Open Access Article
Open Access ArticleDesign, synthesis and evaluation of 3-phenoxypyrazine-2-carboxamide derivatives as potent TGR5 agonists†
Shizhen Zhao‡
 a,
Le Wang‡a,
Jie Wanga,
Chenwei Wanga,
Shaowei Zhenga,
Yajie Fua,
Yunfu Lia,
Wei-Dong Chen
a,
Le Wang‡a,
Jie Wanga,
Chenwei Wanga,
Shaowei Zhenga,
Yajie Fua,
Yunfu Lia,
Wei-Dong Chen ab,
Ruifang Hou*a,
Dongbin Yang*a and
Yan-Dong Wang*c
ab,
Ruifang Hou*a,
Dongbin Yang*a and
Yan-Dong Wang*c
aKey Laboratory of Receptors-Mediated Gene Regulation and Drug Discovery, Hebei Key Laboratory of Liver Disease, People's Hospital of Hebei, School of Medicine, Henan University, Kaifeng, China. E-mail: dr_rfhou@126.com; dongbinyang@126.com
bKey Laboratory of Molecular Pathology, School of Basic Medical Science, Inner Mongolia Medical University, Hohhot, China
cState Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China. E-mail: ydwangbuct2009@163.com
First published on 27th January 2022
Abstract
TGR5 is emerging as an important and promising target for the treatment of non-alcoholic steatohepatitis, type 2 diabetes mellitus (T2DM), and obesity. A series of novel 3-phenoxypyrazine-2-carboxamide derivatives were designed, synthesized and evaluated in vitro and in vivo. The most potent compounds 18g and 18k exhibited excellent hTGR5 agonist activity, which was superior to those of the reference drug INT-777. In addition, compound 18k could significantly reduce blood glucose levels in C57 BL/6 mice and stimulate GLP-1 secretion in NCI-H716 cells and C57 BL/6 mice.
1. Introduction
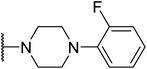
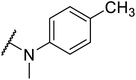
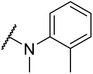
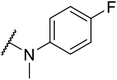
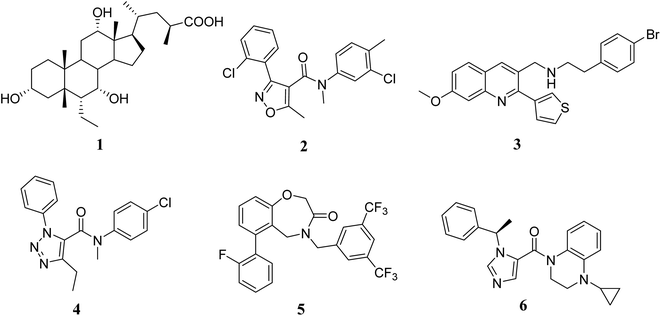
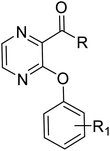
G protein-coupled BA receptor 1 (GPBAR1), also known as Takeda G-protein-coupled receptor 5 (TGR5), is a rhodopsin-like superfamily G-protein-coupled receptor (GPCR) first identified as a bile acid receptor in 2002.1 TGR5 is widely expressed in many tissues including the gall bladder, liver, intestine, pancreas and spleen.2–4 TGR5 activation has been reported to induce glucagon-like peptide-1 (GLP-1) secretion, leading to improve glucose metabolism and energy homeostasis.5,6 Activation of TGR5 can also increase cyclic adenosine monophosphate (cAMP) levels, which is recognized as an important intracellular signaling cascade.7,8 In addition, the TGR5 signaling pathway stimulates energy expenditure and oxygen consumption in brown adipose tissue and muscle.9 Thus, TGR5 might be a considerable therapeutic target for the treatment of metabolic disorders, such as non-alcoholic steatohepatitis, type 2 diabetes mellitus (T2DM), and obesity.10,11Several steroidal and non-steroidal TGR5 agonists have been reported in the literature by pharmaceutical companies (Fig. 1).12,13 The steroidal TGR5 agonists is structurally based on bile acids (BAs), including cholic acid (CA), lithocholic acid (LCA), and their semisynthetic derivatives such as 6α-ethyl-23(S)-methylcholic acid (INT-777, 1).14,15 INT-777, a selective TGR5 agonist, stimulates GLP-1 secretion and improves glucose homeostasis. In addition, many synthetic non-steroidal TGR5 agonists have been reported (2–5), which exhibited excellent TGR5 agonist activity and were orally efficacious in lowering glucose levels in vivo.16–19 However, none of these agonists have been obtained approval and entered clinical application.
We have previously reported the synthesis and in vitro and in vivo evaluation of TGR5 agonistic activities of a new series of 1-benzyl-1H-imidazole-5-carboxamide derivatives.20 Most compounds displayed excellent agonistic activities against hTGR5, which was superior to those of the reference drugs INT-777 and TCA. The chemical scaffold of compound 6 is an attractive lead compound for generating novel TGR5 agonists. To further explore the potent TGR5 agonists, the middle imidazole core of compound 6 was replaced with different heterocycles to get insights into the structure–activity relationships (SARs) and improved TGR5 agonist activity.
2. Results and discussion
2.1 Chemistry
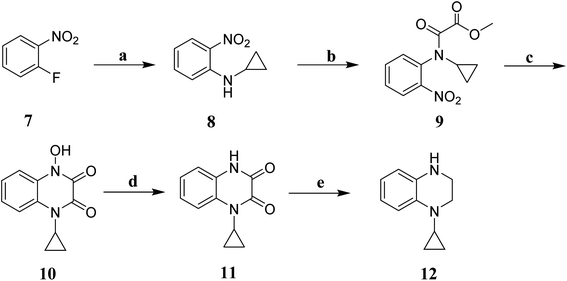
The key intermediate 12 was synthesized according to our previously reported procedure which was outlined in Scheme 1.20 2-Fluoronitrobenzene 7 was used as the starting material, and treated with excess cyclopropylamine to obtain intermediate 8. The intermediate 8 was acylated with methyl oxalyl chloride to form intermediate 9, which was used in a reduction–cyclisation reaction with palladium on carbon to obtain intermediate 10. The intermediate 10 was treated with triphenylphosphine yielded the intermediate 11. Finally, the intermediate 11 was treated with borane–tetrahydrofurane complex by reduction reaction to obtain the key intermediates 12.The synthetic routes of the key intermediates 13–16 are illustrated in Scheme 2. Methyl 1,2,3,4-tetrahydroisoquinoline-3-carboxylate (13), methyl indoline-2-carboxylate (14) or methyl 4H-thieno[3,2-b]pyrrole-5-carboxylate(15) was treated with 4-chlorobenzyl bromide in the presence of NaH by nucleophilic substitution reactions to form intermediate 17–19. Methyl 3-chloropyrazine-2-carboxylate (16) was used as the starting material, and treated with substituted phenol by nucleophilic aromatic substitution (SNAr) reaction to obtain intermediate 16a–i.
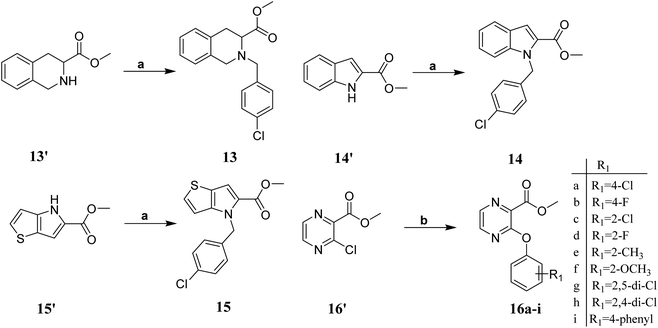 | ||
| Scheme 2 Synthesis of intermediates 13–16. Reagents and conditions:(a) NaH, DMF, 4-chlorobenzyl bromide; (b) K2CO3, substituted phenol. | ||
The syntheses of the target compounds 18a–u were performed according to Scheme 3. The intermediate 13–15, 16a–i were saponified with 2 N NaOH to obtain the key intermediates 17. Then, the intermediates 17 reacted with the key intermediate 12, substituted aniline, substituted N-methylaniline or substituted 1-phenylpiperazine in the presence of HATU, to give target compounds 18a–u.
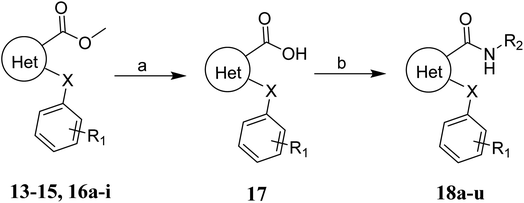 | ||
| Scheme 3 General synthesis of the target compounds 18a–u. Reagents and conditions: (a) NaOH, MeOH/H2O, r.t.; (b) HATU, DIEA, DMF, 60 °C. | ||
2.2 In vitro biological evaluation
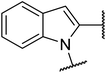
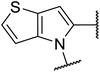
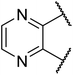
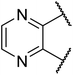
The TGR5 agonist activity of the target compounds were evaluated by assaying cAMP production using homogeneous time resolved fluorescence (HTRF) assay in a recombinant HEK293T cell line expressing human TGR5 (NM_001077191.1) or mouse TGR5 (NM_174985) similar to that reported in the literature.21,22 INT-777 were used as reference drugs.As shown in Table 1, the introduction of different heterocycles rings of target compounds 18a–e had a notable influence on the hTGR5 activity. Many of these modifications showed a decrease agonistic activity at TGR5 compared with the lead compound 6. The heteroatom compounds such as tetrahydroisoquinoline 18a, indoline 18b and thienopyrrole 18c had no TGR5 agonistic activities. Interestingly, pyrazine derivatives 18d and 18e exhibited TGR5 agonistic activities on hTGR5 with EC50 values of 1500 and 442 nM, respectively. This suggested that bicyclic ring is not advantageous for TGR5 agonist activity.
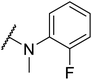
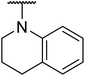
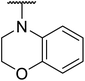
Based on the results above, we selected a scaffold of pyrazine core as our starting point for further modification. Our optimized efforts were directed toward replacing the amides and phenoxy group with various substituents to expand the SAR studies (Table 2). Compounds exhibited excellent hTGR5 agonist activity with EC50 values in the range of 1.4 to 895 nM, which are superior or comparable to those of the reference drug INT-777. Of these, compounds 18g and 18k with 2-Cl and 2,5-di-Cl substituents showed the best hTGR5 agonistic activities, with EC50 values of 1.44 and 0.58 nM on hTGR5, respectively. However, replacing the tetrahydro-quinoxaline group (18k) with N-methyl aniline(18p–s), substituted 1-phenylpiperazine (18m–o), benzomorpholine (18t) and tetrahydroquinoline (18u) resulted in a significant decrease hTGR5 agonist activity.
The weak sequence homology (83%) between the hTGR5 and mTGR5 resulting in different hTGR5 and mTGR5 potencies of reported compounds. Therefore, the most potent compounds (18g, 18j and 18k) were further evaluated for their ability to activate mTGR5 using a similar method (Table 2). The mTGR5 agonism activity of compounds (18g, 18j and 18k) was significantly decreased compared with hTGR5 agonism activity. Among these, the potent compounds 18g and 18k, with EC50 values of 119.6 and 267.2 nM toward mTGR5 in our assay, respectively.
2.3 In vitro human plasma stability assay
The plasma stability of compounds is an important consideration in drug development. Based on their in vitro activities, the most potent compound 18k was selected to evaluate the stability in human plasma. The results were summarized in Table 3. After co-incubation with human plasma for 120 min, compound 18k exhibited excellent metabolic profiles in human plasma, and the half-life value was greater than 289.1 min.| Compd | Time point (min) | % remaining | T1/2 (min) |
|---|---|---|---|
| 18k | 0 | 100.0 | >289.1 |
| 10 | 105.7 | ||
| 30 | 104.4 | ||
| 60 | 101.5 | ||
| 120 | 85.4 |
2.4 In vitro cytotoxicity assay
The cytotoxicity of the most potent compounds18g, 18j and 18k were further evaluated in A549 and HeLa cells using the CCK8 assay. As shown in Table 4, compounds 18g, 18j and 18k showed no cytotoxic activities toward tumor cell lines A549 and HeLa with IC50 > 50 μM.2.5 Oral glucose tolerance test in mice
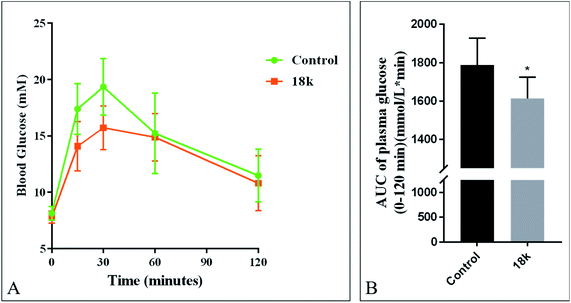
To test the hypoglycemic effects of the most potent compound 18k, an in vivo oral glucose tolerance test (OGTT) were carried out in C57 BL/6 mice (Fig. 2). Mice were fasted overnight before glucose tolerance tests. Mice were orally administered 50 mg Kg−1 of compound 18k. Blood glucose was measured at 0, 15, 30, 60 and 120 min after glucose administration. Meanwhile, the areas under the curve (AUC) for glucose levels were calculated. It was observed that compound 18k caused a significant 9.7% of reduction in blood glucose AUC0–120 min compared with the control group.2.6 GLP-1 secretion
To confirm whether the glucose-lowering effect of compound 18k is dependent on mediated GLP-1 secretion, a further GLP-1 levels of NCI-H716 cells and C57 BL/6 mice were measure by GLP-1(7–36) ELISA assay. As shown in Fig. 3, compound 18k could increase the secretion of GLP-1 compared to control in NCI-H716 cells. Meanwhile, compound 18k could increase GLP-1 secretion compared to control in C57 BL/6 mice. All of these results indicated that compound 18k could stimulate GLP-1 secretion.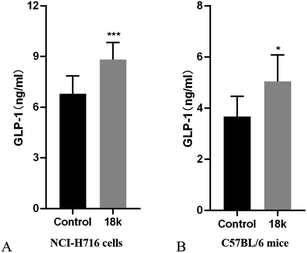 | ||
| Fig. 3 GLP-1 secretion study of compound 18k at 4 μM in NCI-H716 cells and 50 mg kg−1 in C57 BL/6 mice. *P < 0.05; ***P < 0.001 versus control; error bar indicates SEM. | ||
2.7 Molecular docking model analysis of compound 18k in the active site of TGR5
To better understand the binding mode of compound 18k, a highly potent compound, docking simulations were carried out using the Autodock 4.2 software. The published X-ray structure of P395 bound within the active site cavity of TGR5 (PDB ID 7CFM) served as a useful template for generating binding modes.23 Images depicting the proposed binding modes were generated using PyMOL. As shown in Fig. 4, the 2,5-di-Cl phenol moiety of compound 18k formed hydrophobic interaction with Leu74 and Leu166 and pi–pi interactions with Trp75. The 1-cyclopropyl-1,2,3,4-tetrahydroquinoxaline side chain of compound 18k made pi–pi interactions with Phe96 and hydrophobic feature with the surrounding residues Pro92 and Phe 161. The pyrazine group of compound 18k could form hydrogen bond interactions with the Ser240, Thr243 and Ser247. Binding studies may provide a good explanation for the excellent hTGR5 agonistic activity of compound 18k.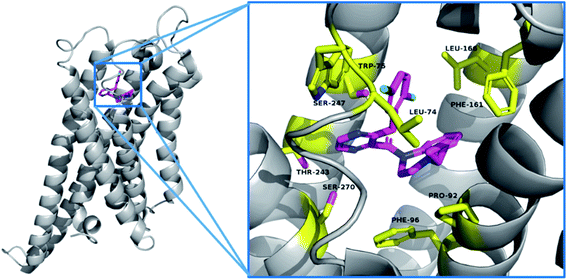 | ||
| Fig. 4 The binding mode of compounds 18k in the active site of TGR5 (PDB ID 7CFM). | ||
3. Conclusion
In conclusion, a series of novel 3-phenoxypyrazine-2-carboxamide derivatives were designed and synthesized as potent TGR5 agonists. Many of pyrazine derivatives exhibited excellent hTGR5 agonist activity with EC50 values in the range of 1.4 to 895 nM. Among these, compounds 18g and 18k with 2-Cl and 2,5-di-Cl substituents displayed the most remarkable agonistic activities against TGR5, which was superior to those of the reference drugs INT-777. In vivo glucose-lowering study indicated that Compound 18k could significantly reduce blood glucose levels in C57 BL/6 mice. In addition, the GLP-1 ELISA assay indicated that compound 18k could stimulate GLP-1 secretion in NCI-H716 cells and C57 BL/6 mice. Further developments of compound 18k are ongoing in our laboratory.4. Experimental section
4.1 General procedure for the synthesis of compounds
Unless otherwise noted, all reagents and solvents were obtained from commercially available sources and were used without purification. TLC analysis was performed on GF254 silica gel plates (Jiangyou, Yantai). Column chromatography was carried out with silica gel (200–300 mesh) from Qingdao Haiyang Chemicals (Qingdao, Shandong, China). Mass spectrometry was performed using ESI mode on an Agilent 1200 LC-MS (Agilent, Palo Alto, CA, USA). High-resolution accurate mass determinations (HRMS) were recorded on a Bruker Micromass Time of Flight mass spectrometer equipped with electrospray ionisation (ESI). Nuclear magnetic resonance (1H-NMR and 13C-NMR) spectra were recorded on a Bruker 400 MHz and 500 MHz NMR spectrometer with TMS as an internal standard. The chemical shifts were reported in parts per million (ppm), the coupling constants (J) were expressed in hertz (Hz). Peak multiplicities were described as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m) and broad (br).4.2 Cyclopropyl-(2-nitro-phenyl)-amine (8)
A solution of cyclopropylamine (12.14 g, 212.61 mmol) and 2-fluoronitrobenzene (15.00 g, 106.31 mmol) in DMF (40 mL) was stirred for 12 h at ambient temperature. After confirming that the reaction was completed by TLC analysis, the solution was extracted with EtOAc and brine. The organic layer was dried over anhydrous Na2SO4 overnight and the solvent was removed under vacuum. The crude product was purified by silica gel column chromatography to give the target product 8.4.3 N-Cyclopropyl-N-(2-nitro-phenyl)-oxalamic acid methyl ester (9)
A solution of cyclopropyl-(2-nitro-phenyl)-amine(15.00 g, 84.18 mmol) and triethylamine (12.78 g, 126.27 mmol) in dichloromethane was cooled to <0 °C using a salted ice bath. Then methyl oxalyl chloride (10.31 g, 84.18 mmol) was added dropwise to the reaction mixture. The resulting mixture was stirred at <0 °C for 30 min, and then at ambient temperature for 12 h. The reaction mixture was extracted with EtOAc and brine. The organic phase was dried over Na2SO4 overnight and the solvent was removed under vacuum to give the target product 9.4.4 1-Cyclopropyl-4-hydroxy-1,4-dihydro-quinoxaline-2,3-dione (10)
Palladium on carbon (0.9 g, 10% w/w) was added to a solution of 9 (18.00 g, 68.12 mmol) in methanol (200 mL). The reaction flask was evacuated and back filled with hydrogen for three times, then allowed to stirred for 18 h under a slight overpressure of hydrogen at ambient temperature. After confirming that the reaction was completed by TLC analysis, the solution was diluted with ethyl acetate (400 mL), filtered and the solvent was concentrated to give the target product 10.4.5 1-Cyclopropyl-1,4-dihydro-quinoxaline-2,3-dione (11)
Triphenylphosphine (23.44 g, 89.36 mmol) was added to a solution of 1-cyclopropyl-4-hydroxy-1,4-dihydro-quinoxaline-2,3-dione (13.00 g, 59.58 mmol)in DMF at 135 °C for 6 h. After confirming that the reaction was completed by TLC analysis, the solution was cooled to room temperature and dichloromethane (400 mL) was added. The suspension was stirred for 30 min, filtered and washed with dichloromethane to give the desired compound 11. 1H NMR (300 MHz, DMSO-d6) δ 11.92 (s, 1H), 7.63 (dd, J = 6.5, 1.8 Hz, 1H), 7.30–7.10 (m, 3H), 2.99–2.86 (m, 1H), 1.30–1.14 (m, 2H), 0.81–0.66 (m, 2H).4.6 1-Cyclopropyl-1,2,3,4-tetrahydro-quinoxaline (12)
Boranete-tetrahydrofurane (1 M) complex (108.8 mL) was slowly added dropwise to a solution of 1-cyclopropyl-1,4-dihydro-quinoxaline-2,3-dione (11.00 g, 54.40 mmol) in THF (200 mL), and the resulting mixture was stirred for 18 h at ambient temperature. After confirming that the reaction was completed by TLC analysis, the solvent was removed under reduced pressure. The residue was extracted with ethyl acetate and washed with brine. The organic layer was dried over anhydrous Na2SO4 overnight and the solvent was removed under vacuum. The crude product was purified by silica gel column chromatography to yield the target product 12 as a white solid. 1H NMR (300 MHz, DMSO-d6) δ 6.90 (d, J = 7.6 Hz, 1H), 6.57–6.32 (m, 3H), 5.47 (s, 1H), 3.29–3.27 (m, 2H), 3.14–3.12 (m, 2H), 2.14–2.12 (m, 1H), 0.76–0.74 (m, 2H), 0.47–0.45 (m, 2H).4.7 Methyl 2-(4-chlorobenzyl)-1,2,3,4-tetrahydroisoquinoline-3-carboxylate (13)
Sodium hydride (83.66 mg, 2.09 mmol, 60%) was slowly added to a solution of methyl 1,2,3,4-tetrahydroisoquinoline-3-carboxylate (200.0 mg, 1.05 mmol) and 1-(bromomethyl)-4-chlorobenzene (214.9 mg, 1.05 mmol) in DMF (20 mL) at ambient temperature. The reaction mixture was stirred at 60 °C for 8 h. After confirming that the reaction was completed by TLC analysis, the mixture was extracted with ethyl acetate and washed with brine. The organic phase was dried over anhydrous Na2SO4 and concentrated. The crude product was purified by silica gel column chromatography to give the target product 13.4.8 Methyl 1-(4-chlorobenzyl)-1H-indole-2-carboxylate (14)
Sodium hydride (228.3 mg, 5.7 mmol, 60%) was slowly added to solution of methyl 1H-indole-2-carboxylate (500.0 mg, 2.85 mmol) and 1-(bromomethyl)-4-chlorobenzene (586.5 mg, 2.85 mmol) in DMF (30 mL) at ambient temperature. The solution was heated to 60 °C for 8 h. After confirming that the reaction was completed by TLC analysis, the mixture was extracted with ethyl acetate and washed with brine. The organic phase was dried over anhydrous Na2SO4 and concentrated. The crude product was purified by silica gel column chromatography to give the target product 14.4.9 Methyl 4-(4-chlorobenzyl)-4H-thieno[3,2-b]pyrrole-5-carboxylate (15)
Sodium hydride (220.7 mg, 5.52 mmol, 60%) was slowly added to a solution of methyl 4H-thieno[3,2-b]pyrrole-5-carboxylate (500.0 mg, 2.76 mmol) and 1-(bromomethyl)-4-chlorobenzene (566.9 mg, 2.76 mmol) in DMF (30 mL) at ambient temperature. The reaction mixture was stirred at 60 °C for 8 h. After confirming that the reaction was completed by TLC analysis, the mixture was extracted with ethyl acetate and washed with brine. The organic phase was dried over anhydrous Na2SO4 and concentrated. The crude product was purified by silica gel column chromatography to give the target product 15.4.10 General procedure for the synthesis of compounds 16a–i
Anhydrous potassium carbonate (2 equiv.) was added to a solution of methyl 3-chloropyrazine-2-carboxylate (1 equiv.) and substituted phenol (1 equiv.) in DMF at ambient temperature. The reaction mixture was stirred at 110 °C for 5 h. After confirming that the reaction was completed by TLC analysis, the mixture was extracted with ethyl acetate and washed with brine. The organic phase was dried over anhydrous Na2SO4 and concentrated. The crude product was purified by silica gel column chromatography to give the target product 16a–i.4.11 General procedure for the synthesis of compounds (17)
2 N sodium hydroxide was added to a solution of intermediate 13–15 or 16a–i (1 equiv.) in methanol at ambient temperature. The reaction mixture was stirred for 4 h and the methanol was concentrated under reduced pressure. The resultant mixture was adjusted to pH = 5–6 with 1 N HCl solution, the precipitated white solid was collected by filtration and dried to give the carboxylic acid intermediate 17.4.12 General procedure for the synthesis of compounds 18a–u
HATU (1.1 equiv.) and DIEA (2 equiv.) were added to a solution of intermediate 12 or substituted aniline (1 equiv.) in anhydrous DMF. The solution was heated to 70 °C for 6–10 h and then cooled to room temperature. The reaction mixture was poured into ice water, and the resulting solid was filtered. The crude product was purified by silica gel column chromatography to give the target product 18a–u.4.13 Cell-based TGR5 agonism assay
The lentivirus vector expressing human TGR5 (NM_001077191.1) and mouse (NM_174985) were provided by Hanbio Biotechnology Co. Ltd. HEK293T cells were transfected with reconstructed lentivirus plasmids and stable cell line was generated by puromycin selection. INT-777 was used as reference compound. 20 mM stock solutions of test and reference compounds were prepared in DMSO. TGR5-mediated cAMP generation was measured using a homogenous time-resolved fluorescence (HTRF) cAMP dynamic 2 assay kit (Cisbio cat #62AM4PEB) following the manufacturer's instructions.4.14 Oral glucose tolerance test (OGTT)
Male C57L/6J mice (6–8 weeks old) were obtained from Beijing Vital River Laboratory Animal Technologies Co. Ltd and were randomly divided into 2 groups (n = 8 in each group). After being fasted overnight, C57L/6J mice were orally administered the vehicle (corn oil) or test compound 18k at 50 mg kg−1, followed by an oral glucose load (4 g kg−1) at 60 min post compound dose. Blood glucose levels were measured via blood drops obtained by clipping the tail of the mice before compound dosing and 0, 15, 30, 60, and 120 min post glucose loading. Blood glucose levels and glucose AUC were carried out in GraphPad Prism.4.15 In vitro GLP-1 secretion assay
Human NCI-H716 cells were grown in high glucose DMEM with 10% fetal bovine serum. Cells were incubated at 37 °C in a humidified chamber containing 5% CO2. 20 μM DPP IV (Vildagliptin) was included in the secretion buffer to prevent GLP-1 degradation. The cells were treated with 0.1% DMSO as solvent control or compound 18k (4 μM) for 2 h. Media was collected and GLP-1 levels was measured using the active GLP-1 ELISA kit according to the manufacturer's instructions.4.16 GLP-1 secretion
To examine the in vivo effect of compound 18k on GLP-1 secretion, all of the overnight-fasted eight week-old C57L/6J mice were received dipeptidyl peptidase IV (DPP IV) inhibitor (3.0 mg kg−1) challenge, 1 h later, vehicle (CMCNa) or test compound 18k at 50 mg kg−1 was administered orally to mice. 4 h later, blood samples were collected and placed into Eppendorf tubes containing the dipeptidyl peptidase IV (DPP IV) inhibitor with a final concentration of 1% blood samples and 25 mg mL−1 EDTA to measure serum active GLP-1[7–36 amide] levels.Author contributions
S. Z. and L. W. contributed equally to this work. W.-D. C., R. H. and S. Z. conceived and designed the project. S. Z. and L. W. wrote the manuscript. S. Z., L. W., J. W., C. W., S. Z., Y. F. and Y. L. carried out the methodology, calculation, prepared the original draft. R. H., D. Y. and Y.-D. W. edited and revised the manuscript. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work were supported by the National Natural Science Foundation of China (Grant No. 81903444, 81970726, 81472232, 81970551 and 81672433), the China Postdoctoral Science Foundation (Grant No. 2018M640674), the Postdoctoral Research Grant in Henan Province (Grant No. 201902027), the Key Program for Science and Technology of Henan Province (Grant No. 192102310145 and 202102310177), College Students' Innovative Entrepreneurial Training Plan Program (No. 20210475084 and 202110475050), Henan Provincial Natural Science Foundation (Grant No. 182300410323), First Class Discipline Cultivation Project of Henan University (Grant No. 2019YLZDYJ19) and Plan for Scientific Innovation Talent of Henan Province (Grant No. 154100510004), Major Scientific and Technological Innovation Project of Hebei, the Fundamental Research Funds for the Central Universities and Research projects on biomedical transformation of China-Japan Friendship Hospital (Grant No. PYBZ1803).References
- H. Duboc, Y. Tache and A. F. Hofmann, Dig. Liver Dis., 2014, 46, 302–312 CrossRef CAS PubMed.
- C. Guo, W. D. Chen and Y. D. Wang, Front. Physiol., 2016, 7, 646 Search PubMed.
- T. Maruyama, Y. Miyamoto, T. Nakamura, Y. Tamai, H. Okada, E. Sugiyama, T. Nakamura, H. Itadani and K. Tanaka, Biochem. Biophys. Res. Commun., 2002, 298, 714–719 CrossRef CAS PubMed.
- X. Q. Zhang, M. Wall, Z. H. Sui, J. Kauffman, C. F. Hou, C. L. Chen, F. Y. Du, T. Kirchner, Y. Liang, D. L. Johnson, W. V. Murray and K. Demarest, ACS Med. Chem. Lett., 2017, 8, 560–565 CrossRef CAS PubMed.
- T. Harach, T. W. H. Pols, M. Nomura, A. Maida, M. Watanabe, J. Auwerx and K. Schoonjans, Sci. Rep., 2012, 2, 430 CrossRef PubMed.
- S. Katsuma, A. Hirasawa and G. Tsujimoto, Biochem. Biophys. Res. Commun., 2005, 329, 386–390 CrossRef CAS PubMed.
- J. Y. L. Chiang, Liver Res., 2017, 1, 3–9 CrossRef PubMed.
- V. Keitel, J. Stindt and D. Haussinger, Handb. Exp. Pharmacol., 2019, 256, 19–49 CAS.
- F. G. Schaap, M. Trauner and P. L. M. Jansen, Nat. Rev. Gastroenterol. Hepatol., 2014, 11, 55–67 CrossRef CAS PubMed.
- Y. P. Xu, J. Med. Chem., 2016, 59, 6553–6579 CrossRef CAS PubMed.
- R. J. Hodge and D. J. Nunez, Diabetes, Obes. Metab., 2016, 18, 439–443 CrossRef CAS PubMed.
- A. Gioiello, E. Rosatelli, R. Nuti, A. Macchiarulo and R. Pellicciari, Expert Opin. Ther. Pat., 2012, 22, 1399–1414 CrossRef CAS PubMed.
- Y. Yun, C. L. Zhang, S. M. Guo, X. Y. Liang, Y. Lan, M. Wang, N. Zhuo, J. P. Yin, H. A. Liu, M. Gu, J. Li, X. Xie and F. J. Nan, J. Med. Chem., 2021, 64, 12181–12199 CrossRef CAS PubMed.
- R. Pellicciari, A. Gioiello, A. Macchiarulo, C. Thomas, E. Rosatelli, B. Natalini, R. Sardella, M. Pruzanski, A. Roda, E. Pastorini, K. Schoonjans and J. Auwerx, J. Med. Chem., 2009, 52, 7958–7961 CrossRef CAS PubMed.
- T. T. Li, S. R. Holmstrom, S. Kir, M. Umetani, D. R. Schmidt, S. A. Kliewer and D. J. Mangelsdorf, Mol. Endocrinol., 2011, 25, 1066–1071 CrossRef CAS PubMed.
- K. A. Evans, B. W. Budzik, S. A. Ross, D. D. Wisnoski, J. Jin, R. A. Rivero, M. Vimal, G. R. Szewczyk, C. Jayawickreme, D. L. Moncol, T. J. Rimele, S. L. Armour, S. P. Weaver, R. J. Griffin, S. M. Tadepalli, M. R. Jeune, T. W. Shearer, Z. B. B. Chen, L. H. Chen, D. L. Anderson, J. D. Becherer, M. De Los Frailes and F. J. Colilla, J. Med. Chem., 2009, 52, 7962–7965 CrossRef CAS PubMed.
- M. R. Herbert, D. L. Siegel, L. Staszewski, C. Cayanan, U. Banerjee, S. Dhamija, J. Anderson, A. Fan, L. Wang, P. Rix, A. K. Shiau, T. S. Rao, S. A. Noble, R. A. Heyman, E. Bischoff, M. Guha, A. Kabakibi and A. B. Pinkerton, Bioorg. Med. Chem. Lett., 2010, 20, 5718–5721 CrossRef CAS PubMed.
- B. W. Budzik, K. A. Evans, D. D. Wisnoski, J. Jin, R. A. Rivero, G. R. Szewczyk, C. Jayawickreme, D. L. Moncol and H. S. Yu, Bioorg. Med. Chem. Lett., 2010, 20, 1363–1367 CrossRef CAS PubMed.
- Q. A. Zou, H. L. Duan, M. M. Ning, J. Liu, Y. Feng, L. M. Zhang, J. J. Zhu, Y. Leng and J. H. Shen, Eur. J. Med. Chem., 2014, 82, 1–15 CrossRef CAS PubMed.
- S. Z. Zhao, X. P. Li, L. Wang, W. J. Peng, W. L. Ye, W. G. Li, Y. D. Wang and W. D. Chen, Bioorg. Med. Chem., 2021, 32, 115972 CrossRef CAS PubMed.
- T. Chen, N. W. Reich, N. Bell, P. D. Finn, D. Rodriguez, J. Kohler, K. Kozuka, L. M. He, A. G. Spencer, D. Charmot, M. Navre, C. W. Carreras, S. Koo-McCoy, J. Tabora, J. S. Caldwell, J. W. Jacobs and J. G. Lewis, J. Med. Chem., 2018, 61, 7589–7613 CrossRef CAS PubMed.
- A. Nakhi, C. M. McDermott, K. L. Stoltz, K. John, J. E. Hawkinson, E. A. Ambrose, A. Khoruts, M. J. Sadowsky and P. I. Dosa, J. Med. Chem., 2019, 62, 6824–6830 CrossRef CAS PubMed.
- F. Yang, C. Y. Mao, L. L. Guo, J. Y. Lin, Q. Q. Ming, P. Xiao, X. Wu, Q. Y. Shen, S. M. Guo, D. D. Shen, R. R. Lu, L. Q. Zhang, S. M. Huang, Y. Q. Ping, C. L. Zhang, C. Ma, K. Zhang, X. Y. Liang, Y. M. Shen, F. J. Nan, F. Yi, V. C. Luca, J. Y. Zhou, C. T. Jiang, J. P. Sun, X. Xie, X. Yu and Y. Zhang, Nature, 2020, 587, 499–504 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08867j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000