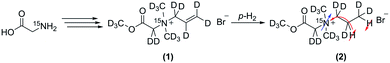Open Access Article
Open Access ArticleHyperpolarization of 15N in an amino acid derivative†
Philip Saul ab,
Salvatore Mamone
ab,
Salvatore Mamone ab and
Stefan Glöggler
ab and
Stefan Glöggler *ab
*ab
aResearch Group for NMR Signal Enhancement, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37 077 Göttingen, Germany. E-mail: stefan.gloeggler@mpibpc.mpg.de; Tel: +49 551 3961 108
bCenter for Biostructural Imaging of Neurodegeneration, Von-Siebold-Straßze 3A, 37 075 Göttingen, Germany
First published on 17th January 2022
Abstract
Hyperpolarization is a nuclear magnetic resonance (NMR) technique which can be used to significantly enhance the signal in NMR experiments. In recent years, the possibility to enhance the NMR signal of heteronuclei by the use of para-hydrogen induced polarization (PHIP) has gained attention, especially in the area of possible applications in magnetic resonance imaging (MRI). Herein we introduce a way to synthesize a fully deuterated, 15N labelled amino acid derivative and the possibility to polarize the 15N by means of hydrogenation with para-hydrogen to a polarization level of 0.18%. The longevity of the polarization with a longitudinal relaxation time of more than a minute can allow for the observation of dynamic processes and metabolic imaging in vivo. In addition, we observe the phenomenon of proton–deuterium exchange with a homogeneous catalyst leading to signal enhanced allyl moeities in the precursor.
Introduction
Nuclear magnetic resonance (NMR) is a versatile tool with possible applications ranging from chemical analysis over the observation of dynamic processes in e.g. biochemical research to medical diagnostics as magnetic resonance imaging (MRI). However, a major drawback, the inherently low sensitivity, still hampers the full potential of NMR. This is especially apparent in MRI, where mostly water is detected and the direct observation of other molecules is more difficult. A way to tackle this problem is hyperpolarization, which can increase the signal to noise by several orders of magnitude.1–9 Several techniques have been explored to generate hyperpolarization of which some have already been shown to be a useful tool in the aforementioned fields of research. One technique, dissolution dynamic nuclear polarization (d-DNP) can be used to generate hyperpolarized contrast agents for MRI.7–12 Another hyperpolarization technique with biomedical applications is spin-exchange optical pumping (SEOP), which can be used to polarize noble gases.13–16 Another possibility to generate hyperpolarization, that has been gaining a lot of attention in recent years, is para-hydrogen induced polarization (PHIP), a technique which allows for the transfer of nuclear spin order from para-hydrogen to other nuclei and thus generates a signal enhancement by several orders of magnitude.17–28 There are two different ways to utilize para-hydrogen in PHIP. One of the possibilities is the signal amplification by reversible exchange (SABRE) which uses dissolved para-hydrogen and allows for magnetization transfer via an intermediate transition metal complex to a substrate.28–33 Another way to make use of para-hydrogen is to hydrogenate an unsaturated bond in a substrate and subsequently transfer the magnetization to a heteronucleus in the same molecule.34–37 Especially in the case of small molecules, attaching an unsaturated sidearm to the compound one desires to polarize can be a facile way to access polarized compounds. After reacting the modified compound with para-hydrogen, the magnetization is transferred to a heteronucleus in the compound and the sidearm can be cleaved off by, e.g. a change in pH to obtain the hyperpolarized compound. For this technique it is essential, that the longitudinal relaxation time (T1) of the precursor and the compound alike, is significantly longer than the time it takes to cleave the sidearm. This method is referred to as para-hydrogen induced polarization by sidearm hydrogenation (PHIP-SAH).38–40 PHIP and PHIP-SAH can polarize metabolites in order to detect metabolic anomalies in medical diagnostics.25,27,41–46 In all cases it is desired to store the magnetization over long periods of time in order to be able to monitor the compounds for a long time. To achieve this, the magnetization is usually transferred to heteronuclei such as 13C or 15N since they usually display long T1. The use of 15N has been shown to be a promising approach, especially in quaternary ammonium compounds, which display long T1.26,47 One class of nitrogen-containing compounds which play a significant role in organisms are amino acids. Not only are they the building blocks of proteins but they also play a role in different metabolic cycles and therefor are an interesting target for hyperpolarization.48,49 To that extend, several techniques have been applied.37,50 Despite having hyperpolarized amino acids or their derivatives with PHIP by SABRE,51,52 PHIP53–55 and PHIP-SAH,50,56 so far no attempts have been conducted to hyperpolarize the 15N of the amino function of perdeuterated, 15N-labelled amino acid derivatives with long T1.Results and discussion
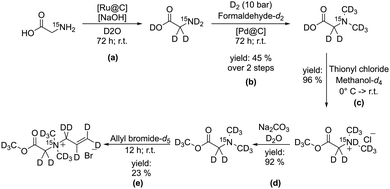
Herein we introduce an approach to obtain a perdeuterated amino acid derivative with residue attached to the amino function which allows for hydrogenation in order to hyperpolarize the 15N. We chose a perdeuterated allyl residue for that matter, which has been shown to be well suited for hydrogenation and consecutive magnetization transfer.47 Furthermore we modified the amino acid in a way to generate a quaternary ammonium function to obtain long T1 for long time monitoring in a potential setting for metabolic studies. This was achieved by adding two deuterated methyl residues to the amine function, which hinders possible relaxation through proton exchange with the solvent and heightens the symmetry around the 15N. We could show that the attached perdeuterated allyl sidearm can be hydrogenated with para-hydrogen and the thus obtained magnetization transferred to the 15N using the ESOTHERIC pulse sequence (Fig. 1).44,45At first we devised a synthetic approach to obtain the precursor molecule N-(2-(methoxy-d3)-2-oxoethyl-1.1-d2)-N,N-bis(methyl-d3)prop-2-ene-1-aminium-d5-N-15N bromide (1) in a multistep synthesis starting from glycine-15N. In a first reaction step we were able to exchange the glycine protons in a base catalyzed reaction over ruthenium on activated charcoal (Fig. 2a), which was reacted with formaldehyde-d2 in an Eschweiler–Clarke analogueous reaction to form N,N-dimethylglycine-15N-d9 (Fig. 2b). In the next steps, N,N-dimethylglycine-15N-d9 was reacted with methanol-d4 and thionyl chloride followed by transformation of the thus obtained chloride in basic conditions in D2O to form the perdeuterated methyl ester (Fig. 2d). In a final step, the N,N-dimethylglycine methyl ester-15N-d11 was reacted with allyl bromide-d5 to form (1).
Polarization experiments of (1) have been conducted in methanol-d4 with a homogeneous rhodium catalyst [Rh(dppb)(COD)][BF4] (dppb: diphenylphosphino butane, COD: cyclooctadiene) at 320 K and a 7 T magnetic field. At first, proton-NMR-experiments were run in order to estimate the coupling constants needed for efficient polarization transfer. Hydrogen gas was enriched in the para spin isomer at 99% by using a custom ordered Sumitomo generator operating at 20 K. Para-hydrogen was bubbled for 10 s at 7 bar directly into the NMR tube containing 2 mM of the catalyst and 1 mM of substrate via an automated console controlled delivery system.44
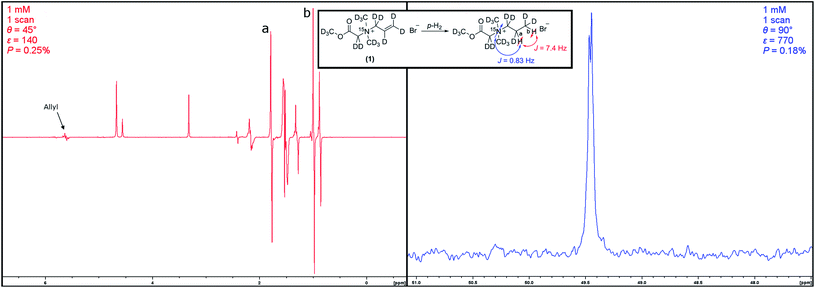
It was apparent, that several different hyperpolarized species were present in the sample after polarization (see Fig. 3). The observation that hyperpolarized anti-phase signal can be observed for the allyl protons, leads us to attribute this to a previously described hydrogenation–dehydrogenation mechanism.57 It should be noted that the mechanism described was observed on surfaces whereby here, a homogeneous catalyst is used. In this proposed model, the unsaturated bond can be hydrogenated and thus hyperpolarized. In another step, the hydrogenated compound can interact with the used catalyst again and be dehydrogenated. The then again unsaturated compound can be hydrogenated again, and so forth. By this mechanism, a proton–deuterium exchange can take place at the allyl function and species with different deuteration degrees can be present in the sample.
Mass spectrometry measurements have been performed on the sample solution before and after hydrogenation gain additional information on the nature of the observed species. Although masses could be detected for (2) as well as the labelled N,N-dimethyl glycine methyl ester for the sample after hydrogenation, the same was true for the sample before hydrogenation, making it rather difficult to definitely tell whether parts of the sample already degrade during the hydrogenation or this effect is solely attributals to the ionization process during the mass spectroscopy measurements. What was apparent though, was that the mass detected for the quaternary ammonium compound were the same before and after hydrogenation, which gives a further indication that the previously discussed hydrogenation–dehydrogenation mechanism takes place, since the molecular mass of (1) is the same than the molecular mass of (2) in which two additional deuterons have been exchanged for protons. The respective mass spectra are shown in the ESI.†
Despite the manifold of signals, we were able to identify the signals of hydrogenated (1) and determine the coupling constants for efficient magnetization transfer from the deuterium decoupled hyperpolarized proton spectrum. The coupling constants have been determined to be 3JH,H = 7.4 Hz with a chemical shift difference of 239 Hz (0.8 ppm) and 3JH,N = 0.84 Hz. Hydrogenation with para-hydrogen then yielded a proton polarization of 0.28%, a signal enhancement of 140-fold at 7 T. Transfer of the magnetization to the 15N was successful with an efficiency of 72%, generating a polarization of 0.18%, corresponding to a signal enhancement of 770-fold at a 7 T field. The relatively low polarization can be attributed to a constant change in the hyperpolarized compounds, leading to a distribution of the polarization. To the same extent the polarization available for transfer to 15N is limited by the same effect. From hyperpolarized (2), the T1 of 15N has been determined by running several scans at low flip angles and was shown to be T1 = 77 ± 30 s. An overview over the obtained data is shown in Table 1.
| ε1H | P1H [%] | ε15N | P15N [%] | T1(15N) [s] |
|---|---|---|---|---|
| 140 | 0.28 | 770 | 0.18 | 77 ± 30 |
Conclusion
In conclusion we were able to demonstrate the synthesis of a perdeuterated glycine derivative with a quaternary amino function by starting from glycine-15N. We could show that hydrogenation with para-hydrogen of the glycine derivative leads to hyperpolarized proton signals with a proton polarization of the hydrogenated compound of 0.28%. Interestingly, we also observed a proton–deuterium exchange with the homogeneous catalyst leading to enhanced allyl signals. The hyperpolarized protons on the alkyl moiety could be transferred efficiently to the 15N by using the ESOTHERIC pulse sequence. The relatively long T1 of the hydrogenated compound of T1 = 77 s gives rise to the possibility of using this and other analogues of amino acids as potential signal enhanced contrast agents. With this publication, we are able to define a starting point for further investigation of the hyperpolarization of 15N in amino acids for the aforementioned application and even their use in a possible MRI setup.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully thank the Max-Planck-Society and the Max-Planck-Institute for Biophysical Chemistry for generous funding and Prof. Christian Griesinger for access to his equipment and facilities. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (Grant agreement No. 949180).Notes and references
- J. H. Ardenkjær-Larsen, B. Fridlund, A. Gram, G. Hansson, L. Hansson, M. H. Lerche, R. Servin, M. Thaning and K. Golman, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10158–10163 CrossRef PubMed.
- K. Golman, R. in't Zandt and M. Thaning, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11270–11275 CrossRef CAS PubMed.
- S. E. Day, M. E. Kettunen, F. A. Gallagher, D.-E. Hu, M. Lerche, J. Wolber, K. Golman, J. H. Ardenkjær-Larsen and K. M. Brindle, Nat. Med., 2007, 13, 1382–1387 CrossRef CAS PubMed.
- S. Jannin, A. Bornet, R. Melzi and G. Bodenhausen, Chem. Phys. Lett., 2012, 549, 99–102 CrossRef CAS.
- X. Ji, A. Bornet, B. V. J. Milani, D. Gajan, A. J. Rossini, L. Emsley, G. Bodenhausen and S. Jannin, Nat. Commun., 2017, 8, 13975 CrossRef CAS PubMed.
- J. Milani, B. Vuichoud, A. Bornet, R. Melzi, S. Jannin and G. Bodenhausen, Rev. Sci. Instrum., 2017, 88, 015109 CrossRef PubMed.
- F. A. Gallagher, M. I. Kettunen, S. E. Day, D.-E. Hu, J. H. Ardenkjær-Larsen, R. in't Zandt, P. E. Jensen, M. Karlsson, K. Golman, M. E. Lerche and K. M. Brindle, Nature, 2008, 453, 940–943 CrossRef CAS PubMed.
- J. Kurhanewicz, D. B. Vigneron, K. M. Brindle, E. Y. Chekmenev, A. Comment, C. H. Cunningham, R. J. DeBernardinis, G. G. Green, M. O. Leach, S. S. Rajan, R. R. Rizi, B. D. Ross, W. S. Warren and C. R. Malloy, Neoplasia, 2011, 13, 81–97 CrossRef CAS PubMed.
- S. J. Nelson, J. Kurhanewicz, D. B. Vigneron, P. E. Z. Larson, A. L. Harzstark, M. Ferrone, M. van Criekinge, J. W. Chang, R. Bok, I. Park, G. Reed, L. Carvajal, E. J. Small, P. Munster, V. K. Weinberg, J. H. Ardenkjær-Larsen, A. P. Chen, R. E. Hurd, L.-I. Odergardstuen, F. J. Robb, J. Tropp and J. A. Murray, Sci. Transl. Med., 2013, 5, 198ra108 CrossRef PubMed.
- K. Golman, R. in't Zandt, M. Lerche, R. Pehrson and J. H. Ardenkjær-Larsen, Cancer Res., 2006, 66, 10855–10860 CrossRef CAS PubMed.
- C. Gabellieri, S. Reynolds, A. Lavie, G. S. Payne, M. O. Leach and T. R. Eykyn, J. Am. Chem. Soc., 2008, 130, 4598–4599 CrossRef CAS PubMed.
- J. J. Miller, J. T. Grist, S. Serres, J. R. Larkin, A. Z. Lau, K. Ray, K. R. Fisher, E. Hansen, R. S. Tougaard, P. M. Nielsen, J. Lindhardt, C. Laustsen, F. A. Gallagher, D. J. Tyler and N. Sibson, Sci. Rep., 2018, 8, 15082 CrossRef PubMed.
- T. G. Walker and W. Happer, Rev. Mod. Phys., 1997, 69, 629–642 CrossRef CAS.
- S. Appelt, A. B.-A. Baranga, C. J. Erickson, M. V. Romalis, A. R. Young and W. Happer, Phys. Rev. A: At., Mol., Opt. Phys., 1998, 58, 1412–1439 CrossRef CAS.
- H. E. Möller, X. J. Chen, B. Saam, K. D. Hagspiel, G. A. Johnson, T. A. Altes, E. E. de Lange and H.-U. Kauczor, Magn. Reson. Med., 2002, 47, 1029–1051 CrossRef PubMed.
- L. Schröder, T. J. Lowery, C. Hilty, D. E. Wemmer and A. Pines, Science, 2006, 314, 446–449 CrossRef PubMed.
- C. R. Bowers and D. P. Weitekamp, Phys. Rev. Lett., 1986, 57, 2645–2648 CrossRef CAS PubMed.
- C. R. Bowers and D. P. Weitekamp, J. Am. Chem. Soc., 1987, 109, 5541–5542 CrossRef CAS.
- T. C. Eisenschmid, R. U. Kirss, P. P. Deutsch, S. I. Hommeltoft, R. Eisenberg, J. Bargon, R. G. Lawler and A. L. Balch, J. Am. Chem. Soc., 1987, 109, 8089–8091 CrossRef CAS.
- K. Golman, O. Axelsson, H. Jóhannesson, S. Måringnsson, C. Olofsson and J. Petersson, Magn. Reson. Med., 2001, 46, 1–5 CrossRef CAS PubMed.
- E. Y. Chekmenev, J. Hövener, V. A. Norton, K. Harris, L. S. Batchelder, P. Bhattacharya, B. D. Ross and D. P. Weitekamp, J. Am. Chem. Soc., 2008, 130, 4212–4213 CrossRef CAS PubMed.
- S. Glöggler, A. M. Grunfeld, Y. N. Ertas, J. McCormick, S. Wagner, P. P. M. Schleker and L.-S. Bouchard, Angew. Chem., Int. Ed., 2015, 54, 2452–2456 CrossRef PubMed.
- O. G. Salnikov, K. V. Kovtunov and I. V. Koptyug, Sci. Rep., 2015, 5, 13930 CrossRef PubMed.
- F. Reineri, T. Boi and S. Aime, Nat. Commun., 2015, 6, 5858 CrossRef CAS PubMed.
- A. B. Schmidt, S. Berner, W. Schimpf, C. Müller, T. Lickert, N. Schwaderlapp, S. Knecht, J. G. Skinner, A. Dost, P. Rovedo, J. Hennig, D. von Elverfeldt and J.-B. Hövener, Nat. Commun., 2017, 8, 14535 CrossRef CAS PubMed.
- J. McCormick, S. Korchak, S. Mamone, Y. N. Ertas, Z. Liu, L. Verlinsky, S. Wagner, S. Glöggler and L.-S. Bouchard, Angew. Chem., Int. Ed., 2018, 57, 10692–10696 CrossRef CAS PubMed.
- E. Cavallari, C. Carrera, M. Sorge, G. Bonne, A. Muchir, S. Aime and F. Reineri, Sci. Rep., 2018, 8, 8366 CrossRef PubMed.
- J.-B. Hövener, A. N. Pravdivtsev, B. Kidd, C. R. Bowers, S. Glöggler, K. V. Kovtunov, M. Plaumann, R. Katz-Brull, K. Buckenmaier, A. Jerschow, F. Reineri, T. Theis, R. V. Shchepin, S. Wagner, P. Bhattacharya, N. M. Zacharias and E. Y. Chekmenev, Angew. Chem., Int. Ed., 2018, 57, 11140–11162 CrossRef PubMed.
- R. W. Adams, J. A. Aguilar, K. D. Atkinson, M. J. Cowley, P. I. P. Elliott, S. B. Duckett, G. G. R. Green, I. G. Khazal, J. Lopez-Serrano and D. C. Williamson, Science, 2009, 323, 1708–1711 CrossRef CAS PubMed.
- T. Theis, M. L. Truong, A. M. Coffey, R. V. Shchepin, K. W. Waddell, F. Shi, B. M. Goodson, W. S. Warren and E. Y. Chekmenev, J. Am. Chem. Soc., 2015, 137, 1404–1407 CrossRef CAS PubMed.
- T. Theis, G. X. Ortiz Jr, A. W. J. Logan, K. E. Claytor, Y. Feng, W. P. Huhn, V. Blum, S. J. Malcolmson, E. Y. Chekmenev, Q. Wang and W. S. Warren, Sci. Adv., 2016, 2, e1501438 CrossRef PubMed.
- P. J. Rayner, M. J. Burns, A. M. Olaru, P. Norcott, M. Fekete, G. G. R. Green, L. A. R. Highton, R. E. Mewis and S. B. Duckett, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E3188–E3194 CrossRef CAS PubMed.
- M. Suefke, S. Lehmkuhl, A. Liebisch, B. Blümich and S. Appelt, Nat. Phys., 2017, 13, 568–572 Search PubMed.
- M. Haake, J. Natterer and J. Bargon, J. Am. Chem. Soc., 1996, 118, 8688–8691 CrossRef CAS.
- J. Natterer and J. Bargon, Prog. Nucl. Magn. Reson. Spectrosc., 1997, 31, 293–315 CrossRef.
- A. S. Kiryutin, G. Sauer, S. Hadjiali, A. V. Yurkovskaya, H. Breitzke and G. Buntkowsky, J. Magn. Reson., 2017, 285, 26–36 CrossRef CAS PubMed.
- A. N. Pravdivtsev, G. Buntkowsky, S. B. Duckett, I. V. Koptyug and J.-B. Hövener, Angew. Chem., Int. Ed., 2021, 60, 23496–23507 CrossRef CAS PubMed.
- E. Cavallari, C. Carrera, T. Boi, S. Aime and F. Reineri, J. Phys. Chem. B, 2015, 31, 10035–10041 CrossRef PubMed.
- E. Cavallari, C. Carrera, S. Aime and F. Reineri, J. Magn. Reson., 2018, 289, 12–17 CrossRef CAS PubMed.
- S. Korchak, M. Emondts, S. Mamone, B. Blümich and S. Glöggler, Phys. Chem. Chem. Phys., 2019, 21, 22849–22856 RSC.
- M. Goldman, H. Jóhannesson, O. Axelsson and M. Karlsson, Compt. Rendus Chem., 2006, 9, 357–363 CrossRef CAS.
- P. Bhattacharya, E. Y. Chekmenev, W. H. Perman, K. C. Harris, A. P. Lin, V. A. Norton, C. T. Tan, B. D. Ross and D. P. Weitekamp, J. Magn. Reson., 2007, 186, 150–155 CrossRef CAS PubMed.
- P. Bhattacharya, E. Y. Chekmenev, W. F. Reynolds, S. Wagner, N. Zacharias, H. R. Chan, R. Bünger and B. D. Ross, NMR Biomed., 2011, 24, 1023–1028 CrossRef CAS PubMed.
- S. Korchak, S. Yang, S. Mamone and S. Glöggler, ChemistryOpen, 2018, 7, 344–348 CrossRef CAS PubMed.
- S. Korchak, S. Mamone and S. Glöggler, ChemistryOpen, 2018, 7, 672–676 CrossRef CAS PubMed.
- S. Berner, A. B. Schmidt, M. Zimmermann, A. N. Pravdivtsev, S. Glöggler, J. Hennig, D. von Elverfeldt and J.-B. Hövener, ChemistryOpen, 2019, 8, 728–736 CrossRef CAS PubMed.
- A. P. Jagtap, L. Kaltschnee and S. Glöggler, Chem. Sci., 2019, 10, 8577–8582 RSC.
- M. Daugherty, B. Polanuyer, M. Farrel, M. Scholle, A. Lykidis, V. de Crécy-Lagard and A. Osterman, J. Biol. Chem., 2002, 277, 21431–21439 CrossRef CAS PubMed.
- W. Wang, Z. Wu, Z. Dai, Y. Yang, J. Wang and G. Wu, Amino Acids, 2013, 45, 463–477 CrossRef CAS PubMed.
- L. Kaltschnee, A. P. Jagtap, J. McCormick, S. Wagner, L.-S. Bouchard, M. Utz, C. Griesinger and S. Glöggler, Chem.–Eur. J., 2019, 25, 11031–11035 CrossRef CAS PubMed.
- S. Glöggler, R. Müller, J. Colell, M. Emondts, M. Dabrowski, B. Blümich and S. Appelt, Phys. Chem. Chem. Phys., 2011, 13, 13759–13764 RSC.
- L. Sellies, R. L. E. G. Aspers, M. C. Feiters, F. P. J. T. Rutjes and M. Tessari, Angew. Chem., Int. Ed., 2021, 60, 26954–26959 CrossRef CAS PubMed.
- P. C. Soon, X. Xu, B. Zhang, F. Gruppi, J. W. Canary and A. Jerschow, Chem. Commun., 2013, 49, 5304–5306 RSC.
- T. Trantzschel, M. Plaumann, J. Bernarding, D. Lego, T. Ratajczyk, S. Dillenberger, G. Buntkowsky, J. Bargon and U. Bommerich, Appl. Magn. Reson., 2013, 44, 267–278 CrossRef CAS.
- J. A. Tang, F. Gruppi, R. Fleysher, D. K. Sodickson, J. W. Canary and A. Jerschow, Chem. Commun., 2011, 47, 958–960 RSC.
- S. Glöggler, S. Wagnerb and L.-S. Bouchard, Chem. Sci., 2015, 6, 4261–4266 RSC.
- O. G. Salnikov, L. M. Kovtunova, I. V. Skovpin, V. I. Bukhtiyarov, K. V. Kovtunov and I. V. Koptyug, ChemCatChem, 2018, 10, 1178–1183 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08808d |
| This journal is © The Royal Society of Chemistry 2022 |