Open Access Article
Open Access ArticlePNA microprobe for label-free detection of expanded trinucleotide repeats†
Narges Asefifeyzabadia,
Grace Durochera,
Kizito-Tshitoko Tshilengeb,
Tanimul Alamb,
Lisa M. Ellerbyb and
Mohtashim H. Shamsi *a
*a
aSchool of Chemical and Biomolecular Sciences, 1245 Lincoln Dr, Southern Illinois University at Carbondale, IL 62901, USA. E-mail: mshamsi@siu.edu
bThe Buck Institute for Research on Aging, 8001 Redwood Blvd, Novato, CA 94945, USA
First published on 10th March 2022
Abstract
We present a PNA microprobe sensing platform to detect trinucleotide repeat mutation by electrochemical impedance spectroscopy. The microprobe platform discriminated Huntington's disease-associated CAG repeats in cell-derived total RNA with S/N 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. This sensitive, label-free, and PCR-free detection strategy may be employed in the future to develop biosensing platforms for the detection of a plethora of repeat expansion disorders.
3. This sensitive, label-free, and PCR-free detection strategy may be employed in the future to develop biosensing platforms for the detection of a plethora of repeat expansion disorders.
Nucleic acid repeats of various types are responsible for almost 50 genetically transferrable diseases.1 These diseases are generally known as repeat expansion disorders and weaken neuromuscular systems. The detection of DNA repeat expansions has been challenging owing to their length and complex structures they can form.2,3 Despite the high sensitivity of the electrochemical techniques, down to attomolar level,4,5 there have been very few efforts to detect expanded repeats. Most of the methods relied on chemical labelling or detecting short target lengths (maximum 10 repeats).6–11 Short DNA probes (18-mer) were recently proposed for label-free detection of G4C2 repeats associated with Amyotrophic lateral sclerosis (ALS) in cell-derived total RNA by measuring the charge transfer resistance (Rct) of the interface.12 Despite the label-free strategy, the sensitivity of the system was lower because difference in the detection signal between abnormal and normal target was only factor of 1.2 and the amount of RNA sample used (1 μg μL−1) was relatively high for electrochemical platform.12 The premise of this work is that how can we improve signal to noise ratio for discrimination of normal and abnormal repeat lengths in a biological sample while employing a label-free electrochemical detection strategy. Lower sensitivity of surface-bound probes in electrochemical biosensing may be a challenge due to limited control of probe orientation and density, and low hybridization efficiency on surface.13,14 Peptide nucleic acid (PNA) probes may mitigate these problems due to the excellent stability and improved hybridization efficiency owing to their electrostatic neutrality and stiff backbone.15–19
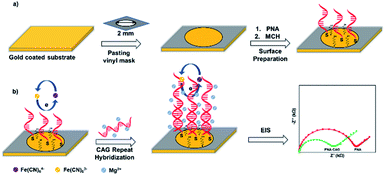
Here, we present a platform comprising PNA microprobes immobilized on gold surface for label-free detection of trinucleotide CAG repeats, associated with Huntington's disease (HD),20 by measuring the charge transfer resistance of the biosensing platform. The PNA probe (CTG-6) was immobilized on gold-coated substrate through Au–S bond followed by immobilizing mercaptahexanol (MCH) filler layer (Scheme 1a). Then, target CAG repeats were hybridized on the surface in presence of high cationic strength (20 mM Mg2+). We observed a decrease in the Rct following the hybridization event (Scheme 1b). The effect following the hybridization event is similar to what we have previously shown, that is addition of metal ion (e.g. Zn2+) into DNA monolayers lowers the Rct signal of the DNA/gold interface, which amplifies the sequence-dependent structural deformation of the DNA film (e.g. single base pair mismatch).21 Mg2+ in this study was used to stabilize DNA duplex formation, however, it also provides a high cationic microenvironment, which reduces the electrostatic repulsion between the biosensing interface and the negatively charged redox probe. As a result, we see decrease in the Rct after exposure to the target. PNA probe CTG-6 (PNA Bio, USA) was thiol modified at 5′-end while the synthetic DNA targets (IDT, USA) were comprised of CAG repeats with 6, 10, 15, and 20 repeat lengths. DNA and RNA sample solutions were prepared in 100 μM Tris containing 20 mM MgCl2 and 200 mM NaCl (pH = 8.5). Total RNA containing 72CAG (HD+) and 21CAG (HD−) were isolated from neural stem cells as described previously.22,23 Extraction details of the RNA samples along with the PCR test to confirm the HD− and HD+ samples (Fig. S1†) are given in the ESI.† RNA samples were diluted to 10 ng·μL−1 concentration for electrochemical detection. For the PNA-based electrochemical detection, gold electrodes (2 mm dia.) were prepared by cutting 1 cm2 pieces from a 100 nm gold coated substrate (Platypus, USA). The small substrates were immersed in an acid piranha H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2 (3
H2O2 (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 10 seconds followed by washing with DI water and 100 μM Tris buffer (pH = 8.5), and drying with N2. Then, a vinyl sheet with a 2 mm diameter hole was pasted on top of the 1 cm2 gold substrate to define the sensing electrode area. Next, a 5 μL aliquot of thiol modified PNA probe ‘CTG-6’ (Table S1†) solution was dropped on to the electrode surface to form a surface-assembled film on the surface at 4 °C. The electrodes were kept in a humid closed container to prevent evaporation. Then, the modified electrodes were washed three times with 10 μL of Tris buffer (pH = 8.5) and incubated with 1 mM mercaptahexanol (MCH) for 30 minutes to block any unmodified surface around the probe. The PNA-MCH modified electrode was later washed again with three times with 10 μL of Tris buffer and dried with N2 before exposing to CAG repeat targets. The hybridized complex was washed again with the buffer three times prior to EIS measurement. For RNA detection, a 5 μL aliquot of the 10 ng·μL−1 cell-extracted total RNA, containing CAG repeats, was placed on the PNA-MCH platform and incubated for 30 min. After exposing the platform to the CAG targets, the platform was washed three times with the Tris buffer followed by EIS measurement. All electrochemical experiments were performed using Autolab (Metrohm, USA) at room temperature in a three-electrode cell using the modified gold electrode as working electrode, Pt wire counter electrode, and Ag/AgCl reference electrode. Electrochemical impedance spectroscopy (EIS) was performed using soluble redox probe, 1 mM K4[Fe(CN)6]/K3[Fe(CN)6] (1
1) for 10 seconds followed by washing with DI water and 100 μM Tris buffer (pH = 8.5), and drying with N2. Then, a vinyl sheet with a 2 mm diameter hole was pasted on top of the 1 cm2 gold substrate to define the sensing electrode area. Next, a 5 μL aliquot of thiol modified PNA probe ‘CTG-6’ (Table S1†) solution was dropped on to the electrode surface to form a surface-assembled film on the surface at 4 °C. The electrodes were kept in a humid closed container to prevent evaporation. Then, the modified electrodes were washed three times with 10 μL of Tris buffer (pH = 8.5) and incubated with 1 mM mercaptahexanol (MCH) for 30 minutes to block any unmodified surface around the probe. The PNA-MCH modified electrode was later washed again with three times with 10 μL of Tris buffer and dried with N2 before exposing to CAG repeat targets. The hybridized complex was washed again with the buffer three times prior to EIS measurement. For RNA detection, a 5 μL aliquot of the 10 ng·μL−1 cell-extracted total RNA, containing CAG repeats, was placed on the PNA-MCH platform and incubated for 30 min. After exposing the platform to the CAG targets, the platform was washed three times with the Tris buffer followed by EIS measurement. All electrochemical experiments were performed using Autolab (Metrohm, USA) at room temperature in a three-electrode cell using the modified gold electrode as working electrode, Pt wire counter electrode, and Ag/AgCl reference electrode. Electrochemical impedance spectroscopy (EIS) was performed using soluble redox probe, 1 mM K4[Fe(CN)6]/K3[Fe(CN)6] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) prepared in 5× PBS buffer (pH 7.2). The following parameters were used to run the EIS: frequency range 100 kHz to 1 Hz, an applied DC potential of 250 mV vs. Ag/AgCl, and AC pulse of 5 mV amplitude. For simulation, Z-view version 3.5d was used to fit the EIS data into a modified Randle's equivalent circuit and extract the values for fitting elements including charge transfer resistance (Rct).
1) prepared in 5× PBS buffer (pH 7.2). The following parameters were used to run the EIS: frequency range 100 kHz to 1 Hz, an applied DC potential of 250 mV vs. Ag/AgCl, and AC pulse of 5 mV amplitude. For simulation, Z-view version 3.5d was used to fit the EIS data into a modified Randle's equivalent circuit and extract the values for fitting elements including charge transfer resistance (Rct).
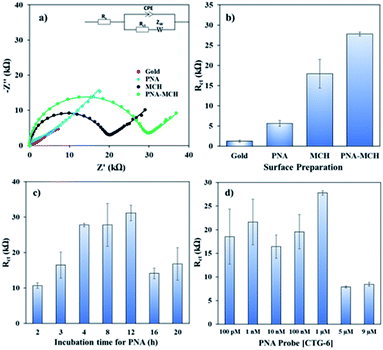
Fig. 1 shows the EIS response of various layers of the sensing platform and optimized conditions for the probe immobilization. The Nyquist form of EIS responses (Fig. 1a) of bare gold, ‘PNA’ probe, ‘MCH’ layer, and the combined ‘PNA-MCH’ layer. The inset shown in the panel is the modified Randel's equivalent circuit that fits the EIS data (extracted fitting values are provided in the ESI Table S2†). The bar graph in Fig. 1b shows that the probe only response (PNA) has a very low Rct which may be due to the absence of electrostatic repulsion between the neutral probe and the negatively charged redox probe. Thus, the resistance to charge transfer from ‘PNA’ is only a physical barrier while there may be a leakage current through empty space present around the probe.24 While the mercaptahexanol blocking layer (MCH) has a higher Rct than the PNA due to its higher film packing and negative charge on the surface due to –OH group. The response of the platform with PNA followed by MCH immobilization (PNA-MCH) has higher Rct than the separate layers with significant reproducibility where MCH blocks the non-specific sites and prevent current leakage through the pinholes. The Rct of PNA-MCH at 4 h immobilization time (Fig. 1c) and at 1 μM PNA concentration (Fig. 1d) was the highest and has the lowest RSD = 1.7% (see EIS curves in Fig. S2†). Based on the results in Fig. 1, the PNA-MCH platform was prepared to detect the CAG repeat targets.
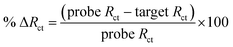 | (1) |
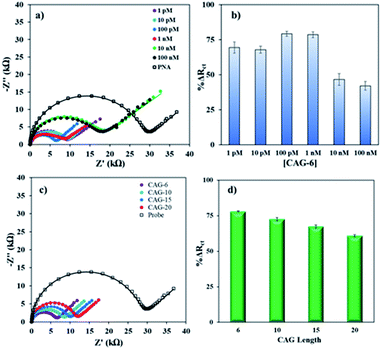
Then, complementary target CAG-6 (6 repeat units same as probe length) was hybridized at 0.25–2 h to optimize the hybridization time. The Rct decreased following hybridization (Fig. S3a in ESI†) due to high cationic strength microenvironment, which reduces electrostatic repulsion between the sensing interface and the negatively charged redox probe. The % ΔRct was calculated using eqn (1) and the results were compared in Fig. S3b.† The hybridization time of 30 min was selected for further experiments where the change in signal has the lowest standard error, i.e. 59.0 ± 0.7%. Then, sensitivity of the PNA microprobes was measured toward target (CAG-6) concentration and target lengths (6–20 CAG repeats) following hybridization at the optimized conditions discussed above (i.e. 1 μM PNA for 4 h incubation, 1 mM MCH for 30 min incubation followed by washing and drying). Fig. 2a represents the Nyquist form of EIS curves of CAG-6 target between 1 pM to 100 nM when hybridized with the optimized PNA microprobe platform for 30 min at room temperature. The % ΔRct with respect to CAG-6 concentration in Fig. 2b shows the highest change in Rct following duplex formation at 100 pM with 79.2 ± 1.7%. Fig. 2c represents the Nyquist plots of 6–20 CAG repeats (100 pM) following surface hybridization at the PNA microprobe platform, which reveals that the increase in repeat length resulted in higher Rct and hence lower % ΔRct (Fig. 2d). Higher resistance to charge transfer at higher repeat length is attributed to steric hindrance caused by size of the target sequence and consequently resisting diffusion of the redox probe through the sensing interface. Nevertheless, the repeat lengths were distinguishable from each other.
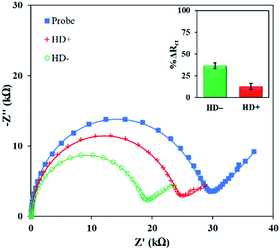
Finally, the PNA microprobe detection platform was applied to discriminate normal and pathogenic CAG repeats in real sample. The total RNA for the detection was extracted from neural stem cells as described in the ESI† and diluted to 10 ng·μL−1 concentration. It is important to note here that CAG expands up to 35 tandem repeats in a normal individual while pathogenicity begins when it expands beyond 38 tandem repeats resulting in Huntington's disease (HD).25 The total RNA sample with normal length (HD−) contains 21CAG repeats while with the pathogenic length (HD+) carries 72CAG repeats. Fig. 3 shows the Rct signals of the PNA microprobe before and after exposed to HD− and HD+ total RNA sample. Evidently, there was a significant difference between the normal and pathogenic repeats. Specifically, there was a higher change in HD− (38.0 ± 3.3%) compared to HD+ (12.7 ± 3.6%). This corroborated the trend with respect to length variation obtained above (see Fig. 2d). There was a 3-fold difference between the average signals of HD− and HD+, which may be the manifestation of the 3.4-fold difference in length between the HD− and HD+ repeats. Moreover, the difference in the absolute responses of the probe and HD+ sample were statistically different from each other as per T-test (see Table S3 in ESI†).
It is important to note that the current genetic testing methods for repeat expansion detection have their own pros and cons. For instance, state-of-the-art nanopore sequencing by Oxford Nanopore Technologies is low-cost and able to read long repeat-sequences (>4.5 kb),26 but the flow-cells with proteins nanopore membranes are prone to clogging and limited shelf-life.27 Single molecule real-time (SMRT) sequencing by PacBio can also read length more than 20 kb with high accuracy, however it relies on fluorescent labels for detection.28 DNA microarray technology relies on fluorescence labels and repeat primed PCR for signal amplification. DNA microarrays have also critical limitations because disease-associated repeats are normally GC-rich and highly repetitive,29–31 and polymerases do not traverse highly repetitive and GC-rich sequences efficiently.32,33 On the other hand, high-throughput whole-genome sequencing technologies are currently limited to ∼150 base pair read lengths.34 The most widely used method to detect repeat expansions is repeat-primed PCR (RP-PCR) with fragment length analysis,35 where the interpretation is usually challenging due to indel in the flanking regions of the repeat, which is prone to both false positives and false negatives.35 While using as a gold standard, Southern blotting requires a significant amount of input DNA (≥10 μg) and size estimates may be imprecise due to somatic heterogeneity.35,36 Electrochemical biosensing strategies are simple, rapid, sensitive, and can be integrated into miniaturized platforms for point-of-need applications.37 We propose that the label-free PNA microprobe-based strategy shown here can potentially be evolved into a simple platform requiring a very low sample amount and volume for discriminations of abnormal expansions of repeat, which may ultimately help rapid testing of a wide range of repeat associated neurodegenerative disorders.
In conclusion, we report here the application of PNA microprobes for label-free detection of length mutation of CAG trinucleotide repeats associated with Huntington's disease. The detection platform was optimized and tested for the detection of CAG repeats in cell-extracted total RNA. Concentration and immobilization time for the probe were optimized to obtain best performance to detect CAG targets of various concentrations and lengths. The conditions with the lowest error were selected to discriminate the normal and abnormal CAG repeats in cell-extracted total RNA. The sensitivity of the PNA microprobe is significantly better than DNA microprobe reported previously for G4C2 repeats,12 by achieving 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (HD−/HD+) signal at 10 ng·μL−1, which is a 2.5-fold signal improvement in 100-fold lower sample concentration. To best of our knowledge, this is the first application of PNA microprobe to discriminate length mutations by simple Rct measurement. We, however, acknowledge that this work is a proof-of-concept that requires further exploration of various sequences and lengths of repeats in total RNA and understand the behaviour of expanded lengths through their impedance responses.
1 (HD−/HD+) signal at 10 ng·μL−1, which is a 2.5-fold signal improvement in 100-fold lower sample concentration. To best of our knowledge, this is the first application of PNA microprobe to discriminate length mutations by simple Rct measurement. We, however, acknowledge that this work is a proof-of-concept that requires further exploration of various sequences and lengths of repeats in total RNA and understand the behaviour of expanded lengths through their impedance responses.
Author contributions
NA contributed to investigation, validation, and writing original draft. GD contributed to investigation. KT, TA, and LME contributed to investigation. MHS contributed to funding acquisition, supervision, writing original draft, editing, and reviewing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MHS acknowledges National Science Foundation EAGER award CBET-1940716 to support this research. MHS also acknowledges Neurodegenerative Disease Research Inc. for its support. LME acknowledges NIH NINDs NS100529.References
- C. Depienne and J.-L. Mandel, Am. J. Hum. Genet., 2021, 108, 764–785 CrossRef CAS PubMed.
- S. M. Mirkin, Curr. Opin. Struct. Biol., 2006, 16, 351–358 CrossRef CAS PubMed.
- A. Crook, A. McEwen, J. A. Fifita, K. Zhang, J. B. Kwok, G. Halliday, I. P. Blair and D. B. Rowe, Amyotrophic Lateral Scler. Frontotemporal Degener., 2019, 20, 310–316 CrossRef CAS PubMed.
- Y. Sun, X. He, J. Ji, M. Jia, Z. Wang and X. Sun, Talanta, 2015, 141, 300–306 CrossRef CAS PubMed.
- N. Asefifeyzabadi, R. Alkhaldi, A. Z. Qamar, A. A. Pater, M. Patwardhan, K. T. Gagnon, S. Talapatra and M. H. Shamsi, ACS Appl. Mater. Interfaces, 2020, 12, 52156–52165 CrossRef CAS PubMed.
- Y. L. Liu, J. Li, G. Chang, R. Z. Zhu, H. P. He, X. H. Zhang and S. F. Wang, New J. Chem., 2018, 42, 9757–9763 RSC.
- X. Q. Zhu, J. Li, H. H. Lv, H. P. He, H. Liu, X. H. Zhang and S. F. Wang, RSC Adv., 2017, 7, 36124–36131 RSC.
- J. Li, Y. Liu, X. Zhu, G. Chang, H. He, X. Zhang and S. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 44231–44240 CrossRef CAS PubMed.
- M. Fojta, L. Havran, M. Vojtiskova and E. Palecek, J. Am. Chem. Soc., 2004, 126, 6532–6533 CrossRef CAS PubMed.
- F. Miroslav, B. Petra, C. Kateřina and P. Petr, Electroanalysis, 2006, 18, 141–151 CrossRef.
- I. V. Yang and H. H. Thorp, Anal. Chem., 2001, 73, 5316–5322 CrossRef CAS PubMed.
- M. Taki, K. J. Rohilla, M. Barton, M. Funneman, N. Benzabeh, S. Naphade, L. M. Ellerby, K. T. Gagnon and M. H. Shamsi, Anal. Bioanal. Chem., 2019, 411, 6995–7003 CrossRef CAS PubMed.
- F. Lucarelli, G. Marrazza, A. P. F. Turner and M. Mascini, Biosens. Bioelectron., 2004, 19, 515–530 CrossRef CAS PubMed.
- J. I. A. Rashid and N. A. Yusof, Sens. Bio-Sens. Res., 2017, 16, 19–31 CrossRef.
- C. Briones and M. Moreno, Anal. Bioanal. Chem., 2012, 402, 3071–3089 CrossRef CAS PubMed.
- S. Barluenga and N. Winssinger, Acc. Chem. Res., 2015, 48, 1319–1331 CrossRef CAS PubMed.
- J. B. Raoof, R. Ojani, S. M. Golabi, E. Hamidi-Asl and M. S. Hejazi, Sens. Actuators, B, 2011, 157, 195–201 CrossRef CAS.
- M. S. Hejazi, M. H. Pournaghi-Azar, E. Alipour, E. D. Abdolahinia, S. Arami and H. Navvah, Electroanal, 2011, 23, 503–511 CrossRef CAS.
- K. Bartold, A. Pietrzyk-Le, K. Golebiewska, W. Lisowski, S. Cauteruccio, E. Licandro, F. D'Souza and W. Kutner, ACS Appl. Mater. Interfaces, 2018, 10, 27562–27569 CrossRef CAS PubMed.
- L. M. Ellerby, Neurotherapeutics, 2019, 16, 924–927 CrossRef PubMed.
- M. H. Shamsi and H.-B. Kraatz, Analyst, 2010, 135, 2280–2285 RSC.
- A. Z. Qamar, N. Asefifeyzabadi, M. Taki, S. Naphade, L. M. Ellerby and M. H. Shamsi, J. Mater. Chem. B, 2020, 8, 743–751 RSC.
- M. C. An, N. Zhang, G. Scott, D. Montoro, T. Wittkop, S. Mooney, S. Melov and L. M. Ellerby, Cell Stem Cell, 2012, 11, 253–263 CrossRef CAS PubMed.
- M. H. Shamsi and H.-B. Kraatz, Analyst, 2011, 136, 3107–3112 RSC.
- N. Zhang, B. J. Bailus, K. L. Ring and L. M. Ellerby, Brain Res., 2016, 1638, 42–56 CrossRef CAS PubMed.
- L. Kinkar, N. D. Young, W.-M. Sohn, A. J. Stroehlein, P. K. Korhonen and R. B. Gasser, PLoS Neglected Trop. Dis., 2020, 14, e0008552 CrossRef CAS PubMed.
- Nanopore sequencing 101 Q&As: 1. How many times can you reuse a flow cell?, https://nanoporetech.com/events/nanopore-sequencing-101-online-event-qa, 2021 Search PubMed.
- S. Ardui, A. Ameur, J. R. Vermeesch and M. S. Hestand, Nucleic Acids Res., 2018, 46, 2159–2168 CrossRef CAS PubMed.
- H. T. Orr and H. Y. Zoghbi, Annu. Rev. Neurosci., 2007, 30, 575–621 CrossRef CAS PubMed.
- M. van Blitterswijk, M. DeJesus-Hernandez and R. Rademakers, Curr. Opin. Neurol., 2012, 25, 689–700 CrossRef CAS PubMed.
- X. Zhao and K. Usdin, DNA Repair, 2015, 32, 96–105 CrossRef CAS PubMed.
- L. J. Chen, A. Hadd, S. Sah, S. Filipovic-Sadic, J. Krosting, E. Sekinger, R. Q. Pan, P. J. Hagerman, T. T. Stenzel, F. Tassone and G. J. Latham, J Mol Diagn, 2010, 12, 589–600 CrossRef CAS PubMed.
- E. W. Loomis, J. S. Eid, P. Peluso, J. Yin, L. Hickey, D. Rank, S. McCalmon, R. J. Hagerman, F. Tassone and P. J. Hagerman, Genome Res., 2013, 23, 121–128 CrossRef CAS PubMed.
- G. Narzisi and M. C. Schatz, Front. Bioeng. Biotechnol., 2015, 3, 8 Search PubMed.
- C. Akimoto, A. E. Volk, M. van Blitterswijk, M. Van den Broeck, C. S. Leblond, S. Lumbroso, W. Camu, B. Neitzel, O. Onodera, W. van Rheenen, S. Pinto, M. Weber, B. Smith, M. Proven, K. Talbot, P. Keagle, A. Chesi, A. Ratti, J. van der Zee, H. Alstermark, A. Birve, D. Calini, A. Nordin, D. C. Tradowsky, W. Just, H. Daoud, S. Angerbauer, M. DeJesus-Hernandez, T. Konno, A. Lloyd-Jani, M. de Carvalho, K. Mouzat, J. E. Landers, J. H. Veldink, V. Silani, A. D. Gitler, C. E. Shaw, G. A. Rouleau, L. H. van den Berg, C. Van Broeckhoven, R. Rademakers, P. M. Andersen and C. Kubisch, J. Med. Genet., 2014, 51, 419–424 CrossRef CAS PubMed.
- V. L. Buchman, J. Cooper-Knock, N. Connor-Robson, A. Higginbottom, J. Kirby, O. D. Razinskaya, N. Ninkina and P. J. Shaw, Mol. Neurodegener., 2013, 8, 12 CrossRef CAS PubMed.
- D. G. Rackus, M. H. Shamsi and A. R. Wheeler, Chem. Soc. Rev., 2015, 44, 5320–5340 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00230b |
| This journal is © The Royal Society of Chemistry 2022 |