insensitive to small fluctuations in instrument variance and fluorescence intensity, having high reproducibility. FP/FA assay is suitable for high-throughput analysis. Conventional immune-FP assays use antibody and dye-labeled small molecules, and they show some limitations in preparing the fluorophore-labeled small molecules.3Aptamers show advantages in facile synthesis, easy labeling, good thermo-stability and low cost.5,6 Aptamers can be selected for various targets, including cells, proteins, small molecules and metals.7 Taking advantages of aptamer and FP/FA technology, a variety of aptamer-based FP/FA assays have been developed for small molecules detection.8–10 As nucleic acid ligands, DNA aptamers can hybridize with its complementary DNA (cDNA). Aptamer structure change can occur due to the competition between aptamer–target binding and aptamer–cDNA hybridization.11–13 This attractive property of aptamer can be applied to develop FA assays by using dye labeled aptamer or cDNA.8–10,14,15 To improve FA signal changes, several signal amplification strategies for small molecule detection have been reported, such as conjugating nanoparticles or biomolecules on DNA.16–20
In this work, we described an aptamer FA assay for detection of small molecules by using fluorescein (FAM)-labeled aptamer at 5′ end as fluorescent probe and antibody conjugated cDNA at 3′ terminal as the signal amplifier. We took aflatoxin B1, a most toxic mycotoxin causing great health risk to health,21–23 and ATP, an important small molecule with many functions in organisms, as two examples of small molecules. FAM-labeled aptamer binds with antibody conjugated cDNA, and a duplex is formed, drawing FAM label into proximity with the large-sized antibody (about 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 in molecular weight) and giving a high FA signal due to the restriction of local rotation. In the presence of small molecule, aptamer binds with target, and the aptamer–cDNA duplex assembly is inhibited. FA signal significantly decreases with addition of small molecules as the FAM-labeled aptamer shows low FA in aptamer–target complex due to the small size of aptamer and rapid local rotation of FAM. This assay enabled detection of 25 pM AFB1 and 1 μM ATP, respectively.
000 g mol−1 in molecular weight) and giving a high FA signal due to the restriction of local rotation. In the presence of small molecule, aptamer binds with target, and the aptamer–cDNA duplex assembly is inhibited. FA signal significantly decreases with addition of small molecules as the FAM-labeled aptamer shows low FA in aptamer–target complex due to the small size of aptamer and rapid local rotation of FAM. This assay enabled detection of 25 pM AFB1 and 1 μM ATP, respectively.
2. Experimental
2.1. Materials and reagents
Aflatoxin B1 (AFB1), ochratoxin A (OTA), fumonisin B1 (FB1), fumonisin B2 (FB2), zearalenone (ZAE) were purchased from Pribolab (Singapore) Co. Ltd. Adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triphosphate (UTP) were ordered from Takara Biotechnology (Dalian, China). Antibody specific for digoxin was purchased from Sigma-Aldrich. White wine and fresh grapes were purchased from local market. Tap water was obtained from the laboratory. Human serum was obtained from Zhongke Chenyu Biotechnology (Beijing, China). All the DNA oligonucleotides (Table S1†) were synthesized and purified by Sangon Biotech (Shanghai, China). All the complementary DNAs (cDNAs) had one digoxin label at the 3′ terminal without any other statement. Ultrapure water was used in this experiment (Ultra Genetic, Purelab Elga Labwater system). Other reagents were of analytical grade. The following binding buffers were used. Buffer A for AFB1 detection contained 10 mM Tris–HCl (7.5), 50 mM MgCl2, 50 mM NaCl, and 0.1% Tween 20. Buffer B for ATP detection contained 50 mM Tris–HCl (7.5), 10 mM MgCl2, 50 mM NaCl, and 0.1% Tween 20.2.2. Aptamer FA detection of targets
First, the digoxin labeled complementary DNA and antibody specific for digoxin were incubated in the binding buffer with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at 4 °C for 15 min to achieve antibody-conjugated cDNA by the immunoreaction between antibody and digoxin. In the assay for AFB1 detection, 1 nM AptAFB -5′FAM and 20 nM antibody conjugated C14AFB for AFB1 were incubated in buffer A for 30 min at 4 °C. In the assay for ATP, 5 nM AptATP-5′FAM and 20 nM antibody conjugated C11ATP for ATP were incubated in buffer B for 10 min at 25 °C. After that, 100 μL of sample solution was added to the wells of the black 96-well microplates (USA, Thermofisher). Duplicate samples were tested. For each sample, FA signals were measured three times by a plate reader Synergy H1 (U.S.A., Biotek), with an excitation at 485 nm and an emission at 528 nm. The average FA values were used. For the three measurements of the same sample, the relative standard deviation (RSD) ranged from 0.4% to 5%. For measurement of duplicate samples, the RSD was less than 3%.
1 at 4 °C for 15 min to achieve antibody-conjugated cDNA by the immunoreaction between antibody and digoxin. In the assay for AFB1 detection, 1 nM AptAFB -5′FAM and 20 nM antibody conjugated C14AFB for AFB1 were incubated in buffer A for 30 min at 4 °C. In the assay for ATP, 5 nM AptATP-5′FAM and 20 nM antibody conjugated C11ATP for ATP were incubated in buffer B for 10 min at 25 °C. After that, 100 μL of sample solution was added to the wells of the black 96-well microplates (USA, Thermofisher). Duplicate samples were tested. For each sample, FA signals were measured three times by a plate reader Synergy H1 (U.S.A., Biotek), with an excitation at 485 nm and an emission at 528 nm. The average FA values were used. For the three measurements of the same sample, the relative standard deviation (RSD) ranged from 0.4% to 5%. For measurement of duplicate samples, the RSD was less than 3%.
2.3. Detection of targets in complex sample matrix
For AFB1 detection, diluted grape juice, white wine and tap water samples were used as complex sample matrix. Grapes without the peel and nucleus were prepared for grape juice through juice extractor. After that, grape juice sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Then the collected supernatant was subsequently filtered with 0.22 μm membrane and ultrafiltration membrane (Millipore, 3 kDa cut-off) at 12
000 rpm for 10 min. Then the collected supernatant was subsequently filtered with 0.22 μm membrane and ultrafiltration membrane (Millipore, 3 kDa cut-off) at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. White wine samples was filtered with 0.22 μm membrane. At last, 20-fold diluted grape juice, 10-fold diluted white wine and 10-fold diluted tap water was obtained by diluting with the binding buffer, used as complex sample matrix. For ATP detection, 50-fold diluted human serum and 20-fold diluted urine samples were used as complex sample matrix. Before dilution, human serum samples were centrifuged at 12
000 rpm for 15 min. White wine samples was filtered with 0.22 μm membrane. At last, 20-fold diluted grape juice, 10-fold diluted white wine and 10-fold diluted tap water was obtained by diluting with the binding buffer, used as complex sample matrix. For ATP detection, 50-fold diluted human serum and 20-fold diluted urine samples were used as complex sample matrix. Before dilution, human serum samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min, and the supernatant was filtered with 0.22 μm membrane. Urine samples were from volunteer in the lab, and filtered with 0.22 μm membrane. The sample preparation and determination were the same to the FA assay procedure described above. Informed consents were obtained from human participants of this study.
000 rpm for 15 min, and the supernatant was filtered with 0.22 μm membrane. Urine samples were from volunteer in the lab, and filtered with 0.22 μm membrane. The sample preparation and determination were the same to the FA assay procedure described above. Informed consents were obtained from human participants of this study.
3. Results and discussion
3.1. Principle of aptamer FA assay for small molecule detection using adjacent antibody to amplify signal change
The principle of the proposed aptamer FA assay using adjacent large-sized antibody as a signal amplifier is shown in Scheme 1. In this design, FAM-labeled aptamer is used as fluorescent probe. Antibody conjugated complementary sequence is employed as the signal amplifier. The antibody conjugated cDNA is obtained by incubating digoxin (Dig) labeled cDNA with anti-digoxin antibody. The hybridization of aptamer and cDNA causes FAM label close to the antibody. The proximity between the large-sized antibody and the FAM label significantly restricts the local rotation of FAM and produces a much higher FA value. The addition of small molecule target in sample solution results in the formation of aptamer and target complex, preventing the assembly of duplex of aptamer probe and cDNA. The FAM label in the aptamer probe shows a lower FA signal. Thus, we can achieve the detection of small molecule by measuring the remarkable FA change upon addition of target.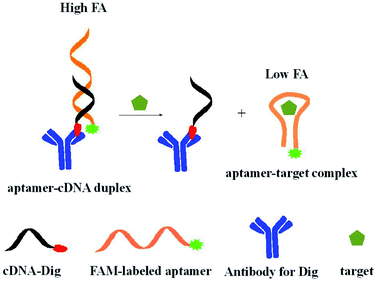 | ||
| Scheme 1 Schematic of aptamer FA assay for small molecules using adjacent antibody as a signal amplifier. | ||
3.2. Aptamer FA assay for AFB1
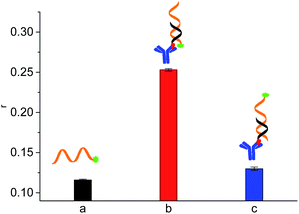
Aflatoxins (AFs) are natural mycotoxins that are produced by Aspergillus parasiticus and Aspergillus flavus.21–23 Many food sources (such as rice, wheat, corn, peanuts and fruits) are easily polluted by AFs. In different types of AFs (including AFB1, AFB2, AFM1, AFM2, AFG1 and AFG2), AFB1 shows most toxic and has been classified as a primary carcinogenic compound by International Agency for Research in Cancer (IARC).22 Developing sensitive and simple method for AFB1 and its analogues is important to ensure food safety and human health.Firstly, we tested the feasibility of our design. FA value of 1 nM 5′FAM-labeled aptamer was 0.116 (Fig. 1a). The antibody-conjugated cDNA hybridized with aptamer, resulting in the proximity of antibody and FAM and the restriction of local rotation of FAM. Thus, the FA response increased to 0.253 (Fig. 1b). When antibody was distant from the FAM label (AptAFB-3′FAM + antibody-conjugated cDNA, Fig. 1c). The DNA hybridization only caused slight FA increase (less than 0.020) due to the fast local rotation of FAM though antibody (about 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 in molecular weight) is much larger than the aptamer probe in size.20 With the addition of AFB1, AFB1 bound to aptamer probe, and the cDNA did not hybridize with the 5′FAM-labeled aptamer probe. Thus, FAM label rotated fast and showed low FA value (0.133, Fig. 2) because the bound AFB1 was small in size. These results demonstrated that our design is feasible for detecting AFB1.
000 g mol−1 in molecular weight) is much larger than the aptamer probe in size.20 With the addition of AFB1, AFB1 bound to aptamer probe, and the cDNA did not hybridize with the 5′FAM-labeled aptamer probe. Thus, FAM label rotated fast and showed low FA value (0.133, Fig. 2) because the bound AFB1 was small in size. These results demonstrated that our design is feasible for detecting AFB1.
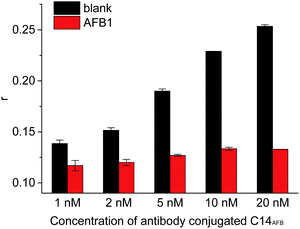 | ||
| Fig. 2 Effects of concentrations of antibody-conjugated C14AFB on the FA values corresponding to the blank sample and the sample containing AFB1 (10 nM). The concentration of AptAFB-5′FAM were 1 nM. | ||
Then, we investigated some important factors for AFB1 detection, such as the concentrations of antibody-conjugated C14AFB and incubation time. As shown in Fig. 2, the FA value of FAM label in formed duplex increased with increasing of antibody-conjugated cDNA. In the presence of 10 nM AFB1, FA value showed slight increase with increasing of antibody-conjugated cDNA. The maximum reduction in FA caused by 10 nM AFB1 increased from 0.021 to 0.120. Further increase of antibody-conjugated cDNA required more antibodies. Thus, we chose 20 nM antibody-conjugated cDNA for further experiments.
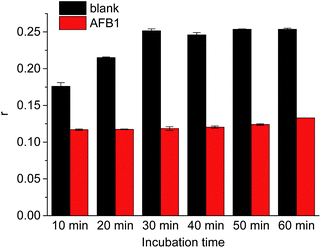
Incubation time shows great effect on aptamer and cDNA hybridization. As shown in Fig. 3, FA value of FAM label in duplex increased with the increase of incubation time from 10 min to 30 min. FA value did not increase with further increase of incubation time. It shows the incubation for 30 min is enough for hybridization between aptamer probe and the antibody conjugated cDNA. In the presence of 10 nM AFB1, similar FA signals were observed in different incubation time. Thus, FA measurement was conducted after 30 min incubation.
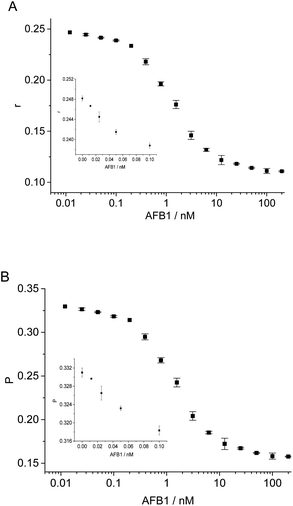
Under optimal conditions, we detected AFB1 by using 1 nM aptamer and 20 nM antibody-labeled cDNA. As shown in Fig. 4, the FA value of FAM label gradually decreased with the increase of AFB1. This method allowed to detect as low as 25 pM AFB1 (∼0.008 μg kg−1) (determined by S/N > 3), with dynamic range from 25 pM to 100 nM. As comparison, the FP (fluorescence polarization) signals were also recorded (Fig. 4B). We compared our method with some FP/FA assays for AFB1 detection in terms of affinity ligand, maximum signal change and detection limit (Table S2†).20,24–27 As shown in Table S2 in ESI,† our assay not only shows larger FA signal change than that in traditional immune-FA assay, but also has lower detection limit than some reported immune-FA assays and aptamer-based FP/FA assays.8–10,20,24,26,27
Then, we tested the selectivity of our proposed assay for AFB1. FA decreased when AFB1 existed in the sample buffer. The FA of duplex in the presence of 200 nM other mycotoxins such as ochratoxin A (OTA), fumonisin B1 (FB1), fumonisin B2 (FB2) and zearalenone (ZAE), was similar to that of blank sample (Fig. S1 in ESI†). Simultaneously existing of other mycotoxins (each of 200 nM) showed little effect on the FA value of FAM label in the presence of AFB1 (10 nM). These results suggest that our proposed assay is highly selective for AFB1 detection.
Our method shows different FA decreases upon other aflatoxin analogues (such as AFB2, AFM1, AFM2, AFG1 and AFG2) by using 1 nM FAM-labeled aptamer and 20 nM antibody conjugated C14AFB (Fig. S2†), which are in agreement with the previous report due to the different affinities between aptamer and these aflatoxins.24 Then, we used the concentrations of analytes corresponding to 50% FA change (EC50) to evaluate the cross-reactivity (CR) of AFB1 and other AFB1 analogues with this method. The respective EC50 values for AFB1, AFB2, AFM1, AFM2, AFG1 and AFG2 were 1.3 nM, 1.6 nM, 4.2 nM, 4.1 nM, 16 nM and 22 nM, respectively (Table S3†). The percentage of CR was obtained by dividing the EC50 of AFB1 with the EC50 of respective aflatoxin species. The cross-reactivity of AFB1, AFB2, AFM1, AFM2, AFG1 and AFG2 were estimated to be 100%, 81%, 32%, 32%, 8% and 6%, respectively (Table S3†). Our method enabled to detect 25 pM AFB1, 25 pM AFB2, 0.39 nM AFM1, 0.39 nM AFM2, 1.6 nM AFG1 and 1.6 nM AFG2, respectively. It shows the aptamer FA method can be used to detect other aflatoxin species. This method shows potential for the detection of total quantity of these aflatoxins (AFB1, AFB2, AFM1, AFM2, AFG1 and AFG2) in some assays.
To evaluate the performance of the aptamer FA assay in complex sample matrix, we tested AFB1 in diluted grape juice, white wine and tap water samples (Fig. S3†). In the tested sample matrix, FA value toward to different concentrations of AFB1 was similar to that in binding buffer. Our method enabled detection of 50 pM (0.016 μg kg−1) AFB1 in complex sample matrix. Our method meets the requirement of the maximum regulation levels of AFB1 setting by European Union (2 μg kg−1) and World Health Organization (WHO, 5 μg kg−1) allowed in food products.21 These results demonstrate that our proposed assay is feasible for AFB1 detection in complex matrix, and it has potential for real sample analysis.
3.3. Aptamer FA detection of ATP
Adenosine triphosphate (ATP), an important small molecule participating in many activities in living organism was chosen for the universal test of our design.28,29 A FAM-labeled aptamer at the 5′ end, AptATP-5′FAM (5 nM; 5′-CCT GGG GGA GTA TTG CGG AGG AAG G-3′), an antibody-conjugated cDNA at the 3′ terminal C11ATP (20 nM; 5′-ACT CCC CCA GG-3′) (Fig. S4†), and 10 min incubation of sample solution at 25 °C (Fig. S5†) were chosen for ATP detection to obtain relatively better FA responses. Under optimal conditions, we successfully achieved detection of ATP (Fig. 5 and S6†). The maximum FA change was 0.06 (the maximum FP change was 0.08). The dynamic detection range was from 1 μM to 1 mM. The detection limit of ATP was 1 μM, which is comparable to that in some previous aptamer-based FP/FA assays for ATP (Table S2†).8–10,18,30–33 The higher detection limits of our FA assays for ATP (1 μM) than that of our FA assay for AFB1 (25 pM) can be attributed to the lower affinity of aptamer against ATP31 and high affinity of aptamer against AFB1.24 This method was selective for ATP detection (Fig. S7†). This strategy also enabled detection of ATP spiked in diluted human serum and urine with a detection limit at 1 μM (Fig. S8†).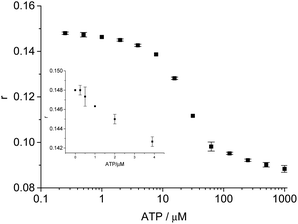 | ||
| Fig. 5 Quantitative detection of ATP. FA responses versus concentrations of ATP. The inset shows the FA signals corresponding to low concentrations of ATP. | ||
4. Conclusions
In this study, a simple and sensitive aptamer FA assay was developed by using large-sized antibody to amplify signal change. We used FAM-labeled aptamer and antibody-conjugated cDNA, which induced the proximity effect between FAM and antibody in duplex. The FA assays allowed for detection of 25 pM AFB1 and 1 μM ATP, showing the method is suitable for aptamers with either high affinity or weak affinity. Moreover, the FA assays were successfully applied to rapid detection of AFB1 and ATP in complex sample matrixes, showing potential in applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thanked the support from National Natural Science Foundation of China (Grant No. 21874146, 22074156).Notes and references
- C. C. Gradinaru, D. O. Marushchak, M. Samim and U. J. Krull, Analyst, 2010, 135, 452–459 RSC.
- W. A. Lea and A. Simeonov, Expert Opin. Drug Discovery, 2011, 6, 17–32 CrossRef CAS PubMed.
- D. S. Smith and S. A. Eremin, Anal. Bioanal. Chem., 2008, 391, 1499–1507 CrossRef CAS PubMed.
- H. Zhang, Q. Wu and M. Y. Berezin, Expert Opin. Drug Discovery, 2015, 10, 1145–1161 CrossRef PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- D. Wilson and J. Szostak, Annu. Rev. Biochem., 1999, 68, 611–647 CrossRef CAS PubMed.
- Q. Zhao, J. Tao, J. S. Uppal, H. Peng, H. Wang and X. C. Le, TrAC, Trends Anal. Chem., 2019, 110, 401–409 CrossRef CAS.
- H. Zhang, S. Yang, K. De Ruyck, N. V. Beloglazova, S. A. Eremin, S. De Saeger, S. Zhang, J. Shen and Z. Wang, TrAC, Trends Anal. Chem., 2019, 114, 293–313 CrossRef CAS.
- O. D. Hendrickson, N. A. Taranova, A. V. Zherdev, B. B. Dzantiev and S. A. Eremin, Sensors, 2020, 20, 7132 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, Chem.–Eur. J., 2004, 10, 1868–1876 CrossRef CAS PubMed.
- X. Su, X. Xiao, C. Zhang and M. Zhao, Appl. Spectrosc., 2012, 66, 1249–1261 CrossRef CAS PubMed.
- J. A. Cruz-Aguado and G. Penner, Anal. Chem., 2008, 80, 8853–8855 CrossRef CAS PubMed.
- Z. Zhu, T. Schmidt, M. Mahrous, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Chim. Acta, 2011, 707, 191–196 CrossRef CAS PubMed.
- Y. Huang, S. Zhao, Z. Chen, M. Shi and H. Liang, Chem. Commun., 2012, 48, 7480–7482 RSC.
- B. Yang, X. Zhang, L. Kang, G. Shen, R. Yu and W. Tan, Anal. Chem., 2013, 85, 11518–11523 CrossRef CAS PubMed.
- J. Tian, Y. Wang, S. Chen, Y. Jiang, Y. Zhao and S. Zhao, Microchim. Acta, 2013, 180, 203–209 CrossRef CAS.
- Y. He, J. Tian, K. Hu, J. Zhang, S. Chen, Y. Jiang, Y. Zhao and S. Zhao, Anal. Chim. Acta, 2013, 802, 67–73 CrossRef CAS PubMed.
- Y. Li and Q. Zhao, Anal. Chem., 2019, 91, 7379–7384 CrossRef CAS PubMed.
- U. P. Sarma, P. J. Bhetaria, P. Devi and A. Varma, Indian J. Clin. Biochem., 2017, 32, 124–133 CrossRef CAS PubMed.
- Y. Q. Dai, K. L. Huang, B. Y. Zhang, L. Y. Zhu and W. T. Xu, Food Chem. Toxicol., 2017, 109, 683–689 CrossRef CAS PubMed.
- I. Y. S. Rustom, Food Chem., 1997, 59, 57–67 CrossRef CAS.
- L. Sun and Q. Zhao, Talanta, 2018, 189, 442–450 CrossRef CAS PubMed.
- H. Huang, J. Qin, K. Hu, X. Liu, S. Zhao and Y. Huang, RSC Adv., 2016, 6, 86043–86050 RSC.
- N. V. Beloglazova and S. A. Eremin, Talanta, 2015, 142, 170–175 CrossRef CAS PubMed.
- Y. Sheng, S. A. Eremin, T. Mi, S. Zhang, J. Shen and Z. Wang, Biomed. Environ. Sci., 2014, 27, 126–129 CAS.
- Z. Liu, Y. Zhong, Y. Hu, L. Yuan, R. Luo, D. Chen, M. Wu, H. Huang and Y. Li, Food Chem., 2019, 270, 573–578 CrossRef CAS PubMed.
- J. T. Dong and M. P. Zhao, TrAC, Trends Anal. Chem., 2016, 80, 190–203 CrossRef CAS.
- J. Liu, C. Wang, Y. Jiang, Y. Hu, J. Li, S. Yang, Y. Li, R. Yang, W. Tan and C. Z. Huang, Anal. Chem., 2013, 85, 1424–1430 CrossRef CAS PubMed.
- Y. Li, L. Sun and Q. Zhao, Talanta, 2017, 174, 7–13 CrossRef CAS PubMed.
- Q. Zhao, Q. Lv and H. Wang, Biosens. Bioelectron., 2015, 70, 188–193 CrossRef CAS PubMed.
- J. Liu, C. Wang, Y. Jiang, Y. Hu, J. Li, S. Yang, Y. Li, R. Yang, W. Tan and C. Z. Huang, Anal. Chem., 2013, 85, 1424–1430 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00843b |
| This journal is © The Royal Society of Chemistry 2022 |