Open Access Article
Open Access ArticleDeactivation of a dimeric DNA-binding peptide through a palladium-mediated self-immolative cleavage†
Jessica Rodríguez‡
 ,
Cibrán Pérez-González‡
,
Cibrán Pérez-González‡ ,
Miguel Martínez-Calvo
,
Miguel Martínez-Calvo ,
Jesús Mosquera and
José L. Mascareñas
,
Jesús Mosquera and
José L. Mascareñas *
*
Centro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CIQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, Rúa Jenaro de la Fuente s/n, Santiago de Compostela, 15782, Spain. E-mail: joseluis.mascarenas@usc.es
First published on 26th January 2022
Abstract
Herein, we describe an approach for the on-demand disassembly of dimeric peptides using a palladium-mediated cleavage of a designed self-immolative linker. The utility of the strategy is demonstrated for the case of dimeric basic regions of bZIP transcription factors. While the dimer binds designed DNA sequences with good affinities, the peptide–DNA complex can be readily dismounted by addition of palladium reagents that trigger the cleavage of the spacer, and the release of unfunctional monomeric peptides.
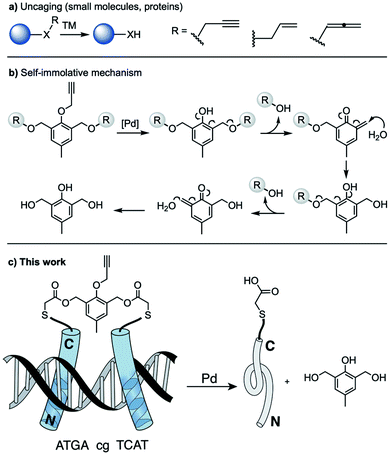
In recent years, there has been a lot of progress in the development of uncaging reactions that release active drugs or biopolymers from inactive precursors.1 The strategy lies on a temporary blocking of a key functional group of an active compound with a caging/protecting group, protection that cancels its biological function. The activity can be eventually recovered by removing the caging pendant, a process that is usually achieved upon application of a light pulse,2 a chemical trigger3 or a metal-based reagent.4 This latter strategy has gained relevance in recent years, owing to its intrinsic catalytic potential and the variety of mechanisms offered by organometallic chemistry. Many of the biological applications of transition metal-mediated bioorthogonal uncaging reactions have relied on the use of allyl or propargyl derivatives as protecting groups, and ruthenium or palladium catalysts (Fig. 1a).5 In particular, palladium-mediated depropargylation reactions have been extensively used for “gain of function” processes based on the release of bioactive products containing amine or alcohol functionalities.6 Surprisingly, metal-promoted processes that lead to “loss of function” properties, which could find important applications in biosupramolecular chemistry, cell biology and/or biomedicine, are essentially unknown.7
Considering our previous work on the deactivation of dimeric DNA-binding peptides using light,8 we wondered whether it would be possible to cleave this type of dimers using a metal-mediated protocol. This would be relevant not only in terms of developing a new methodology to regulate DNA binding processes, but also because of proposing a new type of uncaging approach based on the cleavage of a linker rather than on the release of a caging group. Although there are some precedents on the uncaging of temporary linked systems, they have been essentially centered on the dual release of two active molecules, rather than on the deactivation of biorelevant processes.9
Using as reference a report describing the viability of cleaving benzylic alcohols in ortho to arylether derivatives,10 we reasoned that introducing a propargylic protecting group in this type of phenols might provide for a Pd-mediated release of the pendants (Fig. 1b). Herein we demonstrate the viability of this approach, by describing the synthesis of DNA-binding bZIP-based peptides dimerized through a phenol-like linker that undergoes a self-immolative cleavage after treatment with a palladium reagent. This process results in the release of monomeric peptides and suppression of the DNA binding (Fig. 1c).
Basic leucine zippers (bZIP) are among the most important families of transcription factors (TFs). They are formed by an N-terminal basic region (br) that makes specific interactions with the major groove of the DNA, and a C-terminal leucine zipper domain that mediates dimerization.11 Monomeric basic regions are not able to bind their consensus DNA sites with enough affinity, unless they are connected to other DNA binders,12 or arranged into pre-folded constructs.13 Alternatively, the DNA binding can be rescued when the basic region peptides are engineered as dimeric systems using suitable synthetic connectors.14
To implement our on–off DNA binding system, we selected the GCN4 transcription factor, one of the best-characterized bZIP proteins, which specifically binds to the ATF/CREB (5′-ATGAcgTCAT-3′) or AP1 (5′-ATGA(c/g)TCAT-3′) consensus sequences.15 The DNA-binding peptide fragment (br) was chosen based on previous reports that determined the smallest fragment of the GCN4 basic region that preserves the DNA binding attributes of the full protein when engineered as a disulfide dimer.16
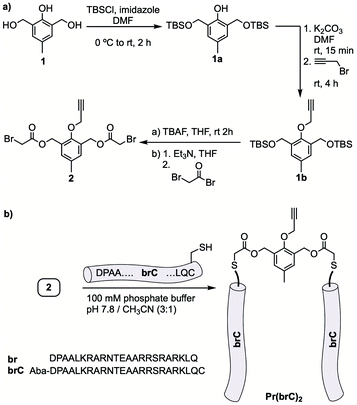
Considering the versatility and wide applicability of cysteine alkylations for the selective bioconjugation of peptides and proteins,17 we selected peptide fragments with a C-terminal cysteine unit. As aryl-containing linking moiety we synthesized product 2, featuring thiol-reactive bromoacetyl units (Fig. 2a). The commercially available precursor 2,6-bis(hydroxymethyl)-p-cresol 1 was first protected as bis-TBS derivative. After reaction of the resulting phenol with propargyl bromide to afford 1b, the TBS group was removed and the diol bis-acylated to give the desired dibromide (overall yield of the synthetic process: 22%).
The chemoselective coupling between this bis-electrophilic linker and the cysteine-containing peptide brC, was carried out in 100 mM aqueous phosphate buffer (pH 7.8) containing 25% of CH3CN, using 2.1 equiv. of the peptide. HPLC-MS analysis of the reaction showed the complete disappearance of the dibromide after 1.5 h at rt, and the formation of a new major species identified as the expected dimeric peptide (Fig. S10†).
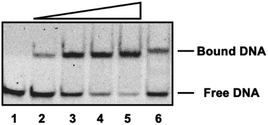
The DNA binding of the resulting dimeric construct Pr(brC)2 was first analyzed by electrophoretic mobility shift assays (EMSA) under non-denaturing conditions18 and using SYBR gold for DNA staining.19 Thus, when a double stranded oligonucleotide containing the target ATF/CREB site (ATF/CREB) was incubated with increasing concentrations of Pr(brC)2 in Tris–HCl buffer at 20 °C, we observed the appearance of a new retarded band consistent with the formation of a specific DNA/Pr(brC)2 complex (Fig. 3). The shift of this band is in line with that observed when the same oligonucleotide is incubated with the reference disulfide dimer (brGCC)2 (Fig. 3, lane 6).14 A control EMSA experiment with an oligonucleotide containing a random sequence does not show retarded bands, thus confirming that Pr(brC)2 is a highly selective DNA binder (Fig. S14†). Fluorescence anisotropy titrations using a FAM-labeled ATF/CREB dsDNA confirmed the high affinity interaction of the peptide dimer Pr(brC)2 with the ds-oligonucleotide, with an apparent KD of 321 nM at 20 °C (Fig. S16†).
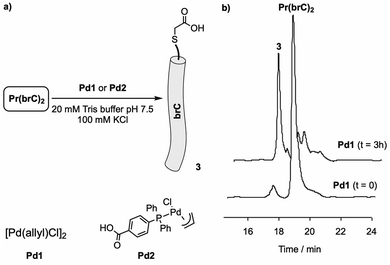
We next studied the cleavage of Pr(brC)2 in the absence of DNA. As palladium reagents we selected commercially available complex [Pd(allyl)Cl]2 (Pd1) and pi-allyl derivative Pd2, both previously shown to mediate depropargylation reactions in water, and even in complex biological settings.6,20 To our delight, incubation of a solution of Pr(brC)2 (10 μM) in 20 mM Tris buffer (pH 7.5) with either Pd1 or Pd2 (100 μM) for 3 h at room temperature led to the disappearance of the starting dimer (peak at 18.7 min, see Fig. 4 and S13,† 81% conversion after 2 h).21 The chromatogram showed a new peak at 17.3 min which was identified as the monomeric fragment 3 by MS (Fig. S12†), which is consistent with the cleavage of the linker and the release of the peptide arms (brC) as terminal acids (Fig. 4).
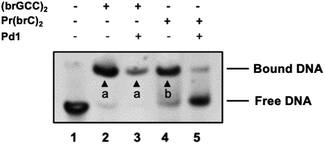
Very importantly, the Pd-promoted process can be performed after formation of the peptide/DNA complex, leading to the expected disassembling. Therefore, when the target DNA (500 nM) was incubated in buffered solution (20 mM Tris buffer pH 7.5, 100 mM KCl) with Pr(brC)2 (5 μM), and the mixture was treated with either Pd1 or Pd2 (50 μM) for 3 h, we could clearly observe by EMSA the disappearance of most of the retarded band corresponding to the Pr(brC)2-DNA complex (band b in Fig. 5, lanes 4–5 and Fig. S15†). A control experiment with the disulfide dimer (brGCC)2 revealed that the DNA binding is retained after incubation with Pd1 or Pd2 (band a in Fig. 5, lanes 2–3 and Fig. S15†), which confirms that the cleavage/disassembly sequence only occurs with our responsive peptide dimer.
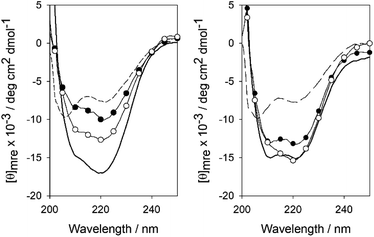
Moreover, in agreement with the EMSA, circular dichroism experiments revealed that addition of 1 equiv. of the dsDNA ATF/CREB to a 5 μM solution of Pr(brC)2 or (brGCC)2 promotes a significant increase in the ellipticity of the negative bands at 208 and 222 nm, consistent with the expected folding of the basic regions into α-helices (Fig. 6).11,14 Treatment of the solution containing the Pr(brC)2–dsDNA complex (5 μM) with Pd1 or Pd2 (50 μM) in 10 mM phosphate buffer (pH 7.5), for 3 h a rt, induced a significant decrease in ellipticity, as expected for single monomers.8a, 13c In contrast, the same addition to the control mixture containing the (brGCC)2 peptide did not promote any change. All these results are in total agreement with a Pd-promoted self-immolative cleavage of the propargylic phenol connector, and subsequent release of the two monomeric basic regions (3), which are not capable to efficiently bind to the ATF/CREB sequence.
In conclusion, we have designed and developed a new palladium-cleavable linker that can work as temporary dimerizing unit. The utility of the strategy was demonstrated for the construction of functional bZIP dimeric peptides which bind the consensus DNA sequences with high affinity. However, the DNA binding can be efficiently suppressed upon incubation with readily available palladium reagents under mild and physiological conditions. Together with its easy synthetic accessibility, these results support the use of the self-immolative linker 2 as an effective dimerizer that can be readily cleaved in aqueous buffers using transition metal reagents. The methodology, which is complementary to other uncaging strategies based on the use of light, should be extendible to switch off other type of functions that depend on the formation of dimers, as well as for releasing active products from inactive bifunctional precursors. Furthermore, the known ability of the palladium catalyst to promote depropargylation reactions in living systems,6a suggest that this type of switches can be translated to in vivo settings.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are thankful for the financial support from the Xunta de Galicia (Centro singular de investigación de Galicia acreditación 2019–2022, ED431G 2019/03) and the European Union (European Regional Development Fund – ERDF). We also thank support given by the Spanish grants PID2019-108624RB-I00, the Xunta de Galicia (grants 2015-CP082, ED431C 2017/19 and 2021-CP054, ED431C-2021/25) and the European Research Council (Advanced Grant No. 340055). MMC and JR thank the MCIN/AEI for their Postdoctoral fellowships (IJCI-2014-19326 and IJC2019-040358-I). Authors would like to thank the use of RIAIDT-USC analytical facilities.Notes and references
- J. Li and P. R. Chen, Development and application of bond cleavage reactions in bioorthogonal chemistry, Nat. Chem. Biol., 2016, 12, 129–137 CrossRef CAS PubMed.
- (a) H. M. Lee, D. R. Larson and D. S. Lawrence, Illuminating the Chemistry of Life: Design, Synthesis, and Applications of “Caged” and Related Photoresponsive Compounds, ACS Chem. Biol., 2009, 4, 409–427 CrossRef CAS PubMed; (b) M. I. Sánchez, O. Vázquez, M. E. Vázquez and J. L. Mascareñas, Light-controlled DNA binding of bisbenzamidines, Chem. Commun., 2011, 47, 11107–11109 RSC; (c) J. Rodríguez, J. Mosquera, J. R. Couceiro, M. E. Vázquez and J. L. Mascareñas, Ruthenation of Non-stacked Guanines in DNA G-Quadruplex Structures: Enhancement of c-MYC Expression, Angew. Chem., Int. Ed., 2016, 55, 15615–15618 CrossRef PubMed; (d) M. Martínez-Calvo, L. Guerrini, J. Rodríguez, R. A. Álvarez-Puebla and J. L. Mascareñas, Surface-Enhanced Raman Scattering Detection of Nucleic Acids Exhibiting Sterically Accessible Guanines Using Ruthenium-Polypyridyl Reagents, J. Phys. Chem. Lett., 2020, 11, 7218–7223 CrossRef PubMed.
- (a) J. Li, S. Jia and P. R. Chen, Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells, Nat. Chem. Biol., 2014, 10, 1003–1005 CrossRef CAS PubMed; (b) B. L. Oliveira, Z. Guo and G. J. L. Bernardes, Inverse electron demand Diels–Alder reactions in chemical biology, Chem. Soc. Rev., 2017, 46, 4895–4950 RSC.
- (a) J. Rodríguez and M. Martínez-Calvo, Transition-Metal-Mediated Modification of Biomolecules, Chem. –Eur. J., 2020, 26, 9792–9813 CrossRef PubMed; (b) P. Destito, C. Vidal, F. López and J. L. Mascareñas, Transition Metal-Promoted Reactions in Aqueous Media and Biological Settings, Chem. –Eur. J., 2021, 27, 4789–4816 CrossRef CAS PubMed; (c) M. van de L'Isle, M. C. Ortega-Liebana and A. Unciti-Broceta, Transition metal catalysts for the bioorthogonal synthesis of bioactive agents, Curr. Opin. Chem. Biol., 2021, 61, 32–42 CrossRef PubMed.
- (a) C. Streu and E. Meggers, Ruthenium-Induced Allylcarbamate Cleavage in Living Cells, Angew. Chem., Int. Ed., 2006, 45, 5645–5648 CrossRef CAS PubMed; (b) G. Y. Tonga, Y. Jeong, B. Duncan, T. Mizuhara, R. Mout, R. Das, S. T. Kim, Y.-C. Yeh, B. Yan, S. Hou and V. M. Rotello, Supramolecular regulation of bioorthogonal catalysis in cells using nanoparticle-embedded transition metal catalysts, Nat. Chem., 2015, 7, 597–603 CrossRef CAS PubMed; (c) S. Learte-Aymami, C. Vidal, A. Gutierrez-Gonzalez and J. L. Mascareñas, Intracellular Reactions Promoted by Bis(histidine) Miniproteins Stapled Using Palladium(II) Complexes, Angew. Chem., Int. Ed., 2020, 59, 9149–9154 CrossRef CAS PubMed; (d) E. Latocheski, G. M. Dal Forno, T. M. Ferreira, B. L. Oliveira, G. J. L. Bernardes and J. B. Domingos, Mechanistic insights into transition metal-mediated bioorthogonal uncaging reactions, Chem. Soc. Rev., 2020, 49, 7710–7729 RSC; (e) J. Wang, S. Zheng, Y. Liu, Z. Zhang, Z. Lin, J. Li, G. Zhang, X. Wang, J. Li and P. R. Chen, Palladium-Triggered Chemical Rescue of Intracellular Proteins via Genetically Encoded Allene-Caged Tyrosine, J. Am. Chem. Soc., 2016, 138, 15118–15121 CrossRef CAS PubMed.
- (a) M. Martínez-Calvo, J. R. Couceiro, P. Destito, J. Rodriguez, J. Mosquera and J. L. Mascareñas, Intracellular Deprotection Reactions Mediated by Palladium Complexes Equipped with Designed Phosphine Ligands, ACS Catal., 2018, 8, 6055–6061 CrossRef PubMed; (b) P. Destito, A. Sousa-Castillo, J. R. Couceiro, F. López, M. A. Correa and J. L. Mascareñas, Hollow nanoreactors for Pd-catalyzed Suzuki–Miyaura coupling and O-propargyl cleavage reactions in bio-relevant aqueous media, Chem. Sci., 2019, 10, 2598–2603 RSC.
- Most deactivation approaches involve light-induced processes, see: (a) B. N. Goguen, A. Aemissegger and B. Imperiali, Sequential Activation and Deactivation of Protein Function Using Spectrally Differentiated Caged Phosphoamino Acids, J. Am. Chem. Soc., 2011, 133, 11038–11041 CrossRef CAS PubMed; (b) J. M. Govan, R. Uprety, J. Hemphill, M. O. Lively and A. Deiters, Regulation of Transcription through Light-Activation and Light-Deactivation of Triplex-Forming Oligonucleotides in Mammalian Cells, ACS Chem. Biol., 2012, 7, 1247–1256 CrossRef CAS PubMed; (c) D. D. Young, M. O. Lively and A. Deiters, Activation and Deactivation of DNAzyme and Antisense Function with Light for the Photochemical Regulation of Gene Expression in Mammalian Cells, J. Am. Chem. Soc., 2010, 132, 6183–6193 CrossRef CAS PubMed.
- (a) J. Mosquera, M. I. Sanchez, M. E. Vázquez and J. L. Mascareñas, Ruthenium bipyridyl complexes as photocleavable dimerizes: deactivation of DNA-binding peptides using visible light, Chem. Commun., 2014, 50, 10975–10978 RSC; (b) J. Rodríguez, J. Mosquera, S. Learte-Aymamí, M. E. Vázquez and J. L. Mascareñas, Stimuli-Responsive DNA Binding by Synthetic Systems, Acc. Chem. Res., 2020, 53, 2286–2298 CrossRef PubMed.
- (a) R. Friedman Ohana, S. Levin, M. G. Wood, K. Zimmerman, M. L. Dart, M. K. Schwinn, T. A. Kirkland, R. Hurst, H. T. Uyeda, L. P. Encell and K. V. Wood, Improved Deconvolution of Protein Targets for Bioactive Compounds Using a Palladium Cleavable Chloroalkane Capture Tag, ACS Chem. Biol., 2016, 11, 2608–2617 CrossRef CAS PubMed; (b) B. J. Stenton, B. L. Oliveira, M. J. Matos, L. Sinatra and G. J. L. Bernardes, A thioether-directed palladium-cleavable linker for targeted bioorthogonal drug decaging, Chem. Sci., 2018, 9, 4185–4189 RSC; (c) X. Wang, Y. Liu, X. Fan, J. Wang, W. S. C. Ngai, H. Zhang, J. Li, G. Zhang, J. Lin and P. R. Chen, Copper-Triggered Bioorthogonal Cleavage Reactions for Reversible Protein and Cell Surface Modifications, J. Am. Chem. Soc., 2019, 141, 17133–17141 CrossRef CAS PubMed; (d) B. L. Oliveira, B. J. Stenton, V. B. Unnikrishnan, C. R. de Almeida, J. Conde, M. Negrão, F. S. S. Schneider, C. Cordeiro, M. G. Ferreira, G. F. Caramori, J. B. Domingos, R. Fior and G. J. L. Bernardes, Platinum-Triggered Bond-Cleavage of Pentynoyl Amide and N-Propargyl Handles for Drug-Activation, J. Am. Chem. Soc., 2020, 142, 10869–10880 CrossRef CAS PubMed.
- C.-C. Song, R. Ji, F.-S. Du and Z.-C. Li, Oxidation-Responsive Poly(amino ester)s Containing Arylboronic Ester and Self-Immolative Motif: Synthesis and Degradation Study, Macromolecules, 2013, 46(21), 8416–8425 CrossRef CAS.
- T. E. Ellenberger, C. J. Brandl, D. Struhl and S. C. Harrison, The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex, Cell, 1992, 71, 1223 CrossRef CAS PubMed.
- (a) J. Rodríguez, J. Mosquera, J. R. Couceiro, M. E. Vázquez and J. L. Mascareñas, The AT-Hook motif as a versatile minor groove anchor for promoting DNA binding of transcription factor fragments, Chem. Sci., 2015, 6, 4767–4771 RSC; (b) J. Rodríguez, J. Mosquera, R. García-Fandiño, M. E. Vázquez and J. L. Mascareñas, A designed DNA binding motif that recognizes extended sites and spans two adjacent major grooves, Chem. Sci., 2016, 7, 3298–3303 RSC; (c) J. Rodríguez, J. Mosquera, M. E. Vázquez and J. L. Mascareñas, Nickel-Promoted Recognition of Long DNA Sites by Designed Peptide Derivatives, Chem. –Eur. J., 2016, 22, 13474–13477 CrossRef PubMed; (d) S. Learte-Aymamí, J. Rodríguez, M. E. Vázquez and J. L. Mascareñas, Metal-Dependent DNA Recognition and Cell Internalization of Designed, Basic Peptides, Chem. –Eur. J., 2020, 26, 8875–8878 CrossRef PubMed; (e) O. Vázquez, M. E. Vázquez, J. B. Blanco, L. Castedo and J. L. Mascareñas, Specific DNA recognition by a synthetic, monomeric Cys2His2 zinc finger peptide conjugated to a minor groove binder, Angew. Chem., Int. Ed., 2007, 46, 6886–6890 CrossRef PubMed; (f) M. E. Vázquez, A. M. Caamaño, J. Martínez-Costas, L. Castedo and J. L. Mascareñas, Design and synthesis of a peptide that binds specific DNA sequences through simultaneous Interaction in the major and in the minor groove, Angew. Chem., Int. Ed., 2001, 40, 4723–4725 CrossRef.
- (a) J. W. Chin and A. Schepartz, Concerted evolution of structure and function in a miniature protein, J. Am. Chem. Soc., 2001, 123, 2929–2930 CrossRef CAS PubMed; (b) J. K. Montclare and A. Schepartz, Miniature Homeodomains:High Specificity without an N -TerminalArm, J. Am. Chem. Soc., 2003, 125, 3416–3417 CrossRef CAS PubMed; (c) S. Learte-Aymamí, N. Curado, J. Rodríguez, M. E. Vázquez and J. L. Mascareñas, Metal-Dependent DNA Recognition and Cell Internalization of Designed, Basic Peptides, J. Am. Chem. Soc., 2017, 139, 16188–16193 CrossRef PubMed.
- (a) R. V. Talanian, C. J. McKnight, R. Rutkowski and P. S. Kim, Minimum length of a sequence-specific DNA binding peptide, Biochemistry, 1992, 31, 6871 CrossRef CAS PubMed; (b) M. Ueno, A. Murakami, K. Makino and T. Morii, Arranging quaternary structure of peptides by cyclodextrin-guest inclusion complex: sequence-specific DNA binding by a peptide dimer with artificial dimerization module, J. Am. Chem. Soc., 1993, 115, 12575 CrossRef CAS; (c) B. Cuenoud and A. Schepartz, Altered Specificity of DNA-Binding Proteins with Transition Metal Dimerization Domains, Science, 1993, 259, 510 CrossRef CAS PubMed; (d) T. Morii, M. Shimomura, S. Morimoto and I. Saito, Sequence-specific DNA binding by a geometrically constrained peptide dimer, J. Am. Chem. Soc., 1993, 115, 1150 CrossRef CAS; (e) J. Mosquera, A. Jiménez-Balsa, V. I. Dodero, M. E. Vázquez and J. L. Mascareñas, Stimuli-responsive selection of target DNA sequences by synthetic bZIP peptides, Nat. Commun., 2013, 4, 1874 CrossRef PubMed.
- (a) W. Keller, P. Kö and T. J. Richmond, Crystal Structure of a bZIP/DNA Complex at 2.2 Å: Determinants of DNA Specific Recognition, J. Mol. Biol, 1995, 254, 657–667 Search PubMed; (b) P. König and T. J. Richmond, The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility, J. Mol. Biol., 1993, 233, 139–154 CrossRef PubMed.
- R. V. Talanian, C. J. McKnight and P. S. Kim, Sequence-Specific DNA Binding by a Short Peptide Dimer, Science, 1990, 249, 769–771 CrossRef CAS PubMed.
- (a) J. Rodríguez, J. Mosquera, O. Vázquez, M. E. Vázquez and J. L. Mascareñas, The ββα fold of zinc finger proteins as a “natural” protecting group. Chemoselective synthesis of a DNA-binding zinc finger derivative, Chem. Commun., 2014, 50, 2258 RSC; (b) S. B. Gunnoo and A. Madder, Chemical Protein Modification through Cysteine, ChemBioChem, 2016, 17, 529–553 CrossRef CAS PubMed.
- L. M. Hellman and M. G. Fried, Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions, Nat. Protoc., 2007, 2, 1849 CrossRef CAS PubMed.
- R. S. Tuma, M. P. Beaudet, X. Jin, L. J. Jones, C.-Y. Cheung, S. Yue and V. L. Singer, Characterization of SYBR Gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators, Anal. Biochem., 1999, 268, 278–288 CrossRef CAS PubMed.
- J. Li, J. Yu, J. Zhao, J. Wang, S. Zheng, S. Lin, L. Chen, M. Yang, S. Jia, X. Zhang and P. R. Chen, Palladium-triggered deprotection chemistry for protein activation in living cells, Nat. Chem., 2014, 6, 352–361 CrossRef CAS PubMed.
- We tested different Pd/dimer molar ratios for the cleavage reaction, and concluded that a 10-fold molar excess of Pd complex over the dimer leads to the best performance (see Fig. S11†). 10 eq. of Pd is the amount commonly used in depropargylation reactions in water, see for example ref. 9b and 20.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra09180h |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |