Open Access Article
Open Access ArticleAcridine-based dyes as high-performance near-infrared Raman reporter molecules for cell imaging†
Jiasheng Du‡
,
Jinming Li‡,
Yuzhan Li,
Dong Wang ,
Hui Cao
,
Hui Cao ,
Wanli He
,
Wanli He and
Yang Zhou
and
Yang Zhou *
*
Department of Materials Physics and Chemistry, School of Materials Science and Engineering, University of Science and Technology Beijing, Beijing 100083, China
First published on 25th January 2022
Abstract
A surface-enhanced Raman scattering (SERS) nanoprobe has been proven to be a promising tool for near-infrared (NIR) biomedical imaging and diagnosis because of its high sensitivity and selectivity. However, the development of NIR SERS reporters has been a bottleneck impeding the preparation of ultrasensitive SERS probes. Herein, we report the design and synthesis of a series of SERS reporters in the NIR region based on 10-methylacridine (AD). The AD nanotags (gold nanostar–AD molecules–BSA, AuNS–AD–BSA) exhibit appreciable SERS signals and can be detected at as low as the sub-picomole level. The results of in vitro imaging experiments show that it can be used in live-cell delineation.
Introduction
Surface-enhanced Raman spectroscopy (SERS) in biomedical applications has attracted growing attention because of its excellent sensitivity and selectivity. To generate highly sensitive Raman signals, it is crucial to choose Raman reporters with the desired optical properties.1 Taking advantage of the chemical enhancement in surface-enhanced resonance Raman scattering (SERRS), the resonance effect between the electronic absorbance band of the Raman reporters and the excitation wavelength of the laser source can be improved, allowing for the increase of SERS signals by 10- to 100-fold.2,3 Furthermore, in the biomedical field, NIR lasers are preferred for SERS-based detection because of their weak autofluorescence and photobleaching in the NIR window.2,4–11However, there are only a few available Raman reporter molecules of which maximum absorption wavelength is resonant with NIR laser.12–16 An effective strategy to redshift the absorption wavelength is to introduce proper chromophores to extend the conjugated structure. Unfortunately, most chromophores lead to high fluorescence background, which may cover the Raman signal. Mahmood et al.17 and Yan et al.18 developed a library of dyes with a N-substituted acridinium moiety exhibiting a moderate or even no emission of fluorescence. Yang et al.19,20 reported that the introduction of a 10-methylacridine substituent accounts for a redshift of the maximum absorption wavelength. Although the acridine compounds induce a redshift in the visible region and enable an intense Raman signal, the dyes cannot produce SERS effect in the NIR region.
In this work, we designed and synthesized a series of polymethine dyes with donor–acceptor–donor (D–A–D) structures based on 10-methylacridine substituents. The dyes (CnAD dyes) were prepared in high yield, which exhibited good stability and maximum absorption wavelengths in the NIR region. Gold nanostar (AuNS) were used to modify the dyes, allowing for an extra enhancement via SERRS and resulting in a strong signal and high sensitivity to a 785 nm excitation. The limits of detection (LOD) of the nanotags can be detected in the sub-picomole range. The SERS images indicate the potential of AuNS–C5AD–BSA (gold nanostar–C5AD dye–BSA) in live cancer cell SERS imaging.
Experimental
Materials
Dichloromethane (DCM), methanol (MeOH), ethyl acetate (EtOAc) and acetic acid were purchased from Peking Reagent. Hydrogen tetrachloroaurate hydrate (HAuCl4·4H2O), ammonia water, and acetic anhydride (Ac2O) were obtained from Sinopharm Chemical Reagent Co., Ltd. Trisodium citrate dihydrate was obtained from Beijing Chemical Works. Zinc chloride (ZnCl2, 98%), acetonitrile (99%) and N,N-diphenylformamidine (98%) were obtained from Macklin. Silver nitrate (AgNO3, 99%) was obtained from GHTECH. Diphenylamine (99%), magnesium sulfate (99.5%), sodium acetate (AcONa, 99%), methyl iodide, N-[5-(phenylamino)-2,4-pentadienylidene]aniline monohydrochloride (98%) and PEG-SH (average molecular weight, 5000 g mol−1) were obtained from J&K science. Ascorbic acid (AA, >99.0%), 3,3′-diethylthiatricarbocyanine iodide (DTTC) and bovine serum albumin (BSA) were obtained from Aladdin. Calcein-AM/PI cell cytotoxicity assay kit were obtained from Beyotime Institute of Biotechnology (China). DMEM, OPTI-MEM and phosphate buffered saline (PBS) were purchased from GIBCO Invitrogen Corp.Instruments
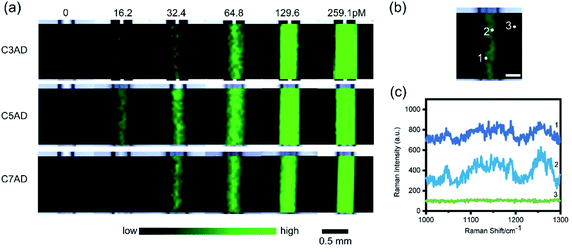
Transmission electron microscopy (TEM, JEM-1200EX, JEOL, Japan) was used to characterize morphologies of the AD nanotags. Dynamic light scattering (DLS) and zeta potential were measured using Zetasizer Nano-ZS (Malvern, UK). Absorption properties of the nanoparticles was measured using a UV-vis-NIR spectrophotometer (V-570, Jasco, Japan). SERS measurements were recorded on a confocal Raman microscope (inVia Reflex, Renishaw, UK) equipped with a 785 nm laser. The Raman measurements of AD and DTTC nanotags were conducted using the following parameters: 50 L × objective lens, 0.85 mW laser power, and 10 s exposure time. The limit of detection (LOD) was evaluated via 50 L × objective lens with 1.7 mW laser power and 10 s exposure time. The SERS images of AD nanotags were acquired under a 5× objective lens with 1.7 mW laser power on the samples, 2 s exposure time and 50 μm step size. The in vitro cell SERS imaging was carried out using a 100× objective lens at 8.5 mW laser power with 5 s exposure time and 3 μm step size. The SERS images of nanotags and cells were created with the intensity of 1277 cm−1 SERS band after removing the baseline via wire 5.1.Reporter library synthesis and properties
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give pure 1 (4.2 g, mmol, 72% yield). δH (500 MHz, CDCl3) 8.29 (2H, d), 8.26 (2H, d, J 8.7), 7.80 (2H, t, J 8.1, 6.6, 1.4), 7.59 (2H, t), 3.16 (3H, s). EI-MS: m/z 194.09.
1) to give pure 1 (4.2 g, mmol, 72% yield). δH (500 MHz, CDCl3) 8.29 (2H, d), 8.26 (2H, d, J 8.7), 7.80 (2H, t, J 8.1, 6.6, 1.4), 7.59 (2H, t), 3.16 (3H, s). EI-MS: m/z 194.09.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give pure C3AD (16.1 mg, 0.0379 mmol, 50.5% yield). δH (600 MHz, DMSO) 8.39 (4H, d, J 7.7), 8.07 (1H, d, J 9.0), 8.00 (4H, d, J 8.8), 7.93 (4H, d, J 7.9), 7.85 (1H, s), 7.58 (4H, t, J 7.6), 7.26 (1H, q, J 7.9, 7.1), 4.13 (6H, s). EI-MS: m/z 425.20 [M]+.
1) to give pure C3AD (16.1 mg, 0.0379 mmol, 50.5% yield). δH (600 MHz, DMSO) 8.39 (4H, d, J 7.7), 8.07 (1H, d, J 9.0), 8.00 (4H, d, J 8.8), 7.93 (4H, d, J 7.9), 7.85 (1H, s), 7.58 (4H, t, J 7.6), 7.26 (1H, q, J 7.9, 7.1), 4.13 (6H, s). EI-MS: m/z 425.20 [M]+.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give pure C5AD (19.3 mg, 0.0428 mmol, 57.1% yield). δH (500 MHz, DMSO) 8.26 (4H, d, J 8.2), 8.05 (1H, t, J 9.0), 7.96 (4H, d, J 8.6), 7.89 (4H, m), 7.58 (1H, t, J 3.7), 7.53 (4H, d, J 7.7), 7.45 (2H, d, J 13.6), 7.10 (1H, t, J 12.3), 4.12 (6H, s). EI-MS: m/z 449.4 [M]+.
1) to give pure C5AD (19.3 mg, 0.0428 mmol, 57.1% yield). δH (500 MHz, DMSO) 8.26 (4H, d, J 8.2), 8.05 (1H, t, J 9.0), 7.96 (4H, d, J 8.6), 7.89 (4H, m), 7.58 (1H, t, J 3.7), 7.53 (4H, d, J 7.7), 7.45 (2H, d, J 13.6), 7.10 (1H, t, J 12.3), 4.12 (6H, s). EI-MS: m/z 449.4 [M]+.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give pure C7AD (14.8 mg, 0.0310 mmol, 41.4% yield). δH (500 MHz, DMSO) 8.22 (4H, m), 7.97 (4H, m), 7.89 (4H, dd, J 7.9, 6.2), 7.53 (4H, m), 7.43 (2H, d, J 13.6), 7.29 (2H, d, J 12.6), 7.19 (1H, dd, J 26.4, 13.3), 6.85 (2H, t, J 12.7), 4.12 (6H, s). EI-MS: m/z 477.4 [M]+.
1) to give pure C7AD (14.8 mg, 0.0310 mmol, 41.4% yield). δH (500 MHz, DMSO) 8.22 (4H, m), 7.97 (4H, m), 7.89 (4H, dd, J 7.9, 6.2), 7.53 (4H, m), 7.43 (2H, d, J 13.6), 7.29 (2H, d, J 12.6), 7.19 (1H, dd, J 26.4, 13.3), 6.85 (2H, t, J 12.7), 4.12 (6H, s). EI-MS: m/z 477.4 [M]+.Preparation of nanotags
Cell culture and biocompatibilities studies
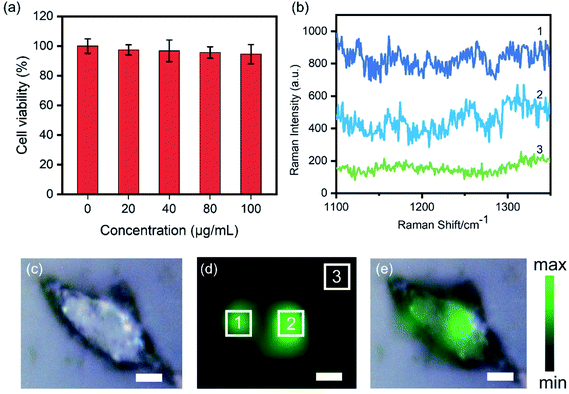
A549 cells were cultured in DMEM medium containing 10% FBS and 1% penicillin/streptomycin at 37 °C in 5% CO2. Cell survival rates of AuNS–C5AD-BSA were evaluated by MTT assays. 1 × 105 cells were seeded into 96-well plates and then allowed to grow for 24 h. The medium was replaced by fresh cell culture medium containing different concentrations (0, 20, 40, 80 and 100 μg mL−1) of AuNS–C5AD-BSA. After incubation for 24 h, the medium was removed and the cells were washed with PBS. Subsequently, MTT was added into 96-well plates. After 4 h, the MTT was removed, 100 μL DMSO was added. Then, the absorbance was measured using a microplate reader.Cellular SERS mapping
1 × 104 A549 cells were seeded onto quartz slides sitting on a 24-well plate overnight. After another 10 h-incubation of 1 mL culture medium containing AuNS–C5AD-BSA ([NP] ∼ 1 × 109 NP mL−1), cells were washed with PBS three times to remove medium and dissociative particles.Results and discussion
Synthesis and characterization of dyes
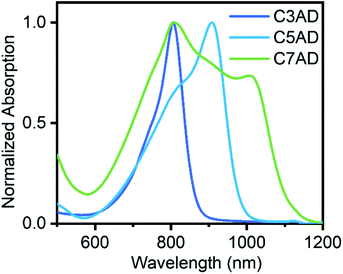
The synthetic routes of AD dyes are shown in Fig. 1a, 9,10-dimethylacridin-10-ium iodide (2, ADI) was synthesized by iodomethane and 9-methylacridine (1, AD), which was prepared using diphenylamine and acetic acid. Acridine dyes were synthesized via a reaction between ADI and electrophile with good yield, ranging from 41.4% to 51.7%. The maximal absorbing wavelengths of C3AD and C5AD in CH2Cl2 are 804 nm and 908 nm, respectively. When the number of methine groups was increased, a redshift of approximately 100 nm of the absorption wavelengths was observed. In chloroform (CHCl3) and ethanol (EtOH), the absorption peak around 804 nm was observed. In addition, there are two characteristic absorption peaks of C7AD in 804 nm and 1006 nm. AD dyes are better coplanar and tend to aggregate face-to-face in the solution (H-aggregation) with the increasing length of the polymethine chain. This phenomenon, with two close-maxima absorption peaks, arises from H-aggregation which leads to weaker fluorescence and facilitates Raman measurement.17,23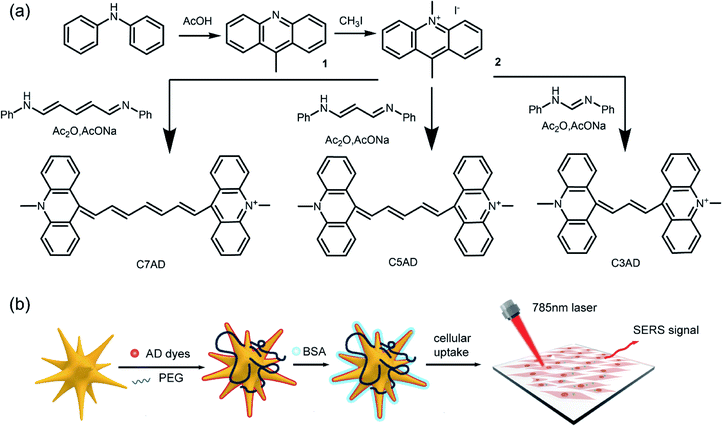 | ||
| Fig. 1 (a) Synthetic routes of AD dyes. (b) Fabrication of NIR Raman reporter molecules modified SERS nanotags. | ||
Synthesis and characterization of nanotags
AuNSs with a localized surface plasmon resonance (LSPR) band at around 730 nm were synthesized by seed-mediated growth.21 Subsequently, ethanol solution containing different Raman reporters (C3AD, C5AD, and C7AD) at a concentration between 2 × 10−9 and 2 × 10−4 M were added to AuNSs, respectively. The reporter molecules were adsorbed to the negatively charged AuNSs surface through electrostatic interaction via the nitrogen atom.24 Then, AuNS–AD (with 2 × 10−5 M dyes) was modified by BSA to improve biocompatibility. After modified with dyes and BSA, plasmon resonance absorption of nanotags was observed to redshift by ≈ 22 nm and ≈ 32 nm (Fig. S2a, d and f†). As shown in Fig. S2b, e and h,† after functionalization and modification, the hydrodynamic diameter increased ∼26 nm and ∼43 nm in the dominant peak compared with the pristine AuNSs. The zeta potential measurements showed that the AuNS–AD was less negatively charged than that of the AuNSs, and that the addition of BSA also resulted in a decrease in negatively charged ions on the surface of AuNSs (Fig. S2c, f and i†). These results indicated that AuNSs were successfully coated with dyes and BSA. Transmission electron microscopy (TEM) was used to reveal the star-like structure of nanotags (Fig. S3†), which exhibited good monodispersity (Table 1).| Dyes | Dye λmax (CH2Cl2) | ε, 104 M−1 cm−1 (CH2Cl2) | Synthesis yield (%) |
|---|---|---|---|
| C3AD | 804 nm | 2.4 | 50.5 |
| C5AD | 908 nm (804 nm) | 2.8 | 51.7 |
| C7AD | 804 nm (1006 nm) | 8.2 | 41.4 |
SERS spectra and SERS nanotags with dyes
The nitrogen atoms in the acridine group provide a binding site for AuNSs. Since these AD dyes have similar structures, the fingerprint information of their Raman spectra highly overlaps. Weak signals of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching and C
N stretching and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching appeared around 1600 cm−1. Signals in the range of 1385–1415 cm−1 was attributed to the acridine stretching. Peaks around 1250–1277 cm−1 arise from the ring deformation of acridinium. Signals for C–H stretching vibrations at 700–900 cm−1 are visible. The signal around 400 cm−1 may refer to the bending of N–C
C stretching appeared around 1600 cm−1. Signals in the range of 1385–1415 cm−1 was attributed to the acridine stretching. Peaks around 1250–1277 cm−1 arise from the ring deformation of acridinium. Signals for C–H stretching vibrations at 700–900 cm−1 are visible. The signal around 400 cm−1 may refer to the bending of N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C.25 These AD dyes, with similar SERS spectra, vary in the intensity of SERS nanotags. This confirms that Raman reporters lead to an extra signal enhancement when the absorption peak is resonant with laser source.
C.25 These AD dyes, with similar SERS spectra, vary in the intensity of SERS nanotags. This confirms that Raman reporters lead to an extra signal enhancement when the absorption peak is resonant with laser source.
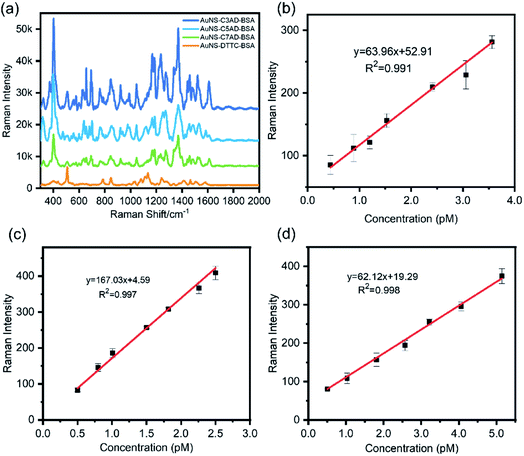
To fully estimate the SERS capabilities of AuNS–AD–BSA nanotags, the LODs was measured and AuNS–AD–BSA nanotags were compared with AuNS–DTTC–BSA nanotags. To study the LOD of the nanotags, AuNSs modified by proper concentration of dye molecules and BSA was synthesized and was diluted until no signal of the nanotags was observed. The peak at 1277 cm−1, which was one of the intense peaks in the spectrum, was used to calculate the LOD. LODs of nanotags were calculated to be approximately 0.4 pM, 0.50 pM, and 0.51 pM, respectively (n = 3). It shows a linear relationship between the concentration of nanotags and Raman intensity with good R2 values (Fig. 2b–d, R2 = 0.991, 0.997, and 0.998), indicating the possibility of quantitative analysis (Fig. 3).
After confirming the SERS sensitivity of the AuNS–AD–BSA nanotags, a commercial Raman reporter DTTC was chosen to substitute the synthesized dyes as the SERS reporter molecule. As shown in Fig. 2a, the contrast ratio between AuNS–AD–BSA nanotags and AuNS–DTTC–BSA nanotags expression was quantified to be approximately 1.7-fold to 4.7-fold based on the same test conditions. This difference may arise from the acridine group of Raman reporters and greater electromagnetic fields due to electrostatic interaction between organic salt and metal surface.26
In vitro experiments
Conclusions
In conclusion, we have designed and synthesized a set of NIR Raman reporters based on 10-methylacridine. The AD dyes are bond to AuNS via nitrogen atoms and LOD of the SERS nanotags (AuNS–CnAD–BSA) is as low as 0.4 pM. The intensity of these nanotags is approximately 5-fold higher than that of a commercial Raman reporter DTTC. The SERS images revealed the promising application of C5AD in biomedical measurement with a 785 nm laser as the excitation source. These significant results demonstrate the great potential of AD dyes as Raman reporters for biomedical imaging and selective detection and accurate resection of cancer cell. The AD dyes can be functionalized with pH responsive substituent and applied in the tracking pH variations and tumor delineation in the future experiments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (51973017, 51773017), the State Key Laboratory for Advanced Metals and Materials (2018Z-06), and the Fundamental Research Funds for the Central Universities (FRF-DF-19-001).Notes and references
- Y. Wang and S. Schlucker, Analyst, 2013, 138, 2224–2238 RSC.
- X. Qian, X. H. Peng, D. O. Ansari, Q. Yin-Goen, G. Z. Chen, D. M. Shin, L. Yang, A. N. Young, M. D. Wang and S. Nie, Nat. Biotechnol., 2008, 26, 83–90 CrossRef CAS PubMed.
- V. Amendola and M. Meneghetti, Adv. Funct. Mater., 2012, 22, 353–360 CrossRef CAS.
- S. Harmsen, M. A. Bedics, M. A. Wall, R. Huang, M. R. Detty and M. F. Kircher, Nat. Commun., 2015, 6, 6570 CrossRef CAS PubMed.
- C. Andreou, V. Neuschmelting, D. F. Tschaharganeh, C. H. Huang, A. Oseledchyk, P. Iacono, H. Karabeber, R. R. Colen, L. Mannelli, S. W. Lowe and M. F. Kircher, ACS Nano, 2016, 10, 5015–5026 CrossRef CAS PubMed.
- X. Jin, B. N. Khlebtsov, V. A. Khanadeev, N. G. Khlebtsov and J. Ye, ACS Appl. Mater. Interfaces, 2017, 9, 30387–30397 CrossRef CAS PubMed.
- C. Song, F. Li, X. Guo, W. Chen, C. Dong, J. Zhang, J. Zhang and L. Wang, J. Mater. Chem. B, 2019, 7, 2001–2008 RSC.
- D. Jimenez de Aberasturi, A. B. Serrano-Montes, J. Langer, M. Henriksen-Lacey, W. J. Parak and L. M. Liz-Marzán, Chem. Mater., 2016, 28, 6779–6790 CrossRef CAS.
- D. Advanced MaterialsJimenez de Aberasturi, M. Henriksen-Lacey, L. Litti, J. Langer and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 1909655 CrossRef.
- Y. Wen, V. X. Truong and M. Li, Nano Lett., 2021, 21, 3066–3074 CrossRef CAS PubMed.
- S. Hua, S. Zhong, A. Hamed, J. He, D. Zhong, D. Zhang, X. Chen, J. Qian, X. Hu and M. Zhou, Adv. Funct. Mater., 2021, 2100468, DOI:10.1002/adfm.202100468.
- M. A. Bedics, H. Kearns, J. M. Cox, S. Mabbott, F. Ali, N. C. Shand, K. Faulds, J. B. Benedict, D. Graham and M. R. Detty, Chem. Sci., 2015, 6, 2302–2306 RSC.
- H. Kearns, M. A. Bedics, N. C. Shand, K. Faulds, M. R. Detty and D. Graham, Analyst, 2016, 141, 5062–5065 RSC.
- R. Javaid, N. Sayyadi, K. Mylvaganam, K. Venkatesan, Y. Wang and A. Rodger, J. Raman Spectrosc., 2020, 51, 2408–2415 CrossRef CAS.
- A. Samanta, K. K. Maiti, K. S. Soh, X. Liao, M. Vendrell, U. S. Dinish, S. W. Yun, R. Bhuvaneswari, H. Kim, S. Rautela, J. Chung, M. Olivo and Y. T. Chang, Angew. Chem., Int. Ed., 2011, 50, 6089–6092 CrossRef CAS PubMed.
- K. K. Maiti, U. S. Dinish, A. Samanta, M. Vendrell, K.-S. Soh, S.-J. Park, M. Olivo and Y.-T. Chang, Nano Today, 2012, 7, 85–93 CrossRef CAS.
- T. Mahmood, A. Paul and S. Ladame, J. Org. Chem., 2010, 75, 204–207 CrossRef CAS PubMed.
- P. Yan, A. Xie, M. Wei and L. M. Loew, J. Org. Chem., 2008, 73, 6587–6594 CrossRef CAS PubMed.
- Y. Zhou, D. Chenchen, D. Jinshuai, Q. Ximei, C. Hui, W. Dong and H. Wanli, China Patent, CN107090190A, 2015 Search PubMed.
- Y. Zhou, D. Jinshuai, D. Yiqiang and Q. Ximei China Patent, CN104387790A, 2017 Search PubMed.
- H. Yuan, C. G. Khoury, H. Hwang, C. M. Wilson, G. A. Grant and T. Vo-Dinh, Nanotechnology, 2012, 23, 075102 CrossRef PubMed.
- L. Scarabelli, A. Sanchez-Iglesias, J. Perez-Juste and L. M. Liz-Marzan, J. Phys. Chem. Lett., 2015, 6, 4270–4279 CrossRef CAS PubMed.
- B. A. Armitage, in DNA Binders and Related Subjects, 2005, ch. 3, pp. 55–76, DOI:10.1007/b100442.
- J. P. G. IKvi, J. P. Marsault and J. Aubard, J. Raman Spectrosc., 1993, 745–752 Search PubMed.
- B. Hao, X. Bu, J. Wu, Y. Ding, L. Zhang, B. Zhao and Y. Tian, Microchem. J., 2020, 155, 104736 CrossRef CAS.
- J. I. Millán, J. V. Garcia-Ramos, S. Sanchez-Cortes and R. Rodríguez-Amaro, J. Raman Spectrosc., 2003, 34, 227–233 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08827k |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |