Open Access Article
Open Access ArticleNosZ gene cloning, reduction performance and structure of Pseudomonas citronellolis WXP-4 nitrous oxide reductase†
Liyong Hu a,
Xiaoping Wanga,
Cong Chena,
Jianmeng Chena,
Zeyu Wangb,
Jun Chen*b,
Dzmitry Hrynshpanc and
Tatsiana Savitskayac
a,
Xiaoping Wanga,
Cong Chena,
Jianmeng Chena,
Zeyu Wangb,
Jun Chen*b,
Dzmitry Hrynshpanc and
Tatsiana Savitskayac
aCollege of Environment, Zhejiang University of Technology, Hangzhou, 310014, China
bKey Laboratory of Pollution Exposure and Health Intervention of Zhejiang Province, Interdisciplinary Research Academy, Zhejiang Shuren University, Hangzhou, 310015, China. E-mail: bec@zjut.edu.cn; Tel: +86-571-88285209
cResearch Institute of Physical and Chemical Problems, Belarusian State University, Minsk, 220030, Belarus
First published on 19th January 2022
Abstract
Nitrous oxide reductase (N2OR) is the only known enzyme that can reduce the powerful greenhouse gas nitrous oxide (N2O) to harmless nitrogen at the final step of bacterial denitrification. To alleviate the N2O emission, emerging approaches aim at microbiome biotechnology. In this study, the genome sequence of facultative anaerobic bacteria Pseudomonas citronellolis WXP-4, which efficiently degrades N2O, was obtained by de novo sequencing for the first time, and then, four key reductase structure coding genes related to complete denitrification were identified. The single structural encoding gene nosZ with a length of 1914 bp from strain WXP-4 was cloned in Escherichia coli BL21(DE3), and the N2OR protein (76 kDa) was relatively highly efficiently expressed under the optimal inducing conditions of 1.0 mM IPTG, 5 h, and 30 °C. Denitrification experiment results confirmed that recombinant E. coli had strong denitrification ability and reduced 10 mg L−1 of N2O to N2 within 15 h under the optimal conditions of pH 7.0 and 40 °C, its corresponding N2O reduction rate was almost 2.3 times that of Alcaligenes denitrificans strain TB, but only 80% of that of wild strain WXP-4, meaning that nos gene cluster auxiliary gene deletion decreased the activity of N2OR. The 3D structure of N2OR predicted on the basis of sequence homology found that electron transfer center CuA had only five amino acid ligands, and the S2 of the catalytically active center CuZ only bound one CuI atom. The unique 3D structure was different from previous reports and may be closely related to the strong N2O reduction ability of strain WXP-4 and recombinant E. coli. The findings show a potential application of recombinant E. coli in alleviating the greenhouse effect and provide a new perspective for researching the relationship between structure and function of N2OR.
Introduction
Nitrous oxide (N2O) is an inert, odorless and non-toxic gas that in particular acts as a powerful greenhouse gas and a significant depleter of ozone. Its global warming potential is more than 300 times that of carbon dioxide (CO2).1,2 N2O is widely emitted due to fuel combustion, nitrogenous fertilizer utilization, and wastewater treatment, and can exist stably in the atmosphere for more than 100 years and its atmospheric concentration level is growing steadily at a rate of 0.2–0.3% per year.3–5 So, the development of N2O control technology is becoming increasingly important.Biodegradation has attracted much attention due to its advantages, such as mild conditions, low cost, and no secondary pollution.6,7 More than two-thirds of the global N2O emissions originate from the soil ecosphere and hydrosphere, and can be reduced to harmless nitrogen gas (N2) at the final step of the microbial denitrification pathway.8–10 Nitrous oxide reductase (N2OR) is the only enzyme that performs the biological denitrification process.11,12 Thus, the efficient utilization of N2OR is essential to the effective control of N2O emission through biological methods.
N2OR is a periplasmic multi-copper enzyme and a head-to-tail homodimer. Each monomer includes two domains: an electron transfer binuclear CuA center at C-terminal and a catalytic tetranuclear CuZ center at N-terminal.13,14 Generally, CuA is liganded by six amino acid residues comprising one methionine, one tryptophan, two cysteines and two histidines; CuZ is coordinated by seven histidines.15,16 On the base of the 3D structure of N2OR, the consensus view of N2O catalytic reduction mechanism is that N2O binds to the catalytically active site of CuZ, then the electron is transferred from CuA to convert N2O into N2.
N2OR is encoded by the nos gene cluster (nosRZDFYL), in which nosZ gene is the structural gene.17 In previous studies, the nosZ gene from various microbial groups has been successfully cloned and expressed in different hosts. Wan et al. reported that the transfer of nosZ gene into tobacco plants resulted in higher N2OR activity and decreased N2O release capacity.18 Jones et al. found that when there were two copies of nosZ in bacillus cells, almost no N2O was produced.19 These results indicate that recombinant N2OR plays an important role in reducing N2O emissions. However, there are few systematic studies on the cloning and heterologous expression of nosZ gene, and its utilization needs to be further excavated.
Recently, a novel facultative anaerobic denitrifying bacteria showing high efficiency N2O degradation, namely, P. citronellolis WXP-4, was isolated from the Qige sewage treatment plant (Hangzhou, China).20 Its reduction efficiency is 100% at an initial N2O concentration of 50 mg L−1 within 2 d (ESI Fig. S1†). Thus, it was intended to deeply understand P. citronellolis WXP-4. The aims of this study are as follows: (1) sequence the whole-genome of strain WXP-4 and identify the key denitrification genes; (2) clone the nosZ gene from strain WXP-4 in E. coli and investigate the N2O removal performance of recombinant E. coli; (3) preliminarily explore the relation between the structure and function of N2OR.
Materials and methods
Materials
P. citronellolis WXP-4 (CCTCC NO. M2018526) was isolated from the wastewater of Qige Sewage Treatment Plant (Hangzhou, China). E. coli BL21(DE3), plasmids pGEM-T and pET28a were purchased from TaKaRa Biotechnology Co., Ltd. PCR primer was purchased from Sangon Biotech (Shanghai, China). EcoR I and Xho I were purchased from Aoqian Biotech Co., Ltd (Hangzhou, China).P. citronellolis WXP-4 and E. coli were grown in a sealed shake flask with LB medium comprising the following (per liter): NaCl (10.0 g), yeast extract (5.0 g), and peptone (10.0 g). The internal space of the sealed shake flask was guaranteed to be completely anaerobic by continuous rinsing with argon gas for 20 min through a sterile thin tube, which was inserted into the bottom of the culture medium. The medium used for N2O degradation performance test was inorganic salt medium without nitrogen source as follows (per liter): sodium succinate (2.36 g), KH2PO4 (1.5 g), Na2HPO4·12H2O (7.715 g), MgSO4·7H2O (0.1 g), agar (20.0 g), and trace element solution (2 mL). The trace element solution comprised the following (per liter): EDTA (5.0 g), FeSO4·7H2O (0.5 g), ZnSO4 (0.43 g), CuSO4·5H2O (0.25 g), CaCl2 (5.5 g), CoCl2·6H2O (0.24 g), MnCl2·4H2O (0.99 g), and H3BO4 (0.014 g). The rotation speed of the sealed shake flask was 160 rpm, the initial pH value of LB medium was set at 7.0, and the initial pH value of the inorganic salt medium without nitrogen source was the natural pH (∼7.5).
The GenBank accession number for the genome sequence of strain P. citronellolis WXP-4 was CP034688 (these data were to be held confidential until: Dec 24, 2022).
De novo sequencing and coding genes prediction of P. citronellolis WXP-4
The genomic DNA of P. citronellolis WXP-4 was extracted by Gentra Puregene Yeast/Bact. Kit (Qiagen, Valencia, CA), and the nanopore library was constructed by using a nanopore sequencing kit. Afterward, second-generation sequencing was conducted by using Illumina high-throughput sequencer, and the third-generation real-time sequencing (single molecule sequencing) was conducted by using GridION X5 sequencer. The genome was assembled by the Unicycler (v0.4.5) software. The genome circle graph of P. citronellolis WXP-4 was drawn by using the online software Circos (v0.69) (http://www.circos.ca/software/download/circos/).The genome annotation was performed by using the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) system. The open reading frame (ORF) prediction was performed by using Prodigal gene-finding software v2.6.3.21 Coding gene's function annotation was aligned with the databases of non-redundant protein (Nr), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups (COG) by BLAST. Metabolic pathways were annotated by KEGG. The proteins encoded by genes were classified via phylogenetic classification by using COG. The composition, structure and location information of denitrification gene clusters in P. citronellolis WXP-4 were analyzed according to the annotation results. The gene structure map was drawn by using the SVG method.
TA cloning of nosZ gene
The genomic DNA of P. citronellolis WXP-4 was used as a DNA template for PCR. PCR was performed in a Mastercycler gradient (Bio-Rad, USA) by using the specific primers designed according to the nosZ gene sequence from strain WXP-4, as follows: nosZ-F (5′-GAATTCATGAACGAGAAGAAACACCCGCAGCCCGAAGACG-3′) and nosZ-R (5′-CTCGAGGGCCTTTTCCACCAGCATGCGGCCGACCATTTCC-3′). The PCR reaction system and reaction cycles refer to the method of Chen et al.22 The PCR reaction system (50 μL) consisted of 25 μL of 2×Phanta Max Buffer, 1 μL of dNTPs Mix (10 μM), 2 μL of nosZ-F (10 μM), 2 μL of nosZ-R (10 μM), 1 μL of Phanta polymerase, 2 μL of template DNA, and 17 μL of double distilled water. The mixture was pre-denatured at 95 °C for 3 min. Then, it was denaturation at 95 °C for 0.5 min, annealed at 55 °C for 0.5 min, and extended at 72 °C for 1.5 min for a total of 33 cycles. Finally, extension was performed at 72 °C for 5 min. The PCR products obtained by SanPrep Rapid Gel DNA Recovery kit were added through “A” tail treatment using flat terminal DNA Fragment Plus “A” kit. Then, they were purified by SanPrep PCR and DNA Fragment Purification kit and ligated into pGEM-T plasmid to form a TA clone at 4 °C overnight. The TA clone was transformed into E. coli DH5α and conformed by colony PCR and restriction enzyme digestion. The positive colony plasmid was extracted by using the SanPrep Rapid Plasmid Extraction kit and sent to Youkang Biotech Co., Ltd (Hangzhou, China) for sequencing.Bioinformatics analysis of nosZ gene and N2OR protein
The sequence similarity of nosZ gene in P. citronellolis WXP-4 was compared with that of other strains by BLAST homology comparison on NCBI. The neighbor joining method in Mega software (v7.0.26) was used to construct the phylogenetic tree. DNAMAN software was used to analyze the length, base composition, and distribution of nosZ gene. The amino acid sequence was translated by gene transcription. The physical and chemical properties of N2OR protein were analyzed by using the ProtParam online website (https://web.expasy.org/protparam/). The basic secondary structure of the protein was analyzed by the predict protein online website (https://www.predictprotein.org/). The Swiss Model online website (https://swissmodel.expasy.org/) was used to predict the 3D structure of the N2OR protein based on the amino acid sequence in the PDB database. A homology model was generated using the N2OR (PDB ID: 3SBP) from P. stutzeri as the template, with which N2OR of strain WXP-4 shared 82.18% sequence identify, and the N2OR protein was edited by using Discovery Studio 4.5 software.Constructure and expression of recombinant E. coli BL21(DE3)-pET28a-nosZ
The pGEM-T-nosZ and pET28a plasmids were respectively extracted from the positive clone colony and E. coli. Then, they were simultaneously digested with EcoR I and Xho I. After the detection of enzyme digestion products by using agarose electrophoresis, the released nosZ gene was ligated into the treated pET28a plasmid at 4 °C overnight to construct the gene expression vector pET28a-nosZ. The constructed expression vector pET28a-nosZ was transformed into E. coli BL21(DE3), which was confirmed by colony PCR and restriction enzyme digestion. The recombinant E. coli BL21(DE3)-pET28a-nosZ was obtained for expression.The recombinant E. coli was incubated in LB liquid medium containing 50 μg mL−1 of kanamycin and then cultured at 37 °C and 160 rpm until the logarithmic growth period. It was induced by IPTG at a final concentration of 1 mM for 5 h at 30 °C and 160 rpm. The total protein of all bacteria was extracted by ultrasonic method (power 400 watts, working for 3 s, intermittently for 2 s, and breaking 99 times) and purified by using His-tagged Protein Purification Kit (soluble protein, ESI S1.1†). SDS-PAGE (ESI S1.2†) was used to determine whether the recombinant N2OR protein was expressed.
N2O reduction performance and its influencing factors of recombinant E. coli
The recombinant E. coli was cultured and induced under the conditions mentioned above, three repeated experiments were set for each bacterial solution. The bacterial cells were harvested by centrifugation at 4 °C and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Then, they were washed twice with 50 mL inorganic salt liquid medium without nitrogen source and transferred to the new 50 mL inorganic salt medium. Pure N2O gas was injected into the headspace of a sealed shake flask with Agilent gas-tight syringe. The final concentrations of N2O inside the shake flask reached 10, 20, 30, 40, and 50 mg L−1. The concentrations of the remaining N2O were measured regularly within 5 d to determine the N2O degradation rate. The effects of factors, such as temperature (30 °C, 35 °C, 40 °C, 45 °C, and 50 °C) and pH (5.0, 6.0, 7.0, 8.0, and 9.0) on the degradation of 10 mg L−1 N2O by recombinant E. coli were investigated. The remaining concentrations of N2O were measured regularly in the shake flask by gas chromatography (Agilent 6890 N, USA).
000 rpm for 5 min. Then, they were washed twice with 50 mL inorganic salt liquid medium without nitrogen source and transferred to the new 50 mL inorganic salt medium. Pure N2O gas was injected into the headspace of a sealed shake flask with Agilent gas-tight syringe. The final concentrations of N2O inside the shake flask reached 10, 20, 30, 40, and 50 mg L−1. The concentrations of the remaining N2O were measured regularly within 5 d to determine the N2O degradation rate. The effects of factors, such as temperature (30 °C, 35 °C, 40 °C, 45 °C, and 50 °C) and pH (5.0, 6.0, 7.0, 8.0, and 9.0) on the degradation of 10 mg L−1 N2O by recombinant E. coli were investigated. The remaining concentrations of N2O were measured regularly in the shake flask by gas chromatography (Agilent 6890 N, USA).
The recombinant E. coli and P. citronellolis WXP-4 were cultured at respectively optimal conditions, then performed as mentioned above and three repeated experiments were set for each bacterial solution. The initial concentration of N2O in the sealed flask was 10 mg L−1. The remaining concentration of N2O was measured regularly in the shake flask by Agilent gas chromatography (Agilent 6890 N, USA).
Analysis methods
Gas samples (250 μL) were regularly collected from the upper space of the shaker by GC-specific pointed gas-tight syringe and detected according to the previous method with slight modification.23 N2O was determined using an Agilent 6890 N GC equipped with an electron capture detector and a column (30 m × 0.32 mm × 0.25 μm) packed with HP-5. The temperatures of the column, injector, and detector were 40 °C, 100 °C, and 300 °C, respectively. The carrier gas was N2 and flowed at a rate of 20 mL min−1. All experiments were performed in triplicate. Data were presented as mean ± standard error and calculated by using the SPSS 13.0 software.Results and discussion
Genome sequence and functional genes annotation of P. citronellolis WXP-4
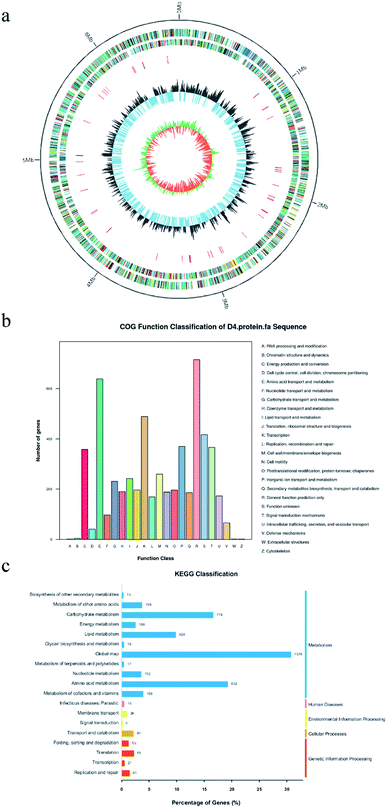
The genomic DNA of P. citronellolis WXP-4 was sequenced by Illumina HiSeq. The sequencing clean data showed that the total genome length of strain WXP-4 was 6672752 bp, and the GC content was 67.40%. Then, the GC skew was calculated, which is the relative content of GC and is a useful tool for marking the start and end points of circular chromosomes. NCBI online database predicted that the genome contained 5757 coding sequences (CDS). Through sequence comparison with the Rfam database, 5756 mRNA, 15 rRNA, 66 tRNA, and 4 ncRNA were found in the genome (Fig. 1a).The coding genes in WXP-4 genome were annotated by Nr, COG, and KEGG databases. Nr is the official protein sequence database of NCBI and typically used for species classification.24 The number of annotated proteins was 5740, which accounted for 99.72% of the total protein number, and 99% of the CDS coding proteins were annotated as Pseudomonas sp. proteins by the database. The statistical results of genome COG showed that the strain WXP-4 contained 24 of 25 categories, and the number of genes responsible for amino acid transportation and metabolism ranked second (Fig. 1b). The metabolic pathways of gene products and compounds in cells and the functions of these gene products were systematically analyzed by KEGG. As shown in Fig. 1c, 3952 genes in the genome were involved in the metabolism of bacteria, accounting for 91.14% of the total KEGG annotation genes. Among these, 718 genes were involved in carbohydrate metabolism; 852 genes were involved in amino acid metabolism; 135 genes were involved in environmental information processing and cellular processes; and 249 genes were involved in the three pathways of the biological system, human disease, and genetic information processing.
Identification of denitrification genes in P. citronellolis WXP-4
The complete denitrification process includes four reductases, namely, nitrate reductase (Nar), nitrite reductase (Nir), nitric oxide reductase (NOR), and N2OR, which were coded by specific genes.25 A total of 25 denitrification genes were found in strain WXP-4. Part of these genes constituted the nar and nos denitrification gene clusters, whereas the other nir and nor genes were not clustered separately. The nar, nir, and nor genes were relatively close to one another on the chromosome. The denitrification gene clusters were arranged in an orderly and relatively compact manner.According to their different locations in the cell, denitrifying bacteria usually express two kinds of Nar, namely, membrane-bound and periplasmic-bound nitrate reductase; their coding genes were usually nar and nap, respectively.26 In P. citronellolis WXP-4, only nar existed and was present in the form of a gene cluster. The nar gene cluster with a total length of 18.02 kb was located on the antisense chain of the genome and encoded 12 ORFs, namely, narL, narQ, narK-2, narK-1, narG, narH, narJ, narI, narG, narH, narJ, and narI, in order (Fig. 2a). Twelve regularly arranged genes were transcribed in the same direction on the chromosome, and no gap was found in the coding region. Among these genes, narGHI was the structural gene of Nar, whereas the others were the auxiliary genes of the enzyme. The largest ORF gene was narG with a length of 3780 bp; it encoded the α subunit of Nar. The narH and narI encoded the β and γ subunits of nitrate reductase, respectively. The narJ encoded the Mo cofactor assembly chaperone of Nar.27
Nir catalyzes the reduction of nitrite to nitric oxide, which is the most important rate-limiting step in the denitrification process. Nir is generally divided into cytochrome cd1-type Nir and Cu-type Nir, which are encoded by nirS and nirK genes, respectively.28,29 However, these two enzymes generally do not coexist in one bacterium. Only one copy of the nirK gene was found in the strain WXP-4, whereas other conventional nir genes did not exist near the upstream and downstream, and nirD genes were found far upstream (Fig. 2b); this might be the evolutionary difference from other bacteria. Possibly, some nir genes have not yet been identified. A search for nir-related proteins in the P. citronellolis WXP-4 genome needs to be performed in the future. All the key genes involved in the nitrite reduction process and their functions need to be identified.
NOR reduces NO to N2O, which can be divided into two types, namely, cNOR and qNOR. cNOR is a heterodimer oligomerase that is encoded by norB and norC, some regulatory genes may be present. qNOR is a monomer enzyme encoded by a single gene, either norB or norZ.30,31 Strain WXP-4 only contains cNOR. Two copies of norB and norR were found in the genome of strain WXP-4, but they were far away. One of the norB genes (1422 bp) was downstream of norC (441 bp). The other norB gene (2279 bp) was downstream of norR (1565 bp), which functions in NOR transcriptional regulation (Fig. 2c).
Compared with other denitrifying gene clusters, the nos gene clusters were very conservative. They were all arranged in the genome with a sequence of nosRZDFYL.32 In P. citronellolis WXP-4, the coding genes of N2OR formed the nos gene cluster, which was transcribed in the same direction, had a relatively compact arrangement, and had no other genes inside. In addition, the total length of the nos gene cluster was 7554 bp and included six ORFs in the following order: nosR, nosZ, nosD, nosF, nosY, and nosL (Fig. 2d). Among them, the longest gene was nosR gene (2115 bp), which encodes an integral membrane protein required for electron delivery and function. The structural nosZ gene with a length of 1914 bp encodes the N2OR protein. The nosDFY genes encode an ABC transporter (NosFY) and a periplasmic interacting protein, NosD, which is presumably required for shuttling a sulfur species into the periplasm for CuZ assembly. The nosL encodes a membrane anchored copper molecular chaperone.33,34
According to the whole genome sequence, four key reductase structure coding genes were involved in the denitrification of WXP-4, as follows: narGHI, nirK, norCB, and nosZ, demonstrating that P. citronellolis WXP-4 was a complete denitrifying bacterium. These results further confirmed the research results of Wang et al.20 According to the position of these four structure genes in the genome, the gene map was drawn and shown in Fig. 2e. The nos, nor, nir, and nar genes were arranged on the genome chain in order.
TA cloning and basic sequence of nosZ gene
The nosZ gene encoding N2OR in strain WXP-4 was amplified by PCR and obtained by TA cloning. The results of the agarose gel electrophoresis of the nosZ gene PCR products demonstrated that the target DNA band was clear (ESI Fig. S2†), indicating the reaction conditions are appropriate. Generally, the number of PCR cycles will be selected between 30–35, which depends on the concentration of template DNA and other substances in the reaction system. Too few cycles will result in insufficient amplification of target DNA, and the more cycles will produce the more non-specific products. The analysis results from DNAMAN software showed that the total length of nosZ gene was 1914 bp, and the ATCG base contained 390, 266, 583, 675, which accounted for 20.4%, 13.9%, 30.5%, and 35.3% of the total base number respectively. The BLAST homology comparison results revealed that the nucleotide sequence of nosZ gene was like that of the encoding N2OR protein in P. aeruginosa HS9. The phylogenetic tree showed that the confidence of nosZ sequence homology between strain WXP-4 and P. aeruginosa was 100 (Fig. 3).The analysis of the nosZ sequence found the presence of 101 restriction endonucleases in the sequence, which could be digested by 49 restriction enzymes. However, the restriction endonucleases of EcoR I and Xho I were absent (ESI Fig. S3†). The sequence could be inserted into other vectors via EcoR I and Xho I double enzyme digestion without sequence self-cutting. The restriction endonuclease site map and electrophoretic simulation of single enzyme digestion of nosZ gene with 49 restriction enzymes were shown in Fig. S3.†
Spatial structure of the N2OR protein
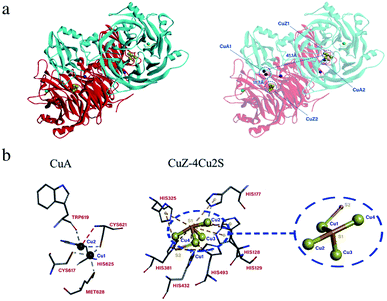
The basic physical and chemical properties of N2OR protein analyzed by ProtParam tool showed that the nosZ protein contained 20 kinds and 637 amino acids, and the predicted molecular mass of the protein was 70.9 kDa. Like the N2OR of W. succinogenes and P. putida HK5, the predicted 3D structure of N2OR protein in P. citronellolis WXP-4 was a head-to-tail homodimer and each monomer contains CuZ center and CuA center.35,36 The distance between the CuA center from one monomer and the CuZ center from the other monomer was approximately 11.7 Å, and the distance between two copper centers in one monomer was about 41 Å, which were too far away for effective electron transfer (Fig. 4a).3 In addition, the CuA center comprising two copper atoms was only coordinated by five conserved amino acid residues (C617, C621, H625, T619, and M628), CuZ center consisting of four copper atoms and two sulfur atoms comprised seven conserved histidine residues (H325, H177, H381, H432, H493, H128, and H129), but the S2 atom only bound one CuI atom (Fig. 4b). These results were different from P. stutzeri and P. nautica, in which the CuA had six amino acid ligands and the S2 atom both bound CuI and CuIV atoms.15,16CuA site is a mixed-valent center that leads to a low reorganization energy, making CuA an efficient electron transfer agent.37,38 Among the ligands of CuA from P. stutzeri N2OR, His583 plays a role in electron gating to CuA. But in the CuA of P. citronellolis WXP-4 N2OR, no similar His583 ligand was found, which may be due to the unbound conformation of H583 or is derived from a different bacterium.39 In previous research, the more labile S2 was found to bridge ions CuI and CuIV in two monomers and prone to be replaced by a water ligand.34 Therefore, the S2 atom only bound one CuI atom in P. citronellolis WXP-4, which seems to be more easily affected by other ligand than when the S2 atom binds two Cu atoms, meaning that N2O may more easily bind to CuZ active sites.
N2OR expression in recombinant E. coli
As shown in Fig. S4,† the growth adaptation period of recombinant E. coli was relatively short. It reached the logarithmic phase after 2 h of inoculation. After inoculation for 10 h, the recombinant E. coli reached a stable period, and OD600 no longer changed significantly. The enhanced expression of N2OR was performed by inducer IPTG, which tightly related to inducer addition time. The addition of inducers in the middle and early logarithmic stages of bacterial growth promoted gene expression and high protein concentrations.10,40 The addition of the inducer during the lag phase makes bacterial growth difficult; its addition during the stabilization phase inhibits the expression of the target gene.41 Generally, the optimal OD600 interval for gene induced expression should be in the range of 0.5–0.8. In this study, the OD600 was selected as 0.6, and the culture time was 2 h. The optimal induce conditions were 1.0 mM IPTG, 5 h, and 30 °C. After protein extraction, isolation, and purification, the expression of the nosZ gene was examined by SDS-PAGE gel electrophoresis, and the results were shown in Fig. 5.The target band appeared at 76 kDa and was congruent with the SDS-PAGE gel electrophoresis results. The theoretical value of the molecular mass of the protein removed by EcoR I and Xho I was about 1.446 kDa. The expression of the remaining protein on pET28a was about 5.230 kDa. The target protein N2OR was about 70.9 kDa. The molecular mass of the fusion protein was about 76.033 kDa. This finding demonstrated that N2OR was successfully expressed in E. coli BL21(DE3)-pET28a-nosZ from one side. The band on the left panel became brighter when the recombinant E. coli was induced by IPTG, which indicated that IPTG promoted the expression of the target protein. Meanwhile, when the N2OR was not diluted, more than five bands were observed on the electrophoresis graph. However, after dilution, the number of electrophoresis bands was significantly reduced, which was probably due to high protein concentration or protein purity.
The reduction performance of recombinant E. coli on N2O
To verify the activity of N2OR protein expressed by recombinant E. coli and the reduction performance of N2O, the N2O degradation amount in the N2O reduction system with different initial concentrations (10, 20, 30, 40, and 50 mg L−1) was determined. As shown in Fig. 6a, the N2O degradation time increased with increasing initial concentration of N2O. When the initial concentration was 10 mg L−1, the effect of complete removal was achieved within 2 d. The results proved that the nosZ gene was transcribed in E. coli BL21(DE3) to activate the N2OR protein. | ||
| Fig. 6 Effect of different conditions on N2O reduction of recombinant E. coli. (a) initial N2O concentration. (b) pH. (c) temperature. | ||
The factors that affect the activity of N2OR, such as pH and temperature, were investigated. When the pH value was 7.0 and 8.0, the reducing effect on N2O was the most significant, reaching 100% and 98% of the relative activity percentage, respectively (Fig. 6b). The results coincided with the findings by Fujita and Dooley, which suggested that the neutral or slightly alkaline environment is conducive to the development of N2OR activity.42 Therefore, the optimum pH for N2OR activity was 7 or 8. The amount of reduced N2O reached the maximum value at 40 °C (Fig. 6c), whereas the activity of N2OR decreased significantly at low and high temperatures. The results were the same as the findings of Ferretti et al.43
The N2O reduction performances of recombinant E. coli and P. citronellolis WXP-4 were compared, as presented in Fig. 7. Under the optimal conditions of pH 7.0 and 40 °C, the recombinant E. coli could reduce 10 mg L−1 of N2O to N2 within 15 h, which corresponded to a reduction rate of 0.67 mg (L h)−1 and was almost 2.3 times that of Alcaligenes denitrificans strain TB (0.29 mg (L h)−1).10 However, the reduction rate of recombinant E. coli was only 80% of that of wild strain WXP-4, and this result may be due to the deletion of one or several genes in the nosRDFYL gene, which encodes the auxiliary protein related to N2OR activity or total enzymes quantity.13,22 Fortunately, heterologous host E. coli maybe has a certain compensation mechanism which can partially compensate for the function of deletion gene, so that the recombinant E. coli can still have strong N2O reduction properties, indicating E. coli is an ideal host for heterologous expression and a highly suitable tool to further investigate the mechanism of N2O reduction.34 Overall, both recombinant E. coli and wild strain WXP-4 have strong N2O reduction ability, which may be closely related to the unique 3D structure of strain WXP-4 N2OR (CuA had only five amino acid ligands and the S2 of CuZ only bound CuI atom), its related mechanism needs to be further studied.
 | ||
| Fig. 7 Comparison of N2O reduction performance between P. citronellolis WXP-4 and recombinant E. coli. | ||
Conclusions
In this work, the whole genome sequence of P. citronellolis WXP-4 was determined for the first time and four key reductase structure coding genes related to complete denitrification were identified. Then single structural coding gene nosZ was cloned and the recombinant E. coli BL21(DE3)-pET28a-nosZ was successfully constructed. It exhibited strong N2O reduction ability and completely reduced 10 mg L−1 N2O to N2 within 15 h under optimal conditions (pH 7.0 and 40 °C), corresponding the N2O reduction rate was almost 2.3 times that of Alcaligenes denitrificans strain TB, but only 80% of that of wild strain WXP-4. It was induced that nos gene cluster auxiliary gene deletion decreased the activity of N2OR. The 3D structure of N2OR was explored, and the predicted results showed that the electron transfer center CuA had only five amino acid ligands, and S2 of the 4Cu:2S structural catalytically active center CuZ only bound one CuI atom. The unique 3D structure was different from the results of previous studies and maybe closely related to the strong N2O reduction ability. These findings provide a potential candidate for decreasing the greenhouse effect and a new perspective for studying the structure–activity relationship of N2OR.Author contributions
Liyong Hu: formal analysis, investigation, writing – review & editing, project administration. Xiaoping Wang: formal analysis, investigation, software, writing – original draft. Cong Chen: validation, data curation, writing – review & editing. Jianmeng Chen: resources, supervision. Zeyu Wang: validation, data curation. Jun Chen: methodology, conceptualization, funding acquisition, writing – review & editing, supervision. Dzmitry Hrynshpan: methodology. Tatsiana Savitskaya: methodology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Primary Research & Development Plan (2018YFE0120300), the National Natural Science Foundation of China (21777142, 22011530015), Zhejiang Provincial Department of Education Research Project (Natural Science), and Zhejiang Shuren University Basic Scientific Research Special Funds (2020XZ012).References
- E. M. Johnston, C. Carreira, S. Dell'Acqua, S. G. Dey, S. R. Pauleta, I. Moura and E. I. Solomon, J. Am. Chem. Soc., 2017, 139, 4462–4476 CrossRef CAS PubMed.
- J. M. Blum, Q. Su, Y. Ma, B. Valverde-Pérez, C. Domingo-Félez, M. M. Jensen and B. F. Smets, Environ. Microbiol., 2018, 20, 1623–1640 CrossRef PubMed.
- D. Richardson, H. Felgate, N. Watmough, A. Thomson and E. Baggs, Trends Biotechnol., 2009, 27, 388–397 CrossRef CAS PubMed.
- J. W. Zou, Y. Y. Lu and Y. Huang, Environ. Pollut., 2010, 158, 631–635 CrossRef CAS PubMed.
- M. S. Zheng, N. Zhou, S. S. He, F. Chang, J. Zhong, S. Xu, Z. Wang and T. Liu, J. Environ. Manage., 2021, 280, 111657 CrossRef CAS PubMed.
- C. Kennes, E. R. Rene and M. C. Veiga, J. Chem. Technol. Biotechnol., 2009, 84, 1419–1436 CrossRef CAS.
- L. R. Bakken, L. Bergaust, B. Liu and Å. Frostegård, Philos. Trans. R. Soc., B, 2012, 367, 1226–1234 CrossRef CAS PubMed.
- K. Lassey and M. Harvey, Water Atmos., 2007, 2, 10–11 Search PubMed.
- D. Fowler, M. Coyle, U. Skiba, M. A. Sutton, J. N. Cape, S. Reis, L. J. Sheppard, A. Jenkins, B. Grizzetti, J. N. Galloway, P. Vitousek, A. Leach, A. F. Bouwman, K. Butterbach-Bahl, F. Dentener, D. Stevenson, M. Amann and M. Voss, Philos. Trans. R. Soc., B, 2013, 368, 20130112 CrossRef PubMed.
- Y. Wang, Z. Y. Wang, Y. K. Duo, X. P. Wang, J. M. Chen and J. Chen, Environ. Pollut., 2018, 239, 43–52 CrossRef CAS PubMed.
- C. Carreira, S. R. Pauleta and I. Moura, J. Inorg. Biochem., 2017, 177, 423–434 CrossRef CAS PubMed.
- M. M. M. Kuypers, H. K. Marchant and B. Kartal, Nat. Rev. Microbiol., 2018, 16, 263–276 CrossRef CAS PubMed.
- S. R. Pauleta, S. Dell'Acqua and I. Moura, Coord. Chem. Rev., 2013, 257, 332–349 CrossRef CAS.
- E. M. Johnston, S. Dell'Acqua, S. Ramos, S. R. Pauleta, I. Moura and E. I. Solomon, J. Am. Chem. Soc., 2014, 136, 614–617 CrossRef CAS PubMed.
- A. Pomowski, W. Zumft, P. Kroneck and O. Einsle, Nature, 2011, 477, 234–237 CrossRef CAS PubMed.
- E. M. Johnston, S. Dell'Acqua, S. R. Pauleta, I. Moura and E. I. Solomon, Chem. Sci., 2015, 6, 5670–5679 RSC.
- L. Q. Yang, X. J. Zhang and X. T. Ju, Sci. Rep., 2017, 7, 43283 CrossRef PubMed.
- S. Wan, T. Greenham, K. Goto, Y. Mottiar, A. M. Johnson, J. Staebler, M. Zaidi, Q. Shu and I. Altosaar, Can. J. Plant Sci., 2014, 94, 1013–1023 CrossRef CAS.
- C. M. Jones, A. Welsh, I. N. Throbäck, P. Dörsch, L. R. Bakken and S. Hallin, FEMS Microbiol. Ecol., 2011, 76, 541–552 CrossRef CAS PubMed.
- X. P. Wang, Y. K. Duo, J. J. He, J. C. Yao, H. F. Qian, D. Hrynsphan, S. Tatsiana and J. Chen, Bioprocess Biosyst. Eng., 2020, 43, 811–820 CrossRef CAS PubMed.
- D. Hyatt, G. L. Chen, P. F. LoCascio, M. L. Land, F. W. Larimer and L. J. Hauser, BMC Bioinf., 2010, 11, 119 CrossRef.
- C. Chen, Y. Wang, H. Liu, Y. Chen, J. C. Yao, J. Chen, D. Hrynsphan and S. Tatsiana, Chemosphere, 2020, 253, 126739 CrossRef CAS PubMed.
- Y. L. Lei, Y. Q. Wang, H. J. Liu, C. W. Xi and L. Y. Song, Appl. Microbiol. Biotechnol., 2016, 100, 4219–4229 CrossRef CAS PubMed.
- A. G. Kennedy, J. Eval. Clin. Pract., 2017, 23, 959–963 CrossRef PubMed.
- W. G. Zumft, Microbiol. Mol. Biol. Rev., 1997, 61, 533–616 CAS.
- L. Philippot, Biochim. Biophys. Acta, 2002, 1577, 355–376 CrossRef CAS.
- J. M. Dow, S. Grahl, R. Ward, R. Evans, O. Byron, D. G. Norman, T. Palmer and F. Sargent, FEBS J., 2014, 281, 246–260 CrossRef CAS.
- S. Henry, D. Bru, B. Stres, S. Hallet and L. Philippot, Appl. Environ. Microbiol., 2006, 72, 5181–5189 CrossRef CAS PubMed.
- C. Sánchez and K. Minamisawa, FEMS Microbiol. Lett., 2018, 365, 1–7 Search PubMed.
- N. Gonska, D. Young, R. Yuki, T. Okamoto, T. Hisano, S. Antonyuk, S. S. Hasnain, K. Muramoto, Y. Shiro, T. Tosha and P. Adelroth, Sci. Rep., 2018, 8, 3637 CrossRef PubMed.
- R. Yamagiwa, T. Kurahashi, M. Takeda, M. Adachi, H. Nakamura, H. Arai, Y. Shiro, H. Sawai and T. Tosha, Biochim. Biophys. Acta, Bioenerg., 2018, 1859, 333–341 CrossRef CAS PubMed.
- L. Philippot, P. Mirleau, S. Mazurier, S. Siblot, A. Hartmann, P. Lemanceau and J. C. Germon, Biochim. Biophys. Acta, 2001, 1571, 436–440 CrossRef.
- V. V. Kadnikov, A. V. Mardanov, O. A. Podosokorskaya, S. N. Gavrilov, I. V. Kublanov, A. V. Beletsky, E. A. Bonch-Osmolovskaya and N. V. Ravin, PLoS One, 2013, 8, e53047 CrossRef CAS PubMed.
- L. Zhang, A. Wüsta, B. Prassera, C. Müllera and O. Einslea, PNAS, 2019, 116, 12822–12827 CrossRef CAS PubMed.
- Z. Chen, K. Matsushita, T. Yamashita, T. Fujii, H. Toyama, O. Adachi, H. D. Bellamy and F. S. Mathews, Structure, 2002, 10, 837–849 CrossRef CAS PubMed.
- S. Dell'Acqua, I. Moura, J. J. G. Moura and S. R. Pauleta, J. Biol. Inorg Chem., 2011, 16, 1241–1254 CrossRef PubMed.
- O. Farver, Y. Lu, M. C. Ang and I. Pecht, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 899–902 CrossRef CAS PubMed.
- P. Kroneck, J. Biol. Inorg Chem., 2018, 23, 27–39 CrossRef CAS PubMed.
- L. Zhang, E. Bill, P. Kroneck and O. Einsle, J. Am. Chem. Soc., 2021, 143, 830–838 CrossRef CAS PubMed.
- X. Y. Ou, F. Peng, X. L. Wu, P. Xu, M. H. Zong and W. Y. Lou, Biochem. Eng. J., 2020, 158, 107573 CrossRef CAS.
- N. Van Noi and Y. C. Chung, Biotechnol. Biotechnol. Equip., 2017, 31, 619–629 CrossRef.
- K. Fujita and D. M. Dooley, Inorg. Chem., 2007, 46, 613–615 CrossRef CAS PubMed.
- S. Ferretti, J. G. Grossmann, S. S. Hasnain, R. R. Eady and B. E. Smith, Eur. J. Biochem., 1999, 259, 651–659 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra09008a |
| This journal is © The Royal Society of Chemistry 2022 |