Open Access Article
Open Access ArticleFacile synthesis of photoactivatable adenosine analogs†
Dawanna S. White‡
a,
Daniel Mongeluzi‡a,
Alyson M. Currya,
Dickson Donua and
Yana Cen *ab
*ab
aDepartment of Medicinal Chemistry, Virginia Commonwealth University, Richmond, VA 23219, USA. E-mail: ceny2@vcu.edu; Tel: +1-804-828-7405
bInstitute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, VA 23219, USA
First published on 14th January 2022
Abstract
Adenosine and its derivatives are important building blocks of the biological system. They serve as the universal energy currency, amplify intracellular signals for various signal transduction pathways, and can also be used as the co-substrates for enzymatic transformations. The synthesis and regulation of adenosine and its analogs rely on the adenosine binding proteins (ABPs). Dysregulated ABP activity contributes to numerous diseases such as cancer, metabolic disorders, and neurodegenerative diseases. Presently, there is intense interest in targeting ABPs for therapeutic purposes. A large fraction of the human ABP family remains poorly characterized. The need for innovative chemical probes to investigate ABP function in the native biological matrix is apparent. In this study, an adenosine analog, probe 1, with a photoaffinity group and biotin tag was synthesized using concise synthetic strategies. This probe was able to label and capture individual recombinant ABPs with good target selectivity. Probe 1 was also evaluated for its ability to label spiked ABP in complex cell lysates. This chemical probe, together with the labeling and enrichment assay, is of great value to interrogate the biological functions of ABPs and to elucidate their diversity under different physiological conditions.
1. Introduction
Adenosine and related nucleosides and nucleotides are important bioactive molecules. They are the major components of genetic materials and serve as energy coupling agents, second messengers in signal transduction pathways, and cofactors in enzymatic reactions. The intracellular levels of ATP, ADP and AMP, the best known adenosine derivatives, are indicative of cellular energy status.1 S-Adenosyl Methionine (SAM) acts as the universal methyl donor for methyltransferases.2 Adenosine 3′,5′-cyclic monophosphate (cAMP) is a key second messenger in multiple signaling pathways that regulate diverse cellular events such as carbon fuel degradation3,4 and reduction of blood platelet aggregation.5 Dysregulation of adenosine levels has been implicated in metabolic disorders,6 inflammatory bowel disease,7 Alzheimer's disease,8 and certain cancers.9–11The intracellular and extracellular concentrations of adenosine and its derivatives are tightly and precisely regulated by adenosine binding proteins (ABPs).12 The activity and expression levels of these proteins have been closely associated with normal physiology as well as pathophysiology.13–16 Given the importance of ABPs in various cellular processes, a great deal of interest exists in understanding their biological functions and expression patterns. However, the identification of ABPs remains challenging, especially on a proteomics scale, and our current inventory of these proteins is far from complete. Thus, innovative and enabling strategies are highly sought after in the investigation of ABPs.
Adenosine analogs carrying photoaffinity groups have emerged as powerful small molecule probes in the proteome-wide interrogation of ABPs.17–19 These probes normally inherited the adenosine or adenine core structure as the “warhead”.17–19 Additionally, a photoactivatable group such as benzophenone, diazirine, or azido was appended to the core structure.17–21 Upon UV irradiation, the photoaffinity group can covalently link to the amino acid(s) in its vicinity, leading to the formation of a protein-probe adduct. Furthermore, a biotin tag or a terminal alkyne can be incorporated into the probes to allow for the subsequent pulldown of the labeled protein via streptavidin–biotin interaction or Cu(I)-mediated “click” conjugation to affinity tags.18,19 These photoaffinity adenosine analogs have been applied to native proteomes to enable the enrichment and proteome-wide analysis of ABPs. In addition to the known ABPs, previously unannotated ABPs have also been revealed in these studies.18,19 These probes greatly expanded our knowledge of ABPs and suggested potential therapeutic targets.
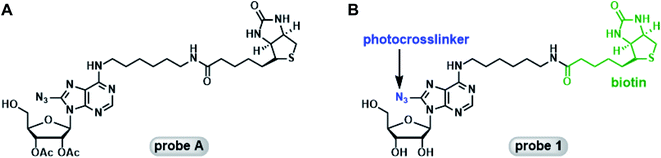
Mahajan et al. reported the synthesis and biochemical characterization of a photoaffinity adenosine probe for the proteomic screening of ABPs (probe A, Fig. 1A).19 It featured the adenosine core structure with a photocrosslinkable azido group attached to the 8-position, and a biotin tag tethered to the C-6 position of the adenine ring through a hexadiamine linker. This probe was able to enrich a panel of ABPs from a mouse neuroblastoma cell proteome.
Our lab has a long-standing interest in using novel chemical probes to investigate biologically important proteins in their native matrix. Probe A appeared to be an ideal candidate for further modifications: the 5′-OH group can be easily elaborated for the generation of 5′-substituted adenosine analogs as selective probes for focused groups of ABPs such as SAM-dependent methyltransferases (MTases) and 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN). In our attempts to repeat the synthetic procedures described by Mahajan et al., we encountered several challenges. First of all, in the original report the 8-azido group was introduced into the core structure using CsN3 in combination with TMSN3.19 CsN3 is no longer readily available from the common chemical vendors. Additionally, several steps in the original synthesis were extremely time-consuming: the bromination took 6 to 7 days, and the introduction of azido group was completed in 3 days. Furthermore, probe A is not a good mimic of naturally occurring adenosine and derivatives due to the presence of the 2′- and 3′-acetyl groups. Herein, we report the convenient synthesis of a photoactivatable adenosine probe, probe 1 (Fig. 1B), using two independent strategies. Subsequently, the probe was assessed for its ability to label recombinant ABPs including adenosine deaminase (ADA) and euchromatin histone lysine methyltransferase 1 (EHMT1). The remarkably simple probe demonstrated good target selectivity. Moreover, the on-target effect was established with competition assays using known cofactors of the target proteins. The easy access to probe 1 and its analogs will facilitate the investigation of the biological functions of ABPs and help elucidate their expression profiles under different physiological conditions.
2. Methods and materials
2.1 Reagents and instruments
All reagents were purchased from Aldrich or Fisher Scientific and were of the highest purity commercially available. NMR spectra were acquired on a Bruker Ultrashield 400 Plus NMR spectrometer and the data were processed using Topspin software. HRMS spectra were acquired with a PerkinElmer AxION 2 TOF Mass Spectrometer. Western blots were imaged on a Bio-Rad ChemiDoc MP imaging system.2.2 Expression and purification of human EHMT1
EHMT1 (2IGQ) was a gift from Cheryl Arrowsmith (Addgene plasmid #25504). Recombinant, N-terminal His-tagged protein was expressed in E coli BL21 (DE3) Codon plus RIL cells (Agilent). The cells were cultured in TB media supplemented with 50 μg mL−1 Kanamycin at 37 °C until OD600 reached 0.8. Protein expression was induced with 1 mM IPTG. The culture was grown overnight at 15 °C before the cells were pelleted and lysed by passing through a Microfluidizer. The protein was purified using a Ni-NTA resin (Thermo Fisher) affinity column and eluted with increasing concentrations of imidazole. The purified protein was dialyzed against a solution of 20 mM Tris–HCl (pH 8.0), 250 mM NaCl, and 15% glycerol. The protein was aliquoted, flash frozen and stored at −80 °C. Protein concentration was determined using Bradford assay. The protein was >95% pure as determined by SDS-PAGE gel electrophoresis.2.3 Photoaffinity labeling of recombinant ABPs
A typical experiment was performed as follows: in a 1.5 mL Eppendorf tube, recombinant ABP (10 μM for EHMT1, 1 μM for ADA) was incubated with probe 1 at 37 °C for 10 min. The sample was transferred to a clear-bottom 96-well plate, placed on ice, and irradiated at 365 nm with a UV-lamp at 4 °C. Subsequently, the sample was resolved on a 10% SDS-PAGE gel and transferred to a Immun-Blot PVDF membrane (Bio-Rad). The blot was blocked with 5% nonfat milk in TBST, probed with an anti-biotin HRP-linked antibody (Cell Signaling Technology), and washed with TBST. The signal was then detected by Clarity™ ECL substrate (Bio-Rad).2.4 Photoaffinity labeling of ADA in E. coli cell lysate
A typical labeling experiment contained 6 μg Rosetta™(DE3) cell lysate, 250 or 500 μM probe 1, and increasing concentrations of ADA. The photoaffinity labeling and western blot analysis were similar to the protocols for labeling recombinant ABPs as described above.2.5 Affinity enrichment of recombinant ABPs
EHMT1 or ADA was incubated with the chemical probe. After the UV irradiation, the unbound free probes were filtered off using an Amicon ultra centrifugal filter unit (Merck Millipore) with a 10 kDa molecular weight cutoff. Subsequently, high-capacity streptavidin beads (Thermo Fisher Scientific) were introduced to the sample to capture the biotinylated proteins. The beads were then rinsed with PBS three times. The captured protein was eluted by incubation with 25 mM biotin and 0.4% SDS in PBS at 95 °C for 5 min. The eluent was concentrated by lyophilization and analyzed by western blot.3. Results and discussion
3.1 Synthesis of probe 1
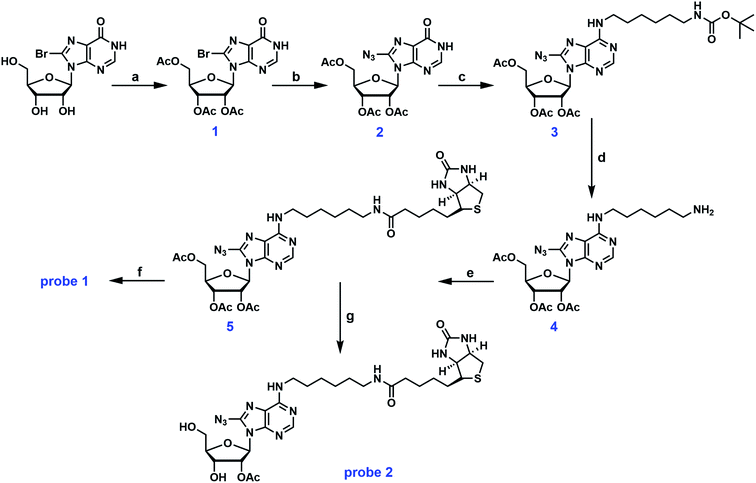
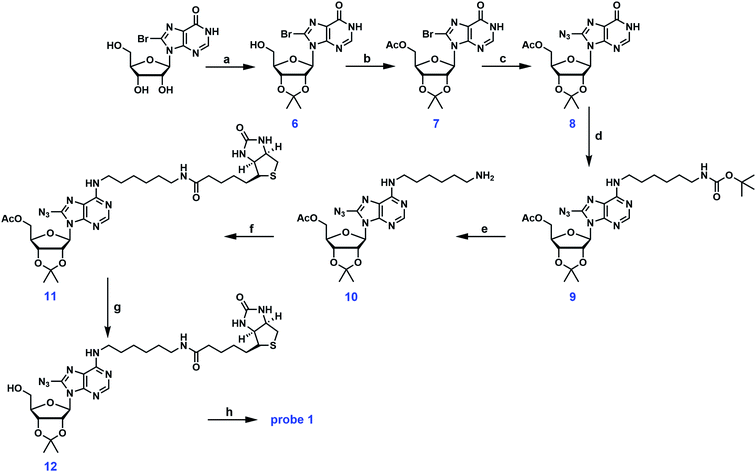
Probe 1 is a structural analog of probe A with free hydroxyl groups at the 2′ and 3′-positions (Fig. 1). To enhance the small molecule–protein interaction, a photoactivatable azido group was introduced at the 8-position of the adenine ring. This group can form an irreversible, covalent adduct with the protein target upon exposure to UV light. Careful inspection of the crystal structures of several ABPs suggested that the exocyclic amino group at the C-6 position of the adenine ring was solvent accessible and can easily accommodate a biotin tag. The first synthetic scheme for probe 1 is illustrated in Fig. 2. Commercially available 8-bromoinosine was protected using acetic anhydride and DMAP in anhydrous CH2Cl2 to generate 2′,3′,5′-O-triacetyl-8-bromoinosine (intermediate 1). Subsequently, the azido group was readily installed at the 8-position with NaN3 in DMF at 80 °C (ref. 22) furnishing intermediate 2. Intermediates 1 and 2 had similar Rf values on TLC plates with various eluent conditions, which posed a significant challenge in separation. Luckily, we made an interesting observation: both compounds were visible at 254 nm, however, only intermediate 2 was visible at 365 nm with a fluorescent blue color. Ultimately, pure intermediate 2 was obtained with 60% isolated yield. Mono-N-Boc-1,6-diaminohexane was coupled to the C-6 position with PyBOP in the presence of DIPEA to generate N6-substituted adenosine analog 3. The deprotection of the N-Boc group was accomplished with TFA yielding intermediate 4 at nearly 80% yield. Biotin was then coupled to the primary amino group using EDCI/DMAP to form the N6-biotinyl-8-azidoadenosine 5. The acetyl groups were removed using ammonia in methanol at 0 °C to afford the photocrosslinkable adenosine derivative probe 1. This synthetic route involved 6 steps from commercially available starting material. Almost all the steps were straightforward with decent to good yield.As mentioned earlier, the 5′-OH group in probe A can be further functionalized to create other adenosine analogs. We have attempted to reproduce probe A using the deprotection strategy reported by Mahajan et al. in which the 5′-O-acetyl group in intermediate 5 was selectively removed with ammonia in methanol at −40 °C.19 However, this step was proven to be unsuccessful in our hands despite numerous trials and modifications. Instead of the mono-deprotected probe A, the mono-protected probe 2 was obtained (Fig. 2). Alternatively, the three hydroxyl groups in 8-bromoinosine can be selectively protected as demonstrated in Fig. 3. The 2′- and 3′-OH groups were protected as an acetonide in a mixture of acetone and 2,2-dimethoxypropane under mild acidic condition. The 5′-OH was then protected with acetic anhydride to afford intermediate 7. The fully protected intermediate 7 was subjected to nucleophilic substitution using NaN3 to form azido-derivative 8. The following steps were similar to those in synthetic scheme 1: intermediate 8 was coupled to mono-N-Boc-1,6-diaminohexane, followed by Boc deprotection and coupling to biotin, leading to the formation of intermediate 11. The selective deprotection of the 5′-O-acetyl group was straightforward using ammonia in methanol at −40 °C. This step resulted in the formation of intermediate 12 in 80% isolated yield. Compound 12 could serve as a versatile precursor for further modifications: the acetonide was readily removed in 50% TFA at 0 °C to afford probe 1 in quantitative yield; alternatively, the 5′-OH can be decorated for 5′-substituted adenosine analogs. Currently, our lab is pursuing the latter strategy for the synthesis of SAM analogs. This synthetic approach involved 8 steps from 8-bromoinosine with a 13.1% overall yield (compared to the 11.2% overall yield of synthetic route 1) with the synthetic flexibility leading to 5′-modified adenosine derivatives.
3.2 Photoaffinity labeling of recombinant ABPs using probe 1
Photoaffinity labeling and the subsequent affinity enrichment of recombinant ABPs with probe 1 served as the model study. Recombinant protein has the advantage of being ample in amount and free of background noise. Important labeling parameters can be easily optimized to enable accurate profiling of ABPs. Technical details were fine-tuned to achieve fast, selective, and efficient labeling.EHMT1 belongs to the SAM-dependent lysine MTase subfamily.23 Extensive studies have revealed its role in epigenetic silencing of gene transcription and promoting DNA damage repair.24–27 It has also been closely associated with familial Alzheimer's disease and various cancers.28–30 The plasmid of EHMT1 was commercially available from Addgene. The protein was recombinantly expressed and purified using a published protocol.31 The identity of the protein was confirmed by tryptic digestion followed by LC-MS/MS analysis performed at the Vermont Biomedical Research Network Proteomics Facility.
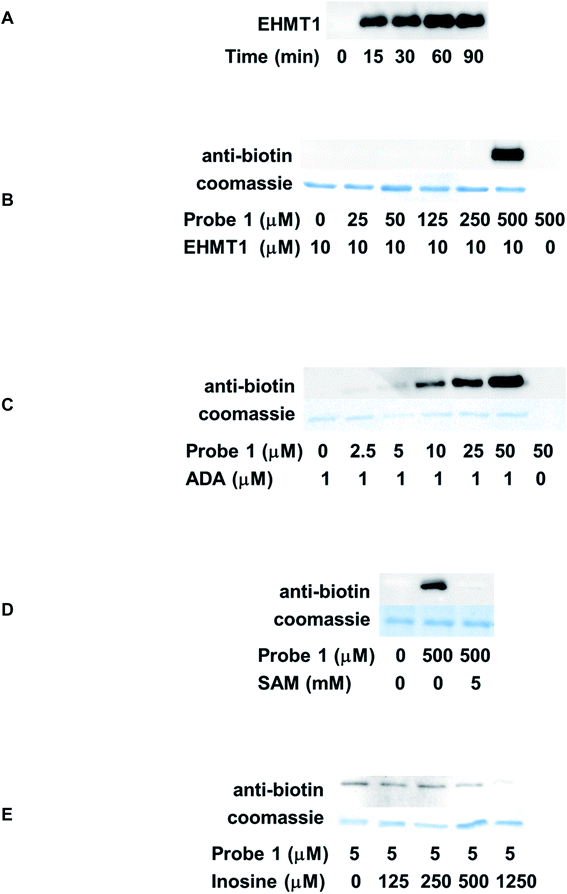
Time-dependent labeling of EHMT1 by probe 1 was investigated first. EHMT1 was incubated with probe 1, followed by UV irradiation at 365 nm for various times. The samples were then resolved by SDS-PAGE and analyzed by western blot using an anti-biotin antibody. The labeling was detected after merely 15 min's irradiation, and the labeling intensity plateaued within 60 min (Fig. 4A). For all the following experiments, a 60 min irradiation time was used.
The dose-dependent labeling of EHMT1 was also evaluated. The labeling was only detected at a high micromolar concentration (500 μM, Fig. 4B). We reasoned that the relatively weak affinity of probe 1 to EHMT1 stemmed from its structural features: it was a close structural mimic of adenosine rather than SAM due to the lack of methionine substitution at the 5′-position. To further assess the selectivity of probe 1, the labeling of ADA was also conducted. ADA is a key enzyme involved in purine salvage pathway.32 It catalyzes the deamination to convert adenosine and 2′-deoxyadenosine to inosine and 2′-deoxyinosine, respectively, in a Zn2+-dependent manner.33 A deficiency of human ADA is known to cause defects in T-lymphocyte development.34 Commercially available ADA was treated with various concentrations of probe 1 before being irradiated and analyzed as described above. As expected, probe 1 demonstrated robust labeling of ADA in a concentration-dependent fashion (Fig. 4C). Even at the lowest probe concentration, 2.5 μM, the labeling could still be detected. Side-by-side comparison of EHMT1 and ADA labeling results suggested that probe 1 was selective for ADA. This selectivity can be attributed to its structural similarity to adenosine, the endogenous substrate of ADA. Therefore, probe 1 has the potential to label other ABPs, especially those targeting adenosine, but this requires further investigation.
One major challenge in photoaffinity labeling is the non-specific binding which may create significant background noise or false positive results. To rule out the possibility of off-target effect, competition analysis was also performed. For EHMT1, the endogenous substrate, SAM, was used as the competitor. The protein was incubated with SAM before the addition of probe 1. Subsequently, the samples were subjected to photoaffinity labeling and western blot analysis. With the introduction of excess amount of SAM (5 mM), the labeling of EHMT1 by probe 1 was almost completely abolished (Fig. 4D), suggesting the on-target activity of the probe. Inosine is a known product from the ADA-catalyzed reaction. With increasing concentrations of inosine, the labeling of ADA by probe 1 was gradually competed off (Fig. 4E).
3.3 Affinity enrichment of recombinant ABPs using probe 1
The biotin moiety in probe 1 not only allowed the labeled proteins to be detected by western blot, but also enabled affinity capture of the labeled proteins using streptavidin–biotin interaction. The schematic representation of the enrichment process is shown in Fig. 5A. ADA was used to establish and optimize the protocol. The protein was incubated with probe 1, followed by UV irradiation to affect the covalent conjugation. The free probes were then filtered off using a centrifugal filter unit with a 10 kDa molecular weight cutoff. Subsequently, streptavidin beads were introduced into the sample to capture the biotinylated ADA. The unbound proteins were separated from the beads. After thorough rinsing, the beads were incubated with an excess amount of biotin to dissociate the proteins from the beads. Finally, the eluted proteins were analyzed by western blot as mentioned previously. The enrichment was detected at 5 μM probe concentration and intensified with increasing dosage of the probe (Fig. 5B). Similar results were obtained for EHMT1, albeit at higher probe concentrations (Fig. S1†). This simple probe exhibited good enrichment efficiency and target selectivity, consistent with the photoaffinity labeling results.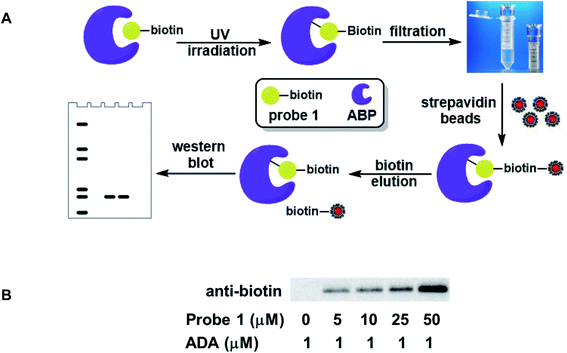 | ||
| Fig. 5 Affinity enrichment of ABPs using probe 1. (A) Schematic representation of the enrichment process; (B) labeling and affinity capture of recombinant ADA with probe 1. | ||
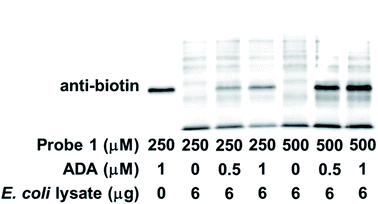
3.4 Labeling ABPs in complex protein mixtures
Encouraged by the robust and dose-dependent labeling of ADA by probe 1, we set out to evaluate the labeling of ADA in protein mixtures. Probe 1 was incubated with E. coli cell lysate with spiked ADA before being subjected to photoaffinity labeling and western blot analysis. As shown in Fig. 6, ADA was preferentially labeled in this protein mixture. The labeling started to be detected with 0.5 μM spiked protein and intensified with increasing amount of exogenously added ADA. As expected, other cellular proteins were also labeled by probe 1, presumably E. coli endogenous ABPs. The identity of these ABPs awaits further investigation. With increasing dosage of probe 1, the labeling intensity increased for both ADA and other targets. In summary, probe 1 not only labeled individual recombinant ABPs, but also demonstrated selective labeling of ABPs in complex proteomes.4. Conclusions
In the current study, we developed facile synthetic strategies to generate the photoaffinity adenosine analog, probe 1. This probe featured an adenosine core structure with a photoactivatable azido group and a biotin tag for affinity enrichment. This probe exhibited robust labeling of recombinant ADA, an enzyme that uses adenosine as the endogenous substrate. In contrast, EHMT1 was only labeled by probe 1 at high micromolar concentrations, owing to the structural difference between probe 1 and SAM, the physiological cofactor for EHMT1. The labeling of these ABPs could be effectively inhibited with the introduction of inosine or SAM, suggesting that probe 1 occupied the nucleoside binding site of its intended targets. The biotin tag allowed the labeled proteins to be captured using streptavidin beads. The photoaffinity labeling and affinity enrichment protocols were established and optimized. Furthermore, probe 1 also demonstrated sensitive labeling of ADA in the presence of excess E. coli cellular proteins. It is expected that probe 1 and its structural relatives will be used to profile ABPs in their native matrix for a better understanding of their roles in different cellular events.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the start-up fund from VCU (to Y .C.). Protein MS analysis reported in this manuscript was performed at the Vermont Biomedical Research Network Proteomics Facility supported by P20GM103449 (NIGMS/NIH).References
- R. L. Veech, J. W. Lawson, N. W. Cornell and H. A. Krebs, J. Biol. Chem., 1979, 254, 6538–6547 CrossRef CAS.
- S. J. Mentch and J. W. Locasale, Ann. N. Y. Acad. Sci., 2016, 1363, 91–98 CrossRef CAS PubMed.
- T. R. Soderling, J. P. Hickenbottom, E. M. Reimann, F. L. Hunkeler, D. A. Walsh and E. G. Krebs, J. Biol. Chem., 1970, 245, 6317–6328 CrossRef CAS.
- E. G. Ezrailson, M. L. Entman and A. J. Garber, J. Biol. Chem., 1983, 258, 12494–12498 CrossRef CAS.
- L. Noe, K. Peeters, B. Izzi, C. Van Geet and K. Freson, Curr. Med. Chem., 2010, 17, 2897–2905 CrossRef CAS PubMed.
- L. B. Sapkota, S. Thapa and N. Subedi, BMJ Open Diabetes Res. Care, 2017, 5, e000357 CrossRef PubMed.
- L. Antonioli, M. Fornai, C. Pellegrini, L. Bertani, Z. H. Nemeth and C. Blandizzi, Front. Immunol., 2020, 11, 1310 CrossRef CAS PubMed.
- A. Rahman, Curr. Neuropharmacol., 2009, 7, 207–216 CrossRef CAS PubMed.
- D. Boison and G. G. Yegutkin, Cancer Cell, 2019, 36, 582–596 CrossRef CAS PubMed.
- D. Vijayan, A. Young, M. W. L. Teng and M. J. Smyth, Nat. Rev. Cancer, 2017, 17, 765 CrossRef CAS PubMed.
- S. Gessi, S. Merighi, V. Sacchetto, C. Simioni and P. A. Borea, Biochim. Biophys. Acta, 2011, 1808, 1400–1412 CrossRef CAS PubMed.
- D. Boison, Pharmacol. Rev., 2013, 65, 906–943 CrossRef CAS PubMed.
- P. A. Borea, S. Gessi, S. Merighi, F. Vincenzi and K. Varani, Physiol. Rev., 2018, 98, 1591–1625 CrossRef CAS PubMed.
- S. Sheth, R. Brito, D. Mukherjea, L. P. Rybak and V. Ramkumar, Int. J. Mol. Sci., 2014, 15, 2024–2052 CrossRef PubMed.
- M. P. Abbracchio, S. Ceruti, R. Brambilla, C. Franceschi, W. Malorni, K. A. Jacobson, D. K. von Lubitz and F. Cattabeni, Ann. N. Y. Acad. Sci., 1997, 825, 11–22 CrossRef CAS PubMed.
- B. B. Fredholm, J. Mol. Med., 2014, 92, 201–206 CrossRef CAS PubMed.
- H. Muranaka, T. Momose, C. Handa and T. Ozawa, ACS Med. Chem. Lett., 2017, 8, 660–665 CrossRef CAS PubMed.
- M. Jelcic, K. Wang, K. L. Hui, X. C. Cai, B. Enyedi, M. Luo and P. Niethammer, Cell Chem. Biol., 2020, 27, 1073–1083.e1012 CrossRef CAS PubMed.
- S. Mahajan, R. Manetsch, D. J. Merkler and S. M. Stevens Jr., PLoS One, 2015, 10, e0115644 CrossRef PubMed.
- K. S. Madhusoodanan and R. F. Colman, Biochemistry, 2001, 40, 1577–1586 CrossRef CAS PubMed.
- N. Williams, S. H. Ackerman and P. S. Coleman, Methods Enzymol., 1986, 126, 667–682 CAS.
- S. J. Hymbaugh Bergman and L. R. Comstock, Bioorg. Med. Chem., 2015, 23, 5050–5055 CrossRef CAS PubMed.
- P. A. Boriack-Sjodin and K. K. Swinger, Biochemistry, 2016, 55, 1557–1569 CrossRef CAS PubMed.
- S. C. Dillon, X. Zhang, R. C. Trievel and X. Cheng, Genome Biol., 2005, 6, 227 CrossRef PubMed.
- M. Tachibana, Y. Matsumura, M. Fukuda, H. Kimura and Y. Shinkai, EMBO J., 2008, 27, 2681–2690 CrossRef CAS PubMed.
- X. Lu, M. Tang, Q. Zhu, Q. Yang, Z. Li, Y. Bao, G. Liu, T. Hou, Y. Lv, Y. Zhao, H. Wang, Y. Yang, Z. Cheng, H. Wen, B. Liu, X. Xu, L. Gu and W. G. Zhu, Nucleic Acids Res., 2019, 47, 10977–10993 CrossRef CAS PubMed.
- V. Ginjala, L. Rodriguez-Colon, B. Ganguly, P. Gangidi, P. Gallina, H. Al-Hraishawi, A. Kulkarni, J. Tang, J. Gheeya, S. Simhadri, M. Yao, B. Xia and S. Ganesan, Sci. Rep., 2017, 7, 16613 CrossRef PubMed.
- Y. Zheng, A. Liu, Z. J. Wang, Q. Cao, W. Wang, L. Lin, K. Ma, F. Zhang, J. Wei, E. Matas, J. Cheng, G. J. Chen, X. Wang and Z. Yan, Brain, 2019, 142, 787–807 CrossRef PubMed.
- Z. Rahman, M. R. Bazaz, G. Devabattula, M. A. Khan and C. Godugu, J. Biochem. Mol. Toxicol., 2021, 35, e22674 CrossRef CAS PubMed.
- N. Saha and A. G. Muntean, Biochim. Biophys. Acta, Rev. Cancer, 2021, 1875, 188498 CrossRef CAS PubMed.
- H. Wu, J. Min, V. V. Lunin, T. Antoshenko, L. Dombrovski, H. Zeng, A. Allali-Hassani, V. Campagna-Slater, M. Vedadi, C. H. Arrowsmith, A. N. Plotnikov and M. Schapira, PLoS One, 2010, 5, e8570 CrossRef PubMed.
- R. Hirschhorn and H. Ratech, Isozymes: Curr. Top. Biol. Med. Res., 1980, 4, 131–157 CAS.
- A. V. Zavialov, X. Yu, D. Spillmann, G. Lauvau and A. V. Zavialov, J. Biol. Chem., 2010, 285, 12367–12377 CrossRef CAS PubMed.
- A. V. Sauer, I. Brigida, N. Carriglio and A. Aiuti, Front. Immunol., 2012, 3, 265 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08794k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |