Open Access Article
Open Access ArticleDeveloping a far-red fluorogenic beta-galactosidase probe for senescent cell imaging and photoablation†
Seung Koo Lee‡
 ,
Zhenhua Shen‡
,
Zhenhua Shen‡ ,
Myung Shin Han and
Ching-Hsuan Tung
,
Myung Shin Han and
Ching-Hsuan Tung *
*
Department of Radiology, Molecular Imaging Innovations Institute, Weill Cornell Medicine, New York, NY 10021, USA. E-mail: cht2018@med.cornell.edu
First published on 4th February 2022
Abstract
A methylene blue (MB)-based beta-galactosidase (β-gal) activatable molecule, Gal-MB, was developed for senescence imaging and light-triggered senolysis. When in contact with LacZ β-gal or senescence-associated β-gal (SA-β-gal), the photoinsensitive Gal-MB becomes fluorescent. Gal-MB also offered selective phototoxicity toward LacZ β-gal expressing cells and drug-induced senescent cells, which express SA-β-gal, after light illumination at 665 nm.
1. Introduction
Beta-galactosidase (β-gal), a glycoside hydrolase that catalyzes the hydrolysis of a terminal β-D-galactose residue, has been commonly applied to monitor gene expression,1–4 transcriptional regulation,5,6 and recently, senescence.7–9 Cellular senescence, a stress-induced irreversible growth arrest, accumulates in aged organisms and contributes to tissue dysfunction thus drives age-related phenotypes.10 The senescent cells express several characteristic senescence biomarkers, such as senescence-associated β-gal (SA-β-gal), reactive oxygen species, p21, and senescence-associated secretory phenotype (SASP) factors.11 SA-β-gal, one of the most accepted hallmarks of senescence,12 is abnormally accumulated in senescent cells because of the increased lysosomal content.13 Although SA-β-gal, encoded by the Galactosidase beta 1 (GLB1) gene, is structurally different from bacterial β-gal, encoded by the LacZ gene, the substrate selectivity is similar to some degree.14 Therefore, the β-gal responsive probes originally developed for bacterial LacZ β-gal, are widely used to detect SA-β-gal, such as the popular X-gal blue staining dye (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside),15 and several fluorogenic probes.16,17As senescent cells gather, they secrete a wide spectrum of SASP, including cytokines, extracellular matrix degrading enzymes, and growth factors, which affect neighboring cells, often prompting low grade inflammation and tissue damage.18 They, in a cell cycle arrested state, survive with upregulated anti-apoptotic pathways. Therefore, senolytics, a new class of drugs that selectively clear senescent cells, are proposed as anti-aging therapeutics. The selective elimination of senescent cells, commonly known as senolysis, decreases chronic inflammation, improves tissue repair capacity, and slows the decline of physical function in aged organisms.19 It showed extend median lifespan, and prevented or attenuated various features of aging and age-related diseases, including osteoarthritis, atherosclerosis, neurodegeneration, and cancer in animals.20,21 Conventional senolytic drugs kill all senescent cells regardless of their types, location, and functionality.22 They can be effective, but their non-selective elimination of all senescent cells may cause systemic side effects.23 A photosensitive senolytic, which can be activated on demand to ablate specific senescent cells in a defined location, avoiding the widespread non-specific killing, may have a far-reaching contribution to study the neighboring effect of senescent cells. Although a number of β-gal activatable fluorescence probes have been reported,8 the β-gal activatable photodynamic agent is still limited.24–26 We thus design a novel β-gal activatable photosensitive probe, named Gal-MB, for senescent cell identification and selective photoelimination, based on a recently developed enzyme activatable methylene blue (MB) construct, gGluMB, which is an activatable photosensitizer sensitive to gamma-glutamyl transpeptidase.27
2. Experimental
2.1. Synthetic materials and methods
All chemicals and solvents were purchased from Sigma-Aldrich (St. Louis, MO) and Thermo Fisher Scientific (Waltham, MA) unless otherwise specified. Starting materials 4-hydroxymethylphenyl(2,3,4,6-tetra-O-acetyl-β-D-galactopyranoside)28 and leucomethylene blue (LMB)27 were synthesized by following the reported procedures. The reaction progress was monitored with liquid chromatography coupled mass spectrometer (LC-MS) (Waters, Milford, MA). Compounds were separated and purified with high performance liquid chromatography (HPLC, preparative C-18 column). 1H and 13C NMR spectra were collected in deuterated methanol (CD3OD) on Ascend-500 spectrometer (Bruker, Billerica, MA). High resolution mass spectroscopy (HRMS) data were collected on a PE Sciex API 100 mass spectrometer (Applied Biosystems, Waltham, MA).2.2. Synthesis of AcGal-MB
Leucomethylene blue27 (LMB, 0.25 mmol) was dissolved in dichloromethane (DCM, 5 mL) and cooled to −10 °C. Phosgene in toluene (caution: toxic, 15% w/w, 0.35 mL, 0.5 mmol) and N,N-diisopropylethylamine (DIPEA, 65 mg, 0.5 mmol) were added to the previous solution slowly. After the solution was stirred at room temperature (RT) for 15 min, the extra phosgene and solvent were dried on a rotary evaporator. The acquired carbamate chloride product was re-dissolved in anhydrous DCM (5 mL) and cooled to 0 °C. To this solution, sodium carbonate (Na2CO3, 53 mg, 0.5 mmol), 4-dimethylaminopyridine (DMAP, 30 mg, 0.25 mmol), and 4-hydroxymethylphenyl(2,3,4,6-tetra-O-acetyl-β-D-galactopyranoside)28 (114 mg, 0.25 mmol) were added. The reaction stirred for 3 h, then was quenched with saturated aqueous ammonium chloride solution. After extraction, the crude product was purified with preparative C-18 column, and AcGal-MB was obtained as a solid (43 mg, yield: 23%). ESI-HRMS: for C38H43N3O12S: expected m/z = 766.2646 [M + H]+; found m/z = 766.2632 [M + H]+; 1.8 ppm error.2.3. Synthesis of Gal-MB
To a solution of AcGal-MB (38 mg, 0.05 mmol) in 10 mL anhydrous MeOH, and cooled to 0 °C. NaOMe in MeOH (25% wt, 46 μL, 0.2 mmol) was added slowly under nitrogen condition. After stirring for 3 h, the reaction was quenched with saturated NH4Cl solution. After removing the solvent, the crude product was purified with preparative C-18 column, and the desired product Gal-MB (17 mg, 58%) was obtained. 1H NMR (500 MHz, CD3OD) δ 7.44 (d, J = 8.9 Hz, 2H), 7.27 (d, J = 8.6 Hz, 2H), 7.08 (d, J = 8.6 Hz, 2H), 7.05 (d, J = 2.5 Hz, 2H), 6.97 (dd, J = 8.9, 2.6 Hz, 2H), 5.15 (s, 2H), 4.86 (s, 1H), 3.90 (d, J = 3.3 Hz, 1H), 3.83–3.71 (m, 3H), 3.71–3.65 (m, 1H), 3.58 (dd, J = 9.7, 3.4 Hz, 1H), 3.05 (s, 12H). 13C NMR (126 MHz, CD3OD) δ 157.83, 154.22, 146.21, 133.24, 131.67, 129.70, 129.29, 127.43, 116.36, 113.69, 113.00, 101.39, 75.63, 73.46, 70.85, 68.84, 67.64, 61.08, 41.60. ESI-HRMS: for C30H35N3O8S: expected m/z = 598.2223 [M + H]+; found m/z = 598.2216 [M + H]+; 1.2 ppm error.2.4. Optical characterization
Gal-MB stock solution (1 mM in water) was stored at −78 °C until use. β-Galactosidase from Escherichia coli (β-gal, Sigma-Aldrich) was stored as 1000 U mL−1 in PBS buffer (pH = 7.4). The optical characterization experiment was carried out using Cary 60 UV-Vis spectrophotometer (Agilent, Santa Clara, CA) and Cary Eclipse fluorescence spectrophotometer (Agilent).2.5. Cell lines
C6 (rat glioma), C6/LacZ (LacZ transfected C6 cell line), MDA-MB231 human breast cancer cell line, and A549 human non-small-cell lung carcinoma cell line were purchased from American Type Culture Collection (ATCC; Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) medium (Mediatech Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), and 1% penicillin/streptomycin in an incubator with humidified air containing 5% CO2 at 37 °C. MDA-MB231 cells were consistently treated with palbociclib (5 μM) for 7 d to have the drug-induced senescent MDA-MB231 cells (MB231_SC). To develop ionizing radiation-induced cellular senescence, A549 cells were irradiated with X-ray using RS 2000 Biological Research X-ray Irradiator (Rad Source Technologies, Buford, GA) and incubated in complete medium for 5 days.2.6. X-gal or senescence β-galactosidase staining
C6 and C6/LacZ cells (2.5 × 104 per well) were seeded on a μ-Plate 96 Well Black (ibidi GmbH, Gräfelfing, Germany) and incubated for 24 h. MDA-MB231 cells (1.0 × 105 per well) were seeded on 6-well plate (Corning Life Sciences, Pittston, PA) and incubated for 24 h. For MB231_SC, MDA-MB231 cells (0.5 × 105 per well) were seeded on 6-well plate, and from next day, palbociclib (5 μM) were added for 7 d. For A549, cells were irradiated with various dose of X-ray (2, 5, 10 Gy) at day 0 and incubated in complete medium. At day 3, A549 and X-ray-irradiated A549 cells (2 × 104 per well) were detached, seeded on a 12-well plate (Corning Life Sciences), and cultured until day 5. Then, C6, C6/LacZ, MDA-MB231, MB231_SC, A549, and X-ray-irradiated A549 cells were stained with Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, Boston, MA) by following the staining protocol. In brief, cells were rinsed with PBS, added 1× fixation solution, and allowed cells to fix for 15 min at RT. Then, cells were washed twice with PBS and incubated overnight at 37 °C with β-gal staining solution containing X-gal in N,N-dimethylformamide (pH 6.0). Cells were checked under a microscope for the development of blue color.2.7. Cellular uptake and location of Gal-MB
C6 and C6/LacZ cells (2.5 × 104 per well) were seeded on a μ-plate 96 Well Black (ibidi GmbH) and incubated for 24 h. MDA-MB 231 cells (5 × 103 per well) were seeded on a 96-well black clear-bottom culture plate (Corning Life Sciences) for 24 h. For MB231_SC, MDA-MB231 cells (2 × 103 per well) were seeded at day 0 and treated with palbociclib (5 μM) for 7 d. For A549, cells were irradiated with various dose of X-ray (2, 5, 10 Gy) at day 0 and incubated in complete medium. At day 3, A549 and X-ray-irradiated A549 cells (3 × 103 per well) were detached, seeded on a 96-well black clear-bottom culture plate (Corning Life Sciences), and cultured until day 5. Then, cells were treated with Gal-MB (5 μM) for 1 h and Hoechst 33342 (Thermo Fisher Scientific) was added for 5 min for nuclei staining. Cells were washed with PBS and the fluorescence images were captured using an EVOS fluorescence microscope (Life Technologies, Carlsbad, CA) in bright field, DAPI channel for nuclei images, and Cy5 for MB images. For the MB relocation from lysosome to nucleus experiment (Fig. S5†), C6/LacZ cells were treated with Gal-MB (20 μM) for 2 h and the cells were illuminated with Cy5-filtered red light (590–650 nm) for 1 min. The cell images were captured before and after light illumination.2.8. Photoablation effect
C6 and C6/LacZ cells (1 × 104 per well) or MDA-MB231 cells (5 × 103 per well) were seeded on a 96-well black clear-bottom culture plate (Corning Life Sciences) for 24 h. For MB231_SC, MDA-MB231 cells (2 × 103 per well) were seeded at day 0 and treated with palbociclib (5 μM) for 7 d. Then, cells were treated with different concentrations of Gal-MB (0, 2.5, 5, 7.5, 10, 15, 20, 30 μM) for 2 h. Cells were illuminated with 665 nm LED light (30 mW cm−2) for 30 min (photo toxicity) or left in dark (dark toxicity). Cells were replaced with fresh medium and cultured for an additional 24 h. The cell viability was further measured by cytotoxicity assay.2.9. Cell viability/cytotoxicity assay
Without or with light illumination, cells (C6, C6/LacZ, MDA-MB231, MB231_SC) were replenished with 10 μL of cell counting kit-8 (CCK-8) solution (Dojindo Molecular Technologies, Rockville, MD) containing medium. Cells were incubated for 3 h and the absorbance of the solution was measured at 450 nm using a plate reader (Tecan, Mannedorf, Switzerland).2.10. Western blot
For western blot analysis, cell extracts (MDA-MB231, MB231_SC-4 d, MB231_SC-7 d) were prepared. Each protein's concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific). The proteins (30 μg per sample) were separated on 4–12% gradient gels (Thermo Fisher Scientific) and transferred to nitrocellulose membrane (Thermo Fisher Scientific) by iBlot 2 Dry Blotting System (Thermo Fisher Scientific). Blots were blocked with blocking buffer (Bio-Rad Laboratories, Hercules, CA) for 30 min and incubated at 4 °C overnight with one of the following primary antibodies: p21Cip1/Waf1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology, #2947), phospho-Rb (Ser807/811, 1
1000 dilution, Cell Signaling Technology, #2947), phospho-Rb (Ser807/811, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology, #9308) Lamin B1 (1
1000 dilution, Cell Signaling Technology, #9308) Lamin B1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology, #13435) and beta-actin (1
1000 dilution, Cell Signaling Technology, #13435) and beta-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Cell Signaling Technology, #4967). The next day, blots were washed 3 times (5 min each) with PBS (0.1% Tween-20). The membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or anti-rabbit IgG secondary antibody (1
1000 dilution, Cell Signaling Technology, #4967). The next day, blots were washed 3 times (5 min each) with PBS (0.1% Tween-20). The membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or anti-rabbit IgG secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, Cell Signaling Technology) for 1 h at RT. The immunoreactive proteins on the membranes were visualized using Supersignal™ West Femto substrate (Thermo Fisher Scientific). Immunoblot signals were developed with a C400 image machine (Azure Biosystems, Dubin, CA) and quantified by ImageJ.
2000 dilution, Cell Signaling Technology) for 1 h at RT. The immunoreactive proteins on the membranes were visualized using Supersignal™ West Femto substrate (Thermo Fisher Scientific). Immunoblot signals were developed with a C400 image machine (Azure Biosystems, Dubin, CA) and quantified by ImageJ.
3. Results and discussion
3.1. Characterization of Gal-MB
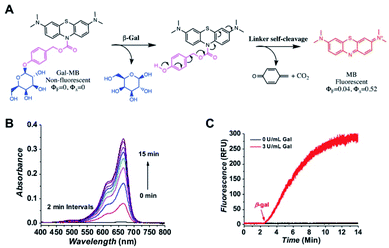
Gal-MB was designed to have three major components, a β-gal recognition domain, a self-immolative linker and a fluorogenic domain (Fig. 1A). To prepare Gal-MB (Fig. S1, ESI†), the fluorescing MB was first reduced to leucomethylene blue (LMB).27 This reduction step broke the conjugating structure of MB and completely blocked its absorption, fluorescence, and 1O2 production. 4-Hydroxymethylphenyl(2,3,4,6-tetra-O-acetyl-β-D-galactopyranoside), prepared by a previously developed protocol,28 was then reacted with LMB under basic condition to have the fully protected AcGal-MB. The subsequent deacetylation by sodium methoxide resulted in the desired product Gal-MB, which is colorless and non-fluorescent.Upon reacting with β-gal, the galactopyranoside will be cleaved, and the exposed linker will go through a spontaneous self-elimination, resulting the original photosensitive MB (Fig. 1A). To confirm the proposed β-gal activation, the absorption and fluorescence spectra of Gal-MB (5 μM) in PBS was collected without β-gal (Fig. S2A, ESI†). As expected, the Gal-MB solution showed no absorbance between 400–800 nm, and no fluorescence when it was excited at 655 nm. Immediately after the β-gal (3 U L−1) addition, rapid absorption (60-fold increase in 15 min) and fluorescence (95-fold increase in 15 min) recovery with the characteristics of MB were observed (Fig. 1B and C), accompanied by an obvious color change from colorless to blue. The released MB had an absorption centered at 665 nm (ε = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1), a fluorescence maximum at 675 nm (ΦF = 0.04 in water), and a high 1O2 production yield (ΦΔ = 0.52). To further confirm the conversion of Gal-MB to MB, Gal-MB (300 μM) solution treated with β-gal (10 U L−1) was tracked by LC-MS (Fig. S2B, ESI†). Gal-MB with a retention time of 5.90 min gradually disappeared (found m/z = 598.5 and calculated m/z = 598.2 [M + H]+), meanwhile a new MB peak at 5.77 min appeared (found m/z = 284.2 and calculated m/z = 284.1 [M]+). All Gal-MB was converted to MB within 60 min. This rapid MB production can be ascribed to effective β-gal activity, self-immolative elimination of the linker, and effortless carbamate bond breakage.
000 M−1 cm−1), a fluorescence maximum at 675 nm (ΦF = 0.04 in water), and a high 1O2 production yield (ΦΔ = 0.52). To further confirm the conversion of Gal-MB to MB, Gal-MB (300 μM) solution treated with β-gal (10 U L−1) was tracked by LC-MS (Fig. S2B, ESI†). Gal-MB with a retention time of 5.90 min gradually disappeared (found m/z = 598.5 and calculated m/z = 598.2 [M + H]+), meanwhile a new MB peak at 5.77 min appeared (found m/z = 284.2 and calculated m/z = 284.1 [M]+). All Gal-MB was converted to MB within 60 min. This rapid MB production can be ascribed to effective β-gal activity, self-immolative elimination of the linker, and effortless carbamate bond breakage.
3.2. Cellular uptake of Gal-MB in β-gal expressing cells
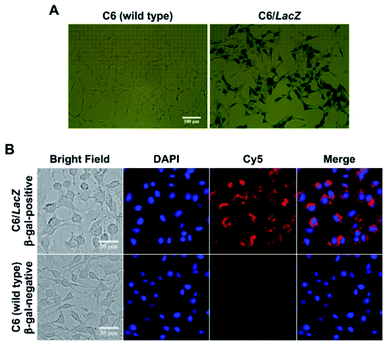
Next, the intracellular β-gal sensitivity was examined using two model cell lines, rat glial tumor C6 cells (control, no β-gal expression) and C6/LacZ cells, which constitutively expresses β-gal. Their β-gal expression in cells were confirmed by standard X-gal staining after fixation (Fig. 2A). The dark blue color, indicating β-gal expression, was only seen in the transfected C6/LacZ cells, but not in the wild type C6 cells. The cells were then incubated with Gal-MB (5 μM) for 1 h and their cellular uptake were investigated with live cell fluorescence imaging. The far-red signal in the Cy5 channel, reporting β-gal enzymatic activity, was only seen in β-gal positive C6/LacZ and not in β-gal negative C6 cells (Fig. 2B), consistent with the X-gal staining results.3.3. β-Gal dependent specific photoablation effect of Gal-MB
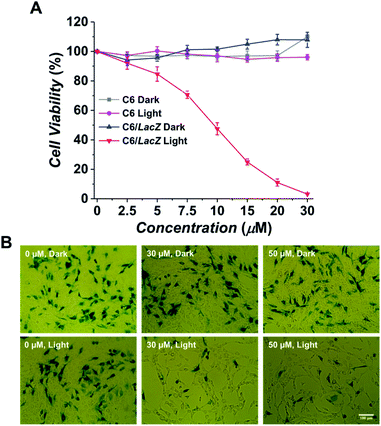
After confirming the specific fluorescence activation in the β-gal expressing cells, its selective phototoxicity was further tested. The C6 and C6/LacZ cells were treated with Gal-MB (0–30 μM) for 2 h, washed with the medium and illuminated using light-emitting diode (LED) lamp (665 nm, 30 mW cm−2) for 30 min. The total light dose of 54 J cm−2 was within the recommended therapeutic light dose (50–200 J cm−2).29 Gal-MB alone group showed no dark toxicity in both cells, up to 30 μM (Fig. 3A). The subsequent light illumination didn't affect the viability of β-gal negative C6 cells, however, it caused significant photodamage toward β-gal expressing C6/LacZ cells. The IC50 of Gal-MB plus light to C6/LacZ cells was about 9 μM. This result strongly supported the β-gal dependent phototoxicity in cells.To further confirm the Gal-MB's β-gal-selectivity, C6 and C6/LacZ cells were cocultured at 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio, treated with Gal-MB (0, 30, 50 μM) for 2 h and then illuminated with light for 30 min (665 nm, 30 mW cm−2). The treated cells were cultured for additional 24 h, fixed and stained by X-gal. The control group of cells, which only treated with Gal-MB but without illumination, was compared side-by-side (Fig. 3B). The X-gal blue staining showed similar density of blue cells after treatment with 0, 30 or 50 μM of Gal-MB, suggesting the Gal-MB is not toxic to the cells, including both C6 and C6/LacZ. In contrast, the light illumination cleared almost all blue cells, which were the β-gal expressing C6/LacZ cells. The survived cells are non-stained β-gal negative C6 cells. This data clearly shows that Gal-MB is capable of discerning cells with different β-gal expression level and its phototoxicity is highly β-gal dependent.
3 ratio, treated with Gal-MB (0, 30, 50 μM) for 2 h and then illuminated with light for 30 min (665 nm, 30 mW cm−2). The treated cells were cultured for additional 24 h, fixed and stained by X-gal. The control group of cells, which only treated with Gal-MB but without illumination, was compared side-by-side (Fig. 3B). The X-gal blue staining showed similar density of blue cells after treatment with 0, 30 or 50 μM of Gal-MB, suggesting the Gal-MB is not toxic to the cells, including both C6 and C6/LacZ. In contrast, the light illumination cleared almost all blue cells, which were the β-gal expressing C6/LacZ cells. The survived cells are non-stained β-gal negative C6 cells. This data clearly shows that Gal-MB is capable of discerning cells with different β-gal expression level and its phototoxicity is highly β-gal dependent.
3.4. Gal-MB's imaging and photoablation effect in senescent cells
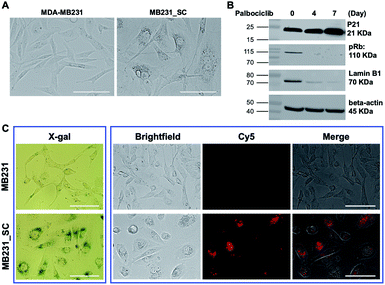
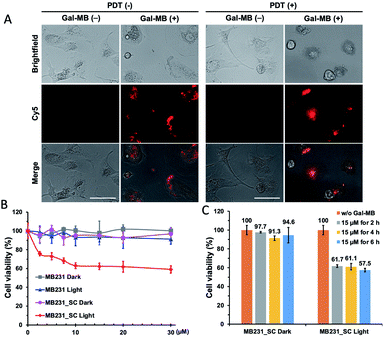
Following the promising selective photoablation results in C6 and C6/LacZ cells, we extended Gal-MB's application to senescent cell imaging and senolysis. The drug-induced senescence model was adopted to verify Gal-MB.23,30 MDA-MB231 breast cancer cells were treated with palbociclib (5 μM), a CDK4/6 inhibitor, for 7 days to induce senescent MDA-MB231 cells (MB231_SC). With this treatment, cells' morphology became flat, irregular and enlarged, which stands for the characteristics of senescent cells (Fig. 4A).31,32 We further examined the changes of representative senescence markers, including p21, phospho-retinoblastoma protein (pRb), and Lamin B1,33 by western blot. Not only increased p21 but also reduced pRb and Lamin B1 expression levels clearly confirmed the successful induction of senescence by palbociclib treatment (Fig. 4B). Although significant morphological changes were seen in cell culture, it might not be obvious in vivo as cell morphology is dictated by the architecture of the tissue. Gal-MB was designed to highlight senescent cells regardless of their shapes, and its far-red signal can offer better imaging sensitivity in vivo. The expression of SA-β-gal, the gold standard of senescence,34 in MB231_SC was also investigated by X-gal staining (Fig. 4C). While the wild type MDA-MB231 cells were colorless, light blue stain was observed in the majority of palbociclib-induced MB231_SC. Similar to the result in C6 and C6/LacZ model cells, Gal-MB signal was not seen in the parent MDA-MB231 cells, but the far-red fluorescent signal in the Cy5 channel was clearly seen in the MB231_SC, supporting that SA-β-gal can also activate the Gal-MB (Fig. 4C). To demonstrate a broad application of Gal-MB in detecting SA-β-gal, a different senescence inducing method, X-ray radiation, was also included.35,36 A second cell line, human non-small-cell lung carcinoma, A549, was irradiated with X-ray (2, 5, or 10 Gy) and then incubated in regular culture media for 5 days. The X-gal staining showed a good correlation between the radiation dose and SA-β-gal level (Fig. S3A, ESI†). Consistent with the X-gal staining results (Fig. S3A, ESI†), a similar dose response was observed with Gal-MB, confirming the utility of Gal-MB in senescence imaging (Fig. S3B and C, ESI†).The selective photoablation effect toward senescence cells was quickly confirmed by microscopic imaging. The photodynamic killing of senescent cells was only seen in the Gal-MB (15 μM, 2 h) treated and illuminated MB231_SC (Fig. 5A), while sparing the control MDA-MB231 cells (Fig. S4, ESI†). Without the combination of Gal-MB and light, the cells remain alive, and their morphology were intact (Fig. 5A). We noticed that the MB fluorescence signal relocated from cytoplasm to nucleus after illumination. It is not unique for the MB231_SC, the same light-triggered MB redistribution is also observed in the C6/LacZ cells (Fig. S5, ESI†), and previously in the gGluMB treated glioma cells.27 As seen in Fig. S5 (ESI†), a large number of dark dots were visualized in the bright field, suggesting a strong aggregation of the hydrophobic MB released by β-gal reaction. The initial intracellular fluorescence signal was relatively low due to the aggregation-induced quenching. Upon a strong continuous light illumination, the aggregated MB absorbed and converted the light energy to thermodynamic energy. Thereafter, MB dispersed and relocated to the nucleus. The freed MB was highly fluorescent and photosensitive, and ultimately caused photodamage to the cells.27,37
To verify the dose and time-dependent photo-senolytic effect of Gal-MB, the cell viability of MDA-MB231 and MB231_SC was determined after various treatments. Around 40% killing in MB231_SC was observed via Gal-MB (15 μM, 2 h incubation) and light illumination (30 min, 665 nm LED, 30 mW cm−2). However, longer incubation time (4 h–6 h) and higher concentration (20–30 μM) of Gal-MB showed little benefit in senescent cell killing (Fig. 5B and C). This result was different from the C6/LacZ data (Fig. 3A) which achieved around 70% of cell killing effect under identical Gal-MB and light illumination conditions. Complete elimination of C6/LacZ cells was obtained by 30 μM of Gal-MB. We postulate that low SA-β-gal level may play a role in this photoablation difference (70% vs. 40%) between C6/LacZ and MB231_SC. While the stably transfected C6/LacZ cells expressed high level of β-gal, the palbociclib-induced MB231_SC merely produce moderate level of SA-β-gal. The color of X-gal staining clearly showed the different β-gal expression levels between these two cell lines (Fig. 2A vs. 4C).
4. Conclusions
In summary, we have developed a β-gal responsive far-red fluorogenic probe, Gal-MB, suitable for β-gal imaging and photoablation. Gal-MB is based on an FDA-approved photosensitizer, MB. The fluorescence and photosensitivity of Gal-MB are prohibited by the disrupted conjugating double bonds. Importantly, the lost optical properties can be re-claimed by β-gal. Cell ablation assay showed that Gal-MB can induce phototoxicity specifically toward β-gal-expressing cells, while sparing β-gal non-expressing cells. This selectivity also applied to the senescent cells. Outstanding senescence imaging efficiency of Gal-MB was confirmed by correlating it with the standard X-gal staining of senescence cells. Presumably due to low expression of SA-β-gal in the palbociclib-induced MB231_SC cells, Gal-MB's phototoxicity is lower than it in the constitutive β-gal expressing C6/LacZ cells. Taken together, Gal-MB could be a promising SA-β-gal specific probe for senescence diagnosis and imaging, and a sub-optimal photodynamic agent for senolytic application.Conflicts of interest
There are no conflicts to declare.Notes and references
- I. G. Serebriiskii and E. A. Golemis, Anal. Biochem., 2000, 285, 1–15 CrossRef CAS PubMed.
- C. M. Ghim, S. K. Lee, S. Takayama and R. J. Mitchell, BMB Rep., 2010, 43, 451–460 CrossRef CAS PubMed.
- A. M. Broome, N. Bhavsar, G. Ramamurthy, G. Newton and J. P. Basilion, Mol. Pharm., 2010, 7, 60–74 CrossRef CAS PubMed.
- C. H. Tung, Q. Zeng, K. Shah, D. E. Kim, D. Schellingerhout and R. Weissleder, Cancer Res., 2004, 64, 1579–1583 CrossRef CAS PubMed.
- Y. Fu and W. Xiao, EMBO Mol. Med., 2006, 313, 257–264 CAS.
- D. J. Spergel, U. Kruth, D. R. Shimshek, R. Sprengel and P. H. Seeburg, Prog. Neurobiol., 2001, 63, 673–686 CrossRef CAS PubMed.
- G. P. Dimri, X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj and O. Pereira-Smith, et al., Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9363–9367 CrossRef CAS PubMed.
- B. Lozano-Torres, J. F. Blandez, F. Sancenon and R. Martinez-Manez, Anal. Bioanal. Chem., 2021, 413, 2361–2388 CrossRef CAS PubMed.
- Y. Yao, Y. Zhang, C. Yan, W. H. Zhu and Z. Guo, Chem. Sci., 2021, 12, 9885–9894 RSC.
- W. Wei and S. Ji, J. Cell. Physiol., 2018, 233, 9121–9135 CrossRef CAS PubMed.
- E. Fitsiou, A. Soto-Gamez and M. Demaria, Semin. Cancer Biol., 2021 DOI:10.1016/j.semcancer.2021.03.021.
- M. Safir Filho, P. Dao, M. Gesson, A. R. Martin and R. Benhida, Analyst, 2018, 143, 2680–2688 RSC.
- E. Gonzalez-Gualda, A. G. Baker, L. Fruk and D. Munoz-Espin, FEBS J., 2021, 288, 56–80 CrossRef CAS PubMed.
- X. K. Li, W. J. Qiu, J. W. Li, X. Chen, Y. L. Hu, Y. Gao, D. L. Shi, X. M. Li, H. L. Lin, Z. L. Hu, G. Q. Dong, C. Q. Sheng, B. Jiang, C. L. Xia, C. Y. Kim, Y. Guo and J. Li, Chem. Sci., 2020, 11, 7292–7301 RSC.
- K. Itahana, Y. Itahana and G. P. Dimri, Methods Mol. Biol., 2013, 965, 143–156 CrossRef CAS PubMed.
- B. Lozano-Torres, J. F. Blandez, I. Galiana, A. Garcia-Fernandez, M. Alfonso, M. D. Marcos, M. Orzaez, F. Sancenon and R. Martinez-Manez, Angew. Chem., Int. Ed. Engl., 2020, 59, 15152–15156 CrossRef CAS PubMed.
- H. Ito, Y. Kawamata, M. Kamiya, K. Tsuda-Sakurai, S. Tanaka, T. Ueno, T. Komatsu, K. Hanaoka, S. Okabe, M. Miura and Y. Urano, Angew. Chem., Int. Ed. Engl., 2018, 57, 15702–15706 CrossRef CAS PubMed.
- A. Hernandez-Segura, T. V. de Jong, S. Melov, V. Guryev, J. Campisi and M. Demaria, Curr. Biol., 2017, 27, 2652–2660 CrossRef CAS PubMed.
- Y. Cai, H. Zhou, Y. Zhu, Q. Sun, Y. Ji, A. Xue, Y. Wang, W. Chen, X. Yu, L. Wang, H. Chen, C. Li, T. Luo and H. Deng, Cell Res., 2020, 30, 574–589 CrossRef CAS PubMed.
- J. M. van Deursen, Science, 2019, 364, 636–637 CrossRef CAS PubMed.
- M. Borghesan, W. M. H. Hoogaars, M. Varela-Eirin, N. Talma and M. Demaria, Trends Cell Biol., 2020, 30, 777–791 CrossRef CAS PubMed.
- R. Di Micco, V. Krizhanovsky, D. Baker and F. d'Adda di Fagagna, Nat. Rev. Mol. Cell Biol., 2021, 22, 75–95 CrossRef CAS PubMed.
- D. Munoz-Espin, M. Rovira, I. Galiana, C. Gimenez, B. Lozano-Torres, M. Paez-Ribes, S. Llanos, S. Chaib, M. Munoz-Martin, A. C. Ucero, G. Garaulet, F. Mulero, S. G. Dann, T. VanArsdale, D. J. Shields, A. Bernardos, J. R. Murguia, R. Martinez-Manez and M. Serrano, EMBO Mol. Med., 2018, 10, e9355 Search PubMed.
- Y. Ichikawa, M. Kamiya, F. Obata, M. Miura, T. Terai, T. Komatsu, T. Ueno, K. Hanaoka, T. Nagano and Y. Urano, Angew. Chem., Int. Ed., 2014, 53, 6772–6775 CrossRef CAS PubMed.
- Y. Koide, Y. Urano, A. Yatsushige, K. Hanaoka, T. Terai and T. Nagano, J. Am. Chem. Soc., 2009, 131, 6058–6059 CrossRef CAS PubMed.
- M. Chiba, M. Kamiya, K. Tsuda-Sakurai, Y. Fujisawa, H. Kosakamoto, R. Kojima, M. Miura and Y. Urano, ACS Cent. Sci., 2019, 5, 1676–1681 CrossRef CAS PubMed.
- Z. Shen and C. H. Tung, Chem. Commun., 2020, 56, 13860–13863 RSC.
- N. H. Ho, R. Weissleder and C. H. Tung, Chembiochem, 2007, 8, 560–566 CrossRef CAS PubMed.
- N. Mohanty, M. Jalaluddin, S. Kotina, S. Routray and Y. Ingale, J. Clin. Diagn. Res., 2013, 7, 1254–1257 Search PubMed.
- S. Vijayaraghavan, C. Karakas, I. Doostan, X. Chen, T. Bui, M. Yi, A. S. Raghavendra, Y. Zhao, S. I. Bashour, N. K. Ibrahim, M. Karuturi, J. Wang, J. D. Winkler, R. K. Amaravadi, K. K. Hunt, D. Tripathy and K. Keyomarsi, Nat. Commun., 2017, 8, 15916 CrossRef CAS PubMed.
- Z. Chen, K. Jiang, Z. Zou, X. Luo, C. T. Lim and C. Wen, Biomicrofluidics, 2020, 14, 034106 CrossRef CAS PubMed.
- L. Yin, Y. Wu, Z. Yang, C. A. Tee, V. Denslin, Z. Lai, C. T. Lim, E. H. Lee and J. Han, Lab Chip, 2018, 18, 878–889 RSC.
- A. Hernandez-Segura, J. Nehme and M. Demaria, Trends Cell Biol., 2018, 28, 436–453 CrossRef CAS PubMed.
- A. Biran, L. Zada, P. Abou Karam, E. Vadai, L. Roitman, Y. Ovadya, Z. Porat and V. Krizhanovsky, Aging Cell, 2017, 16, 661–671 CrossRef CAS PubMed.
- H. Q. Nguyen, N. H. To, P. Zadigue, S. Kerbrat, A. De La Taille, S. Le Gouvello and Y. Belkacemi, Crit. Rev. Oncol. Hematol., 2018, 129, 13–26 CrossRef PubMed.
- E. Fletcher-Sananikone, S. Kanji, N. Tomimatsu, L. F. M. Di Cristofaro, R. K. Kollipara, D. Saha, J. R. Floyd, P. Sung, R. Hromas, T. C. Burns, R. Kittler, A. A. Habib, B. Mukherjee and S. Burma, Cancer Res., 2021, 81, 5935–5947 CAS.
- W. Zhang and C. H. Tung, Chemistry, 2018, 24, 2089–2093 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting figures. See DOI: 10.1039/d2ra00377e |
| ‡ S. K. Lee and Z. Shen contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2022 |