Open Access Article
Open Access ArticleGold nanoparticle-based optical nanosensors for food and health safety monitoring: recent advances and future perspectives
Nguyen Ha Anh†
*a,
Mai Quan Doan† a,
Ngo Xuan Dinh
a,
Ngo Xuan Dinh a,
Tran Quang Huy
a,
Tran Quang Huy ab,
Doan Quang Tric,
Le Thi Ngoc Loand,
Bui Van Hao
ab,
Doan Quang Tric,
Le Thi Ngoc Loand,
Bui Van Hao e and
Anh-Tuan Le
e and
Anh-Tuan Le *ae
*ae
aPhenikaa University Nano Institute (PHENA), Phenikaa University, Hanoi 12116, Vietnam. E-mail: anh.nguyenha@phenikaa-uni.edu.vn
bFaculty of Electric and Electronics, Phenikaa University, Hanoi 12116, Vietnam
cAdvanced Institute for Science and Technology (AIST), Hanoi University of Science and Technology (HUST), 1st Dai Co Viet Road, Hanoi, Vietnam
dFaculty of Natural Sciences, Quy Nhon University, Quy Nhon 55113, Vietnam
eFaculty of Materials Science and Engineering, Phenikaa University, Hanoi 12116
First published on 7th April 2022
Abstract
Modern society has been facing serious health-related problems including food safety, diseases and illness. Hence, it is urgent to develop analysis methods for the detection and control of food contaminants, disease biomarkers and pathogens. As the traditional instrumental methods have several disadvantages, including being time consuming, and having high cost and laborious procedures, optical nanosensors have emerged as promising alternative or complementary approaches to those traditional ones. With the advantages of simple preparation, high surface-to-volume ratio, excellent biocompatibility, and especially, unique optical properties, gold nanoparticles (AuNPs) have been demonstrated as excellent transducers for optical sensing systems. Herein, we provide an overview of the synthesis of AuNPs and their excellent optical properties that are ideal for the development of optical nanosensors based on local surface plasmon resonance (LSPR), colorimetry, fluorescence resonance energy transfer (FRET), and surface-enhanced Raman scattering (SERS) phenomena. We also review the sensing strategies and their mechanisms, as well as summarizing the recent advances in the monitoring of food contaminants, disease biomarkers and pathogens using developed AuNP-based optical nanosensors in the past seven years (2015–now). Furthermore, trends and challenges in the application of these nanosensors in the determination of those analytes are discussed to suggest possible directions for future developments.
1. Introduction
Never in the history of humanity have food and health not been major global concerns. In the Christian Bible, the Four Horsemen of the Apocalypse are figures representing four heart-struck fears of human: Pestilence, War, Famine and Death. Nowadays, by virtue of the rapid developments of agriculture and food processing industry, the worries of famine are less serious in most areas of the world, instead, people have to deal with the threat of unsafe food and drinks. There have been incidents of heavy metal contaminants in wheat,1 melamine in milk,2 fipronil in chicken eggs and poultry products,3 Cholera outbreaks,4 etc. Food contamination may lead to serious gastrointestinal infections,5 malnutrition,6 and even cancers.6,7 Moreover, humanity has been fighting against diseases such as cancer and diabetes for decades. Besides, the continuous appearance of new pathogenic viruses such as Severe Acute Respiratory Syndrome coronaviruses (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) keeps challenging every medical system. Recently, the COVID-19 outbreak has proved that human beings have never escaped from the threat of fatal diseases and pathogens. All of these problems have been putting human health in serious dangers. Therefore, it is essential to develop rapid and accurate tools and techniques to detect food contaminants, as well as diseases and pathogens.The traditional methods such as high performance liquid chromatography (HPLC),8 mass spectrometry (MS),9 liquid chromatography-MS,10 gas chromatography-MS,11 and polymerase chain reactions (PCR)12 are powerful techniques to detect different kinds of chemical compounds and biological elements. Despite their high sensitivity, accuracy and stability, they are time consuming, laborious and require expensive instruments.13,14 Therefore, sensing systems have been investigated to develop rapid, convenient, effective and less expensive detecting tools. In general, a sensor requires a target recognition element to ensure the selectivity and a transducer element to signal the binding events and manage the sensitivity of the detection.15 Antibodies,16 enzymes17 and aptamers18 are obviously effective for specific recognitions. A useful transducer element has to possess the ability to convert physical and chemical changes into detectable or measurable signals.15 Gold nanoparticles (AuNPs) have been attracting considerable attention as excellent scaffolds for the development of advanced sensing systems.19 With the sizes in the range of ∼1–100 nm, AuNPs possess unique physical and/or chemical properties in comparison with either the bulk state or the atomic level. Stable AuNPs can be easily prepared in either chemical or physical approaches.20,21 Moreover, their excellent biocompatibility and high surface-to-volume ratio allow the binding of various organic and biological ligands.22 Furthermore, their unique electrical, electrochemical, catalytic and especially, optical properties are remarkable.21,23,24 Every modification of size, shape, analyte binding, aggregation, etc. leads to change(s) in optical behaviors of AuNPs such as wavelength shift, color change, and enhancement of Raman scattering.25,26 They are detectable response signals that researchers are always looking for in an appropriate transducer. Therefore, it is not surprising that AuNPs-based optical sensors have attracted their interest for several years. Fig. 1 demonstrates the statistics of publications about AuNPs-based optical sensors over the last seven years (2015 – February 2022) according to Scopus data. Even after many years of investigation, it is still a well-concerned topic of researchers all around the world without any sign of stop. Possessing the properties of local surface plasmon resonance (LSPR), fluorescence quenching and surface-enhanced Raman scattering (SERS), AuNPs have been employed to develop different kinds of optical sensing systems including LSPR, colorimetric, fluorescence resonance energy transfer (FRET)-based and SERS-based sensors (Fig. 2). In this review, we focus on the strategies to develop those sensors, especially for the applications in food safety monitoring, and in vitro disease diagnosis. Recent advances in optical nanosensors for those analytes will be highlighted, focusing on the use of mono-metallic AuNPs, however, potential of multi-metallic gold-based optical nanosensors will be also introduced. Trends and challenges will be discussed to consider the future perspectives.
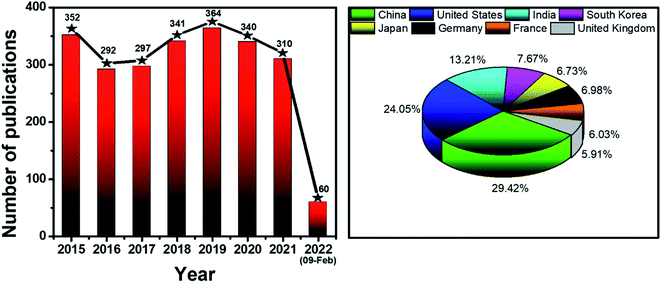 | ||
| Fig. 1 Statistics of publications about AuNPs-based optical sensors (2015 – February 2022). Data source: Scopus. Keywords: gold nanoparticles optical sensors. | ||
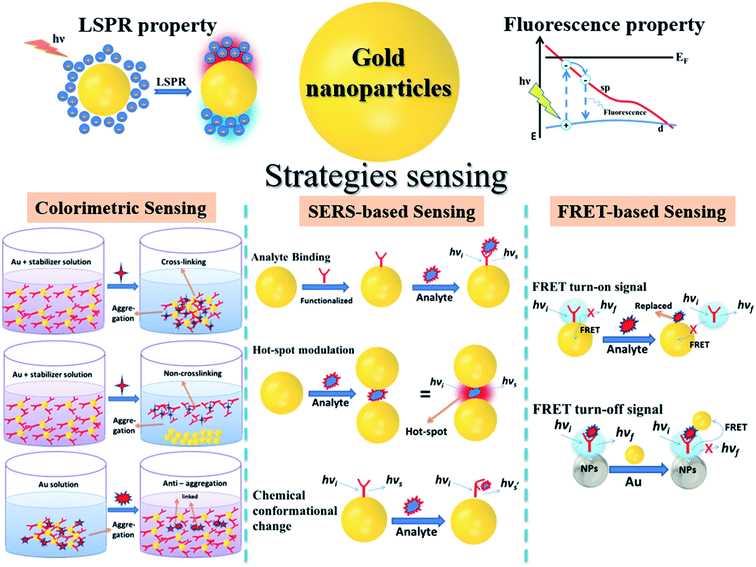 | ||
| Fig. 2 Illustration of gold nanoparticles, their optical properties and strategies to develop AuNPs-based optical nanosensors. | ||
Recently, several reviews relating AuNPs-based sensors for food safety14,27 and disease diagnosis28–30 have been established. However, most of them only focused on colorimetric sensors.14,27–30 Other types of AuNPs-based optical sensors were mentioned in a few reviews but only as an introduction without full discussion.28–30 This review exhibits a systematic look of AuNPs-based optical sensors for food and health safety monitoring with three kinds of sensors including colorimetric, FRET and SERS sensors. Further discussion about the sensing strategies developed for each kind of those optical sensors would provide readers more choices when considering designing a AuNPs-based optical sensor for their desired analyte. On the other hand, instead of focusing on the sensing mechanism for each kind of analytes as in the literature,28–30 this review shows several comparisons of sensing strategies for different selection of analyte for one identical target, for example, nucleic acid or protein biomarker for cancer detection; nucleic acid, capsid protein or antibody for virus determination; etc., which is useful for researchers when selecting a suitable analyte for their study. Therefore, this review would give researchers who are working on AuNPs-based sensors some suggestions to develop their sensing strategies to their designed targets in the future.
2. Gold nanoparticles
2.1. Synthesis and modifications
Analogously to other nanomaterials, AuNPs can be prepared by both “top-down” and “bottom-up” methods. In general, the “top-down” fabrication of AuNPs involves breaking down the bulk Au into nanoparticles, while the “bottom-up” method is based on the assembly of precursor molecules or atoms. For the “top-down” approach, various techniques have been developed, such as chemical etching, lithography, pyrolysis, laser ablation, sputtering, etc.20,21,31 Despite their advantages of fast and simple fabrication procedures, these “top-down” methods commonly encounter the limitations in controlling the particle size and shape.32 In addition, some of these synthesis procedures require extreme conditions, including high vacuum, high temperature, and large amount of energy as well as expensive instruments.21In contrast, the “bottom-up” methods commonly involve the reduction of Au ions into Au atoms, followed by the assembly of the atoms to form NPs. In this case, the formation of AuNPs normally follows two steps: the nucleation and the subsequent growth.32,33 In the first step, under certain circumstance, Au atoms are assembled to form small clusters, which function as nuclei. The addition of the incident atoms to the surface of the nuclei leads to the formation of AuNPs in the second step. An isotropic growth normally results in spherical nanoparticles. In order to tailor the particle morphology, surfactants such as cetyltrimethylammonium bromide (CTAB),34 sodium dodecylsulfonate (SDS),35 and poly(vinylpyrrolidone) (PVP)36 are usually added. This is due to the fact that surfactants may suppress the growth in certain directions and promote the growth in the other directions, resulting in the anisotropic growth of the NPs.37,38 Therefore, by choosing an appropriate surfactant, the particle size and shape can be controlled. This is a great advantage of the “bottom-up” methods over the “top-down”. Nevertheless, the use of toxic chemicals and solvents is a major drawback of this approach.39
The era of size- and shape-controlled synthesis of AuNPs started with the method developed by Turkevich in 1951, in which AuNPs were synthesized by reducing hydrogen tetrachloroaurate (HAuCl4) in the presence of sodium citrate.40 This method was further improved by Frens et al. in 1957, who demonstrated the size control of AuNPs by varying the ratio between HAuCl4 and sodium citrate.41 Although this protocol requires a high dilution of the Au salt that leads to the low AuNP concentration, its simplicity is a great advantage that retains it as the first choice of researchers for the synthesis of spherical and monodispersed AuNPs with diameters in the range of 10–20 nm. In 1994, Brust and co-workers developed a novel approach for the synthesis of ultra-small AuNPs, namely two-phase liquid–liquid reduction of AuCl4− by sodium borohydride in the presence of alkanethiol, which is nowadays known as the Schiffrin-Brust method.42 In this method, HAuCl4 was transferred from the aqueous solution to toluene using tetraoctylammonium bromide as the phase-transfer reagent. The addition of dodecanethiol and aqueous sodium borohydride facilitated the reduction of AuCl4−. In this process, the growth of Au clusters was accompanied by the simultaneous attachment of self-assembled thiol monolayers on Au surface. This provided an excellent control of the particle size, which allowed the synthesis of AuNPs with diameters in the range of 1–3 nm.42 These are the pioneering works that paved the way for the tremendous development in AuNPs synthesis in the last several decades.42–44
At the same time, the shape-controlled growth has also received enormous attention due to the shape-dependent properties of AuNPs.45,46 In this regard, the most common approach is utilizing surfactants to promote the anisotropic growth of AuNPs. A diversity of AuNPs such as nanorods, nanostars, nanoflowers, and many others has been fabricated by adding CTAB as a surfactant.37,38 PVP has been reported to be useful to control the formation of star-like Au nanoplates.47 Recently, surfactant-free techniques have been developed to control the shape of AuNPs. Wall et al. demonstrated the fabrication of AuNPs with different morphologies, including nanostars, nanospheres, nanorods, and nanoplates without using any surfactant. Instead, the morphology was controlled by varying the ratio between the reactants, pH, temperature and reaction time.48
As the environmental issues raise the concerns, chemical solvents are gradually replaced by other environmentally benign reducing and capping agents such as biomolecules like lactic acids and chitosan,49 or other biological agents such as extracts from Citrus maxima fruit or alfalfa plants.50 Alternative approaches used physical agents to assist the reduction of Au ions, such as UV irradiation or electrical current in the electrochemical and photochemical methods.33,51,52 Laser ablation is another popular physical method of synthesis as it can be employed to synthesize AuNPs in both aqueous and organic solvents.53,54 Moreover, this process does not require the removal of excess reagents after the synthesis. It also helps avoid the contamination on the surface of the nanoparticles and reduce the toxicity of the prepared nanoparticles. Vinod et al. demonstrated a successful synthesis of AuNPs by pulsed laser ablation.55 The obtained AuNPs exhibited excellent SERS and photothermal properties.55 Electron-beam-induced reduction method has also been employed for the synthesis of AuNPs with a diameter of less than 20 nm that can be used as bioimaging probes in mammalian cells.56 However, in comparison with the chemical synthesis methods, the physical approaches are less convenient and provide a lower yield.
For technological sensing applications, AuNPs are usually modified by appropriate chemical or biological elements for target recognition. Binding a self-assembled monolayer (SAM) to the AuNPs surface is a common technique to functionalize AuNPs. In general, SAM involves the adsorption of organic molecules onto the solid surface.57,58 Thiol, amides and carboxylic acids are ideal candidates for SAM due to their affinities to the AuNPs surface. Among them, thiols are most used because the Au–S bond is the strongest (∼40 kcal mol−1) in comparison with Au–N (∼8 kcal mol−1) and Au–COO (∼2 kcal mol−1).59 Thanks to these interactions, a wide range of molecules can be anchored to AuNPs. For example, peptides or proteins can be attached.60 Kumar and coworkers used cysteinylated peptide to modified AuNPs and developed a sensor for Newcastle disease virus.61 Similarly, thiolated aptamers were bound to AuNPs in a colorimetric sensor for interleukin-6, a biomarker for acute inflammation.62 Thanks to this great biocompatibility, AuNPs can be the scaffold for many kinds of recognition elements, leading to the ability to sense various targets, from inorganic to organic and from molecules to cells.
2.2. Optical properties of gold nanoparticles for sensing applications
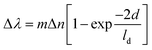 | (1) |
Due to their strong LSPR absorption in the green region, AuNP solutions usually exhibit a characteristic ruby red color. In most cases, an LSPR wavelength-shift can lead to a change in color of the solution. For example, an aggregation of AuNPs that caused a red shift of the LSPR spectra was also observed as a red-to-purple/blue change in the solution color.69 Therefore, the changes in LSPR spectra can be detected by either using an UV-visible spectrophotometer or bare-eyes. Hence, from the LSPR characteristics of AuNPs, two typical types of optical sensors have been developed: LSPR-based and colorimetric sensors. Importantly, the shift of LSPR spectra is strongly correlated with the aggregation of AuNPs. This correlation has been utilized in many sensing systems that are related to the aggregation of AuNPs (Fig. 3).
The aggregation state of AuNPs can be triggered through either crosslinking or non-crosslinking strategies. In the crosslinking method, ligand-functionalized AuNPs are assembled in the addition of a target element. Herein, the ligand acts as recognition probes for the target compound, which binds to this compound, decreases the interparticle distance and triggers aggregation of the AuNPs (Fig. 2). For instance, in the study of Megarajan and Veerapan, the aggregation of N-lauryltyramine (NLTA) capped AuNPs was induced in the addition of Al3+, which led to a pink-to-purple change in the solution color as well as a red-shift from 538 nm to 670 nm of the LSPR peak. The mechanism was suggested to be the chelation effect between NLTA and Al3+.70 In contrast, the non-crosslinking approach, which is also known as de-protection method, is based on the removal of stabilizing agent on the surface of AuNPs due to the presence of the target element. In this strategy, AuNPs are well-protected with the stabilizing agent to avoid aggregation. However, in the addition of the target element, which has higher affinity to the stabilizing agent, the AuNPs lose their coating agent (i.e. they are “de-protected”). This loss of electrosteric stabilizations results in AuNP aggregation. Aptamer-anchored AuNPs are widely used to develop colorimetric sensors based on this mechanism. In the addition of salts and the target elements, the aptamers are disassociated from AuNPs to “capture” the targets while high-salt condition induced the aggregation of the AuNPs. Wu et al. have applied this strategy and designed aptasensors for detection of Escherichia coli O157:H7 and Salmonella typhimurium with the specificity of 100%.71
On the contrary, a change of color from blue to red is observed in another strategy of colorimetric sensing called anti-aggregation. In this method, a linking molecule (or linker) induces aggregation of NPs by crosslinking mechanism. However, as the target element has a higher affinity to the linker, their interaction inhibits the binding of the linker to the ligand on AuNP surface and consequently prevents the aggregation of those AuNPs. For example, Gao's group introduced a sensing system for Ag+, in which thiamazole played the role of a linking molecule to induce aggregation of AuNPs.72 The presence of Ag+ ions triggered the formation of preferential combination between the pyrimidic nitrogen of thiamazole and Ag+ ions. Moreover, thiol group of thiamazole was also oxidized by Ag+. Therefore, the aggregation of AuNPs was inhibited, which resulted in a blue-to-red change in the solution color72 (Fig. 4). The same strategy has been utilized to detect other heavy metals, food additives, etc.14,73,74
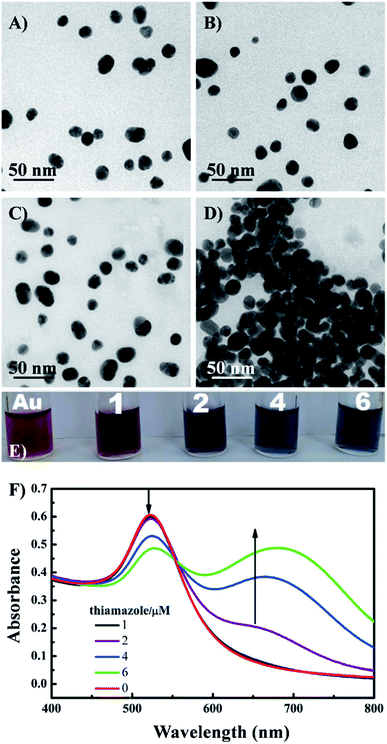 | ||
| Fig. 4 TEM images of the AuNPs under different conditions: (A) AuNPs, (B) AuNPs + Ag+ ions (1 nM), (C) AuNPs + thiamazole (2 μM) + Ag+ (1 nM), (D) AuNPs + thiamazole (2 μM); (E) photographs and (F) UV-VIS absorption spectra of AuNPs and AuNPs (200 μL) mixed with different concentrations of thiamazole (1–6 μM). Reprinted from ref. 72 Copyright 2021, with permission from Elsevier. | ||
Mainly concerning the aggregation of AuNPs (either formation or destruction), all strategies mentioned above do not require any specific shape of the AuNPs. Therefore, any kind of AuNPs can be employed to develop these colorimetric sensors, however, spherical AuNPs are the most selected in practice.75 The choice of isotropic AuNPs helps simplify the fabrication procedures of the sensors, consequently, it is more time- and cost-effective to develop these sensors. Brief information about AuNPs-based colorimetric sensors and their particle shapes can be found in Table 1 and 2, and further discussion about those sensors will be presented in the following sections.
| Target | Nanomaterials | Sensing principle | LOD | Linear range | Real sample | Ref. |
|---|---|---|---|---|---|---|
| Heavy metals | ||||||
| Hg2+ | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 3.4 nM | 5 nM to 10 μM | Tap water | 200 |
| Hg2+ | Aptamer-spherical AuNP | FRET | 60 nM | 0.2–20 μM | Tap water, milk | 201 |
| Hg2+ | cDNA-spherical AuNP | FRET | 7.5 × 10−13 M | 1.3 × 10−12 to 2.4 × 10−5 M | Fish | 145 |
| Hg2+ | ssDNA-spherical AuNP | SERS (hot-spot formation) | 0.45 pg mL−1 | 0.001–0.5 ng mL−1 | Drinking water | 117 |
| Hg2+ | DNAzyme contained ssDNA-spherical AuNP | Colorimetric (anti-aggregation) | 59.59 nM | 0–1 nM | Tap water | 148 |
| 1–15 nM | ||||||
| Cr3+ | 4-Amino hippuric acid (PAH)-spherical AuNP | Colorimetric (crosslinking) | 1.17 μM | 5.0–120 μM | Liquid milk, milk power | 202 |
| Cd2+ | 2,6-Dimecartopurine-spherical AuNP | Colorimetric (crosslinking) | 32 nM | 0.75–3.0 μM | Milk, honey | 203 |
| Cu2+ | Dipicolylamine-spherical AuNP | SERS (change in ligand) | 5 × 10−8 M | 5 × 10−8 to 5 × 10−4 M | White wine | 120 |
| Pb2+ | Citrate-spherical AuNP | Colorimetric (crosslinking) | 0.5 μM | 0.5–100 μM | Water | 204 |
| Pb2+ | DNAzyme-spherical AuNP | Colorimetric (anti-aggregation) | 8.0 nM | 0–500 nM | Tap water | 205 |
| Pb2+ | Maleic acid-spherical AuNP | Colorimetric (crosslinking) | 0.5 μg L−1 (∼2.4 nM) | 0.0–10.0 μg L−1 (∼0.0–48 nM) | Breast milk | 206 |
| Pb2+ | GSH-spherical AuNP | LSPR (change in refractive index) | 5 × 10−11 M | 10−10 to 10−5 M | Preserved egg | 207 |
| As3+ | Citrate-spherical AuNP | Colorimetric (crosslinking) | 1.8 ppb | 4–100 ppb | Drinking water | 152 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Pesticides | ||||||
| Phorate | Hyaluronan-tyrosine-spherical AuNP | Colorimetric (non-crosslinking) | 0.005 μg mL−1 (∼0.006 mg kg−1) | 0.001–10 μg mL−1 | Tomato | 155 |
| Organophosphorus pesticides | Spherical AuNP | Colorimetric (AuNP growth) | 0.7 ppb (∼2.4 nM) | 25–65 ppb | Tap water, apple washing solution | 157 |
| Dichlorvos | Citrate-spherical AuNP | Colorimetric (crosslinking) + enzyme inhibitor | 0.0120 mg kg−1, 0.0224 mg kg−1, 0.0106 mg kg−1 | 0–1000 μg L−1 | Pear, Chinese cabbage | 208 |
| Trichlorfon | ||||||
| Paraoxon | ||||||
| Triazophos | Fluorophore-ssDNA-spherical AuNP | FRET | 0.007 μg L−1 | 0.01–20 μg L−1 | Homogenized rice, wheat, cucumber, cabbage, apple | 158 |
| Parathion | 0.009 μg L−1 | 0.05–50 μg L−1 | ||||
| Chlorpyrifos | 0.089 μg L−1 | 0.5–1000 μg L−1 | ||||
| Parathio-methyl | Spherical AuNP on sticky side of adhesive tape | SERS | 2.60 ng cm−2 | — | Vegetable, cucumber, orange, apple | 161 |
| Thiram | 0.24 ng cm−2 | |||||
| Chlorpyrifos | 3.51 ng cm−2 | |||||
| Parathio-methyl | C14PDB-snowflake-like AuNP | SERS (analyte binding) | 0.026 ng cm−2 | — | Cucumber, apple, mango, green pepper, tomato peel | 159 |
| Triazophos | 0.031 ng cm−2 | |||||
| Phosmet | 0.032 ng cm−2 (apple peel) | |||||
| Malathion | Citrate-spherical AuNP | Colorimetric (anti-aggregation) | 11.8 nM | 0.05–0.8 μM | Tap water, apple juice, vegetable juice | 209 |
| Chlorsulfuron | Acetamiprid-spherical AuNP | Colorimetric (anti-aggregation) | 0.025 mg L−1 | 0.1–100 mg L−1 | Tap water | 210 |
| Thiram | Citrate-spherical AuNP | SERS | 5 × 10−10 M | 10−8 to 10−5 M | Tap water, orange juice | 114 |
| Thiram | Multi-branched Au nanostar | SERS | 10−10 M | 0.24 ng cm−2 to 2 μg cm−2 | Apple peel | 112 |
| 0.24 ng cm−2 (apple peel) | ||||||
| Carbendazim | Citrate-spherical AuNP | SERS (analyte binding) | 0.1 ppm | — | Oolong tea | 160 |
| Dimethoate | Citrate-spherical AuNP | FRET | 0.004 ppm | 0.005–1.0 ppm | Water, fruit | 211 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Herbicides | ||||||
| Paraquat | Citrate-spherical AuNP | SERS (analyte binding) | 0.1 μg mL−1 | — | Apple juice | 212 |
| Metsulfuron-methyl | Melamine-spherical AuNP | Colorimetric (anti-aggregation) | 0.05 mg L−1 | 0.1–100 mg L−1 | Tap water | 125 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Farm animal drugs | ||||||
| Ractopamine | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 10 ng mL−1 | 10–400 ng mL−1 | Beef | 163 |
| Ractopamine | Spherical AuNP | AuNP growth (aggregate or not) | 0.35 ng mL−1 | 2–512 ng mL−1 | Sheep urine | 164 |
| Kanamycin | DNA-spherical AuNP | Colorimetric (crosslinking) | 10 pM | 20 pM to 5 nM | Milk, honey | 213 |
| Chloramphenicol tetracycline | Multifunctional aptamer-spherical AuNP | Colorimetric (crosslinking) | 7.0 nM | 0.05–1.8 μM | Chicken, milk | 132 |
| 32.9 nM | 0.05–3.0 μM | |||||
| Chloramphenicol | Citrate-AuNP | SERS | 5.5 × 10−8 M | 10−7 to 10−5 M | Beef, shrimp | 114 |
| Dopamine | Aptamer-spherical AuNP | FRET | 2 nM | 26–2.90 × 103 nM | Swine feeds, chicken livers | 91 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Bacteria | ||||||
| E. coli | Aptamer-spherical AuNP | FRET | 3 CFU mL−1 | 5–106 CFU mL−1 | Tap/pond water, milk | 187 |
| Shigella flexneri | Aptamer-spherical AuNP | Colorimetric (non-crosslinking) | 80 CFU mL−1 | 102–106 CFU mL−1 | Salmon | 191 |
| E. coli | 4-MPBA-spherical AuNP | Colorimetric (anti-aggregation) | — | 104 to 107 CFU mL−1 | Drinking water | 192 |
| S. pollorum | ||||||
| S. aureus | ||||||
| E. faecalis | ||||||
| S. mutants | ||||||
| S. aureus | Vancomycin-spherical AuNP | FRET | 10 CFU mL−1 | 20 to 108 CFU mL−1 | Milk, orange juice | 195 |
| S.aureus | Teicoplainin-spherical AuNP | FRET | 2 CFU mL−1 | 10 to 5 × 108 CFU mL−1 | Milk, orange juice | 190 |
| Vibrio parahaemolyticus | Citrate-spherical AuNP | Colorimetric (AuNP growth) | 10 CFU mL−1 | 10–106 CFU mL−1 | — | 186 |
| P. aeruginosa | Chimeric M13 phage-spherical AuNP | Colorimetric (anti-aggregation) | 102 CFU mL−1 | — | Drinking water, nonfat bovine milk | 196 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Toxins | ||||||
| Aflatoxin B1 | DNA-spherical AuNP | FRET | 61 pM | 61 pM to 4.0 μM | Wine, beer | 95 |
| Aflatoxin M1 | (Aptamer + cDNA)-spherical AuNP | Colorimetric (anti-aggregation) | 30 ng L−1 | 300–75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng L−1 000 ng L−1 |
Milk | 214 |
| Aflatoxin M1 | (Aptamer + cDNA)-spherical AuNP | Colorimetric (anti-aggregation) | 1.8 nM in milk | — | Milk | 215 |
| T-2 toxin | Aptamer-spherical AuNP | Colorimetric (non-crosslinking) | 57.8 pg mL−1 (0.124 nM) | 0.1–5000 ng mL−1 (0.21435–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 717.5 nM) 717.5 nM) |
Wheat, corn | 197 |
| Zearalenone | (Aptamer + horseradish peroxidase)-spherical AuNP | Indirect colorimetric (nanozyme) | 0.08 ng mL−1 | 20–80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng L−1 000 ng L−1 |
Corn oil | 216 |
| Fumonisin B1 | Cysteamine-spherical AuNP | Colorimetric (crosslinking) | 0.90 μg kg−1 | 2–8 μg kg−1 | Corn | 198 |
| Saxitoxin | Cysteine-spherical AuNP | SERS (analyte binding) | 1 × 10−7 M | — | — | 199 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Adulterants | ||||||
| Melamine | (PEG + citrate)-spherical AuNP | Colorimetric (crosslinking) | 1.05 nM | 1.05 nM to 1 mM | Raw milk | 170 |
| Melamine | Spherical AuNP (plant extract) | Colorimetric (crosslinking) | 1.82 μM | 1.80 to 2.60 μM | Milk | 171 |
| Melamine | Spherical AuNP | Colorimetric (crosslinking) | 2.38 × 10−7 M | 3.90 × 10−7 to 3.97 × 10−6 M | Liquid milk | 217 |
| 5.56 × 10−6 M (in milk) | ||||||
| Melamine | Triton X-100-spherical AuNP | Colorimetric (crosslinking + de-protection) | UV-vis: 5.1 nM | 0–2.5 μM | Milk | 174 |
| Paper: 1.0 μM | 1–1.8 μM | |||||
| Melamine | p-Chlorobenzenesulfonic acid-spherical AuNP | Colorimetric (crosslinking) | 2.3 nM | 6.0 × 10−7 to 1.5 × 10−6 M | Liquid milk, milk powder | 175 |
| Melamine | Thymine derivative-spherical AuNP | Colorimetric (crosslinking) | 3.5 nM | 0.75–5.00 μM | Milk | 172 |
| Melamine | 1,4-Dithiothreitol (DTT)-spherical AuNP | Colorimetric (crosslinking) | 2.4 × 10−8 M | 8 × 10−8 to 6 × 10−7 M | Milk | 173 |
| 6 × 10−7–1.5 × 10−6 M | ||||||
| Melamine | Terpyridyl zinc complex-spherical AuNP | Colorimetric (crosslinking) | 2.4 ppb | 0.1–0.45 μM | Milk | 218 |
| Melamine | Citrate-spherical AuNP | FRET | Green: 8 ng mL−1 | 20–100 ng mL−1 | Milk | 219 |
| Red: 11 ng mL−1 | ||||||
| Melamine | Citrate-spherical AuNP | FRET | 1.7 nM | 5 nM to 1.9 μM | Milk powder | 176 |
| Sodium dodecylbenzenesulfonate | Citrate-spherical AuNP | Colorimetric (anti-aggregation) | 23 μg mL−1 | 23–300 μg mL−1 | Milk | 220 |
| Commercial anionic detergents | 92 μg mL−1 | 92–900 μg mL−1 | ||||
| Sibutramine | Citrate-spherical AuNP | Colorimetric (crosslinking) | 1.15 μM | 5–15 μM | Dietary supplement products | 221 |
| Urea | FITC-aptamer-spherical AuNP | FRET | 20 mM | 20–150 mM | Milk | 222 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Preservatives | ||||||
| Formaldehyde | Resorcinol-spherical AuNP | SERS (analyte binding) | 17 μM | 25–1000 μM | — | 179 |
| Formaldehyde | Citrate-spherical AuNP | SERS (purge-trap device) | 1 × 10−4 μg mL−1 | 1 × 10−4 to 3 × 10−3 μg mL−1 | Duck blood, rice flour | 180 |
| Thiabendazole | CTAB-Au nanorod | SERS (analyte binding) | Lemon: 149 μg L−1 | — | Lemon, carrot, mango juice | 181 |
| Carrot: 216 μg L−1 | ||||||
| Mango: 179 μg L−1 | ||||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Food dyes | ||||||
| Allura red sunset yellow | CTAB-Au nanorod | SERS (analyte binding) | 0.1 mg L−1 | — | Beverages | 113 |
| Malachite green | Aptamer-AuNP | Colorimetric (non-crosslinking) | 15.95 nM | 20–300 nM | Fish | 182 |
| Malachite green | Citrate-spherical AuNP | Indirect colorimetric (nanozyme) | 1.8 nM | 10–500 nM | Fresh water | 183 |
| Target | Nanomaterials | Sensing principle | LOD | Linear range | Real sample | Ref. |
|---|---|---|---|---|---|---|
| Molecule/peptides | ||||||
| Glucose | GOx-spherical AuNP (on a tapered fiber structure) | LSPR (change in refractive index) | 322 μM | 0–10 mM | — | 230 |
| Glucose | 4-Cyanophenyl boronic acid (CPBA)-b-cyclodextrin (b-CD)-spherical AuNP | Colorimetric (crosslinking) | — | 1–20 mM | Serum | 280 |
| Glucose | D-Glucose-spherical AuNP | Colorimetric (AuNP growth) | 0.65 mM | 1.25–20 mM | Human serum | 234 |
| Glucose | Spherical AuNP | Colorimetric (AuNP growth) | 0.03 mM | 0.1–10 mM | Serum | 281 |
| Glucose | Spherical AuNP | Colorimetric (AuNP growth) | 0.081 mM | 0.3–5.0 mM | Saliva | 282 |
| Glucose | Multibranched AuNP | Colorimetric (etching) | 0.4 mg dL−1 | — | Saliva | 77 |
| 1.4 mg dL−1 (real samples) | ||||||
| Glucose | CTAB-Au nanorod | Colorimetric (etching) | LSPR: 0.1 μM | 0.3–1.0 μM | Serum | 66 |
| 1.0–10 μM | ||||||
| Naked eye: 3 μM | 3–30 μM | |||||
| Glucose | Chitosan-spherical AuNP | Indirect colorimetric (nanozyme) | 3 μM | — | 60% serum | 81 |
| Glucose | Citrate-spherical AuNP | FRET (donor: GOQD) | 0.65 μM | 2.5–75 μM | Serum | 232 |
| Glucose | Citrate-spherical AuNP | Colorimetric | 0.043 μM | 0–40 μM | Urine | 283 |
| Glucose (through H2O2) | 3-Mercaptophenylboronic acid (3-MPBA)-spherical AuNP | SERS (change in ligand) | H2O2: 70 nM | 70 nM to 150 μM | Serum, urine | 231 |
| Glucose: — | 0.5–5 mM | |||||
| Glycated hemoglobin | Spherical AuNP | Colorimetric (AuNP growth) | 0.124% of total hemoglobin | — | Whole blood | 236 |
| Urea | Citrate-spherical AuNP | Indirect colorimetric (nanozyme) | 5 μM | 0.02–0.4 mM | Human urine | 284 |
| Dopamine | Citrate-spherical AuNP | Colorimetric (crosslinking) | 22 nM | 0.1–4 μM | Human plasma, urine | 285 |
| Dopamine | Citrate-spherical AuNP | Colorimetric (crosslinking) | 1.85 μM | 0–300 μM | Urine | 225 |
| FRET | 0.29 μM | 0–80 μM | ||||
| Glutathione | Spherical AuNP (plant extract) | Indirect colorimetric (nanozyme) | 0.013 μM | 1–40 μM | Urine | 82 |
| Glutathione | Citrate-spherical AuNP | FRET | 0.21 μM | 3.8–415.1 μM | Serum | 97 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Protein markers | ||||||
| Interleukin-6 | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 1.95 μg mL−1 | 3.3–125 μg mL−1 | — | 62 |
| Hemoglobin | 2,6-Diaminopurine-spherical AuNP | Indirect colorimetric (nanozyme) | 0.96 nM | — | Urine | 80 |
| CA15-3 | Antibody-Au nanorod | Colorimetric (crosslinking) | — | — | Human serum of breast cancer | 256 |
| MUC4 | (Antibody + SERS reporter)-spherical AuNP | SERS | — | — | — | 257 |
| p53 mutant | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 5 nM | — | — | 242 |
| CEA | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 3 ng mL−1 | 0.1–120 ng mL−1 | Human serum | 260 |
| Erα | Aptamer-spherical AuNP | Colorimetric (de-protection) | 2.4 ng mL−1 | 10 ng mL−1 to 5 μg mL−1 | Cellular extract | 245 |
| HER2 | Aptamer-spherical AuNP | Colorimetric (de-protection) | 10 ng mL−1 | 0–99 ng mL−1 | 10% serum | 244 |
| AFP | Antibody-spherical AuNP | FRET | 400 pg mL−1 | 0.5–45 ng mL−1 | Human serum | 246 |
| AFP | Spherical AuNP | SERS (hot-spots with antibody-AgNPs) | 5 ng mL−1 | 0.05–10 μg mL−1 | Fetal bovine serum | 258 |
| AFP-L3 | Antibody-spherical AuNP | SERS (conformational change in linker) | 05 ng mL−1 | 0.5–1000 ng mL−1 | Human serum | 122 |
| PSA | Aptamer-spherical AuNP | Colorimetric (crosslinking) | 20 pg mL−1 | 0.1–100 ng mL−1 | Human serum | 259 |
| CA-125 | DNA-spherical AuNP | FRET | 1.5 fg mL−1 | 5 fg mL−1 to 50 ng mL−1 | Human serum | 261 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Acid nucleic markers | ||||||
| p53 | Aptamer-spherical AuNP | FRET (donor: FAM) | 1.6 pM | 5 pM–1 nM | — | 241 |
| BRCA1 mutation | Aptamer-spherical AuNP on 2D material (GO, Bi2Se3) | Indirect colorimetric (nanozyme) | 10−18 M | 10−18 to 10−12 M | Extracted from patients' blood | 243 |
| PCA3 | Citrate-spherical AuNP | PCR + colorimetric (anti-aggregation) | 31.25 ng | — | Urine | 247 |
| 3 sequences containing melanoma DNA mutations | ssDNA-spherical AuNP | SERS (+PCR) | 0.1% mutation (10 copies) | — | — | 250 |
| PCA3 mimic DNA | ssDNA-hollow AuNP | SERS | 2.7 fM | 1 fM to 100 nM | — | 251 |
| microRNA | Unfunctionalized spherical AuNP | Colorimetric (de-protection) | 0.6 nM | 0.5 nM to 1 μM | Total RNA extracted from cancer cells (MCF-7) and normal cells (HEK 293) | 252 |
| FRET (donor: Aptamer-AgNC) | 0.4 pM | 1 pM–5 252μM | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Other markers | ||||||
| Tumor extracellular vesicles | Aptamer-spherical AuNP | SERS (creating hot-spots by organizing around an aptamer-agarose bead) | 2.44 pg μL−1 | 0.043–1240 ng μL−1 | Human serum | 264 |
| Exosome | Antibody-spherical AuNP | Colorimetric (plasmonic + immunoassay) | 8.54 × 105 exosomes per μL | 0–1.44 × 108 exosomes per μL | — | 286 |
| MCF-7 breast cancer cell | 150 nm spherical AuNP | NIR SERS | — | — | Human serum, plasma | 128 |
| MCF-7 breast cancer cell | Aptamer-Au nanorod | Colorimetric (crosslinking) | 100 cells per mL | 102–105 cells per mL | — | 265 |
| MDA-MB-231 breast cancer cell | Au nanostar | SERS | — | 10–500 cells per mL | — | 287 |
| Red blood cell | 2,6-Diaminopurine-spherical AuNP | Indirect colorimetric (nanozyme) | 1.6 × 106 cells per L | — | Urine | 80 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Infectious diseases | ||||||
| E. coli | Chimeric phage-spherical AuNP | Colorimetric (crosslinking) | ∼102 CFU | — | — | 169 |
| V. cholera | ||||||
| P. aeruginosa | ||||||
| Tuberculosis (DNA) | Citrate-spherical AuNP | Colorimetric (de-protection) | 1.95 × 10−2 ng mL−1 | 1.95 × 10−2 to 1.95 × 101 ng mL−1 | Sample from 01 patient | 268 |
| Tuberculosis (ManLAM lipoglycan) | Antibody-spherical AuNP | SERS | 0.8 ng mL−1 (46 pM) | — | Serum | 269 |
| H3N2 influenza A virus | Antibody-spherical AuNP | Colorimetric (crosslinking) | 7.8 HA units | 10–80 HA units | Clinical sample | 272 |
| Influenza viruses | Glycans-spherical AuNP | Colorimetric (crosslinking) | 8 HA units | — | Clinical sample | 273 |
| Tamiflu-resistant influenza virus | Oseltamivir hexylthiol-spherical AuNP | Colorimetric (crosslinking + lateral flow assay) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) × ×![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 PFU mL−1 104 PFU mL−1 |
— | — | 271 |
| H1N1 influenza A virus | Positive charged spherical AuNP | Indirect colorimetric (nanozyme) | 10.79 pg mL−1 | 10 pg mL−1 to 10 μg mL−1 | Human serum | 270 |
| H3N2 influenza A virus | 11.62 PFU mL−1 | 10–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PFU mL−1 000 PFU mL−1 |
||||
| Newcastle disease virus (viral particle) | Peptide–Cys-spherical AuNP | Colorimetric (crosslinking) | 0.125 HA units | — | — | 61 |
| DEVN WVN (viral particle) | Citrate-spherical AuNP | SERS (hot-spot: organization of AuNP on viral particles) | 10 PFU mL−1 | — | — | 116 |
| MERS-CoV (upE gene) | Citrate-spherical AuNP | Colorimetric (crosslinking) | 1 pmol μL−1 | — | Real sample after PCR | 274 |
| SARS-CoV-2 influenza B | Sialic-spherical AuNP | Colorimetric (crosslinking) | — | — | Throat swab | 275 |
| MERS-CoV (viral particle) | ||||||
| SARS-CoV-2 (viral particle) | (3 types of antibodies)-spherical AuNP | Colorimetric (crosslinking) | — | — | Throat swab | 224 |
| SARS-CoV-2 (viral particle) | ACE2 protein/mAbs-spherical AuNP | Colorimetric (plasmonic + immunoassay) | 370 vp mL−1 | 0–107 vp mL−1 | Pseudovirus | 276 |
| SARS-CoV-2 (IgM antibody) | (Anti-human IgM)-spherical AuNP | Colorimetric (plasmonic + immunoassay) | — | — | Serum | 86 |
| SARS-CoV-2 (IgG antibody) | Virus epitope-spherical AuNP | Colorimetric (crosslinking) | 3.2 nM | — | Real plasma samples (including patients) | 277 |
| SARS-CoV-2 Spike protein | Aptamer-spherical AuNP | Colorimetric (anti-aggregation) | 16 nM3540 | — | — | 279 |
| Heat-inactivated virus | Genome copies per μL | |||||
| SARS-CoV-2 (protease) | Citrate-spherical AuNP | Colorimetric (aggregation) | 10 pM | 0.01–0.5 nM | — | 288 |
Based on the aggregation of AuNPs, the colorimetric sensing strategies mentioned above are fast and simple with only a few steps of preparation and testing. Moreover, the obvious color change can be observed with bare-eye without any complicated instrument. However, the main limitation of these approaches is that AuNPs can be aggregated due to numerous factors, including pH, temperature and ionic forces that can lead to false positive/negative results. Therefore, researchers have investigated AuNPs-based sensors applying non-aggregation mechanisms, including etching, growth and nanozyme.76 Etching techniques focus on the effect of enchants such as H2O2 and I− ions to change the sizes and shapes of AuNPs, which leads to a shift in LSPR peak and a subsequent color change of reaction solution. Thus, etching is the only strategy, in which anisotropic NPs, such as Au nanorods and multibranched AuNPs, are preferable when designing colorimetric sensors.66,77 An example of this strategy is a urine glucose sensor established by Zhang et al., in which glucose oxidase enzymatic reaction led to the release of H2O2 that oxidized I− to I2 rapidly.66 Au nanorods were etched by I2, especially at their tips, thus, a longitudinal LSPR blue-shift and a blue-to-red color change were observed for glucose detection.66 Besides, tetracycline, one of the over-used antibiotics for farm animals, could reduce Au3+ ions to trigger the growth of AuNPs and introduce the characteristic red color to the solutions. Hence, it has become the principle to develop tetracycline sensing systems.78 However, it is worth mentioning that the selectivity of this method is low among other antibiotics in the same family (with tetracycline) due to their similar phenolic structures.78 In another approach, the peroxidase-like activity of AuNPs allows them to oxidize TMB in the presence of H2O2 to achieve the colorless-to-blue color change in solutions. Hence, target elements that enhance the catalytic activity such as Hg2+ (ref. 79) and Fe2+,80 or induce the formation of H2O2 such as glucose81 (in the interaction with glucose oxidase) can be detected. In another study, the color change was reversed upon the addition of glutathione, which suggested another principle to sense this disease marker.82 Furthermore, AuNPs can mimic the catalytic activity of several other enzymes such as glucose oxidase and peroxidase.76,83 With the highest surface area-to-volume ratio, there is no surprise that spherical AuNPs are the most selected material for this approach (see Tables 1 and 2).
AuNPs-based sensors using non-aggregation approaches are more complicated than those using aggregation or anti-aggregation methods. For instance, to detect zearalenone, a mycotoxin, the sensors fabricated based on the catalytic activity of AuNPs to convert 4-nitrophenol (yellow) into 4-aminophenol (colorless) needed to be functionalized with two kinds of single stranded DNA (ssDNA), including one aptamer and its complementary strand. Moreover, the detecting procedure required two steps of adding exonuclease III and 4-nitrophenol.84 Nevertheless, these novel methods have helped researchers to escape from the “red-to-blue” trap as with every kind of target element, the aggregation methods only exhibit the colors in that range. The novel indirect colorimetric approaches introduce new color ranges which would be useful to develop multi-target detecting systems.
In several immunoassays, AuNPs play the role of a chromatic substrate, in which the characteristic red color of AuNPs would be a reporting signal for the detection. It is convenient to generate immune testing strips.85–87
Similarly, different kinds of organic dyes, fluorescent proteins, quantum dots, and unconventional NPs have been utilized as donors, from which excited energy is transferred to AuNPs within a short distance.89,96 AuNPs quench the fluorescence signal, so the system is in “turn-off” state. Hence, the key strategy to “turn-on” a FRET sensor is to elongate the distance between the donor and the acceptor, leading to the recovery of fluorescent signal. Therefore, the target element should bind to either the donor or the acceptor in a competitive mechanism. For example, Dong et al. reported a “turn-on” fluorescent sensor to detect glutathione (GSH) based of FRET effect between N,S dual-doped carbon dots (N,S-CDs) and AuNPs. Positively charged N,S-CDs bound to negatively charged AuNPs via electrostatic interaction.97 Fluorescence intensity of CDs was quenched by AuNPs due to FRET effect. In the addition of GSH, the strong Au–S bond between GSH and the NPs leads to the release of N,S-CDs from the NPs surface, so fluorescence recovery occurs (Fig. 5). The limit of detection (i.e., LOD) of this method was as low as 0.21 μM. As the concentration of GSH in human serum is up to mM levels, the sensing system was expected to be able to determine GSH in diluted real samples.97 This example demonstrates the high sensitivity and accuracy of AuNPs-based FRET sensors that take advantage of the fluorescence efficiency of the donors and the excellent quenching ability of AuNPs. However, further studies are required to achieve the optimal conditions for FRET sensing. Each of donors possess its own advantages and disadvantages. For example, organic dyes are small, simply-prepared and cost-effective, but they have high toxicity, high photobleaching rate and low stability; fluorescent proteins are nontoxic, but they display wide emission spectrum as well as large sizes that can elongate the distance to the acceptors; QDs exhibit high quantum yield, narrow emission spectrum and high photostability, but they are highly toxic, etc.89 Therefore, researchers need to balance out these advantages and disadvantages to choose the suitable donors for FRET sensors based on the desired target elements and the sensing environment. Moreover, other NPs have been investigated as the promising candidates for the role of donors. For instance, unconventional nanoparticles (UCNPs) can emit fluorescence under the excitation wavelengths in the near infrared region, which is less harmful for biological samples.89,98 Recently, graphene NPs have been regarded as a potential choice as they can be fabricated easily at large scale of production. Moreover, their cost-efficiency, high solubility and biocompatible are advantages. However, the structure of this nanomaterial should be further studied to improve their wide emission spectra.89,99
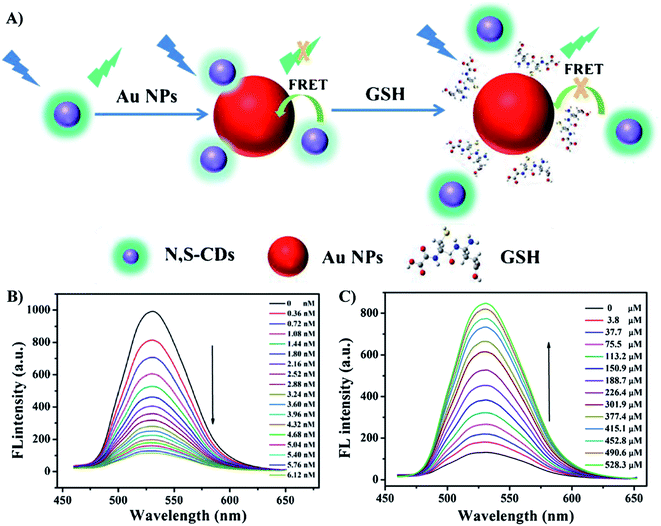 | ||
| Fig. 5 (A) Schematic principle for the determination of GSH based on FRET of N,S-CDs and AuNPs; (B) The fluorescence spectra of N,S-CDs (19 μg mL−1) in presence of different concentrations of AuNPs from 0 to 6.12 nM; (C) the fluorescence spectra of the N,S-CDs/Au NPs in the presence of different concentrations of GSH (0 μM to 528.3 μM). Reprinted by permission from Springer Nature: ref. 97 Copyright 2019. | ||
In general, AuNPs are non-luminescent materials, however, ultrasmall AuNPs (<3 nm), which are known as gold nanoclusters (AuNCs), have molecular-like properties, including photoluminescence.100 Therefore, they have been studied to design fluorescence sensors for many different targets. However, in this review, we only focus on AuNPs. The fluorescent sensing mechanisms using AuNCs have been reviewed in detail by Halawa et al.100
| P = αE | (2) |
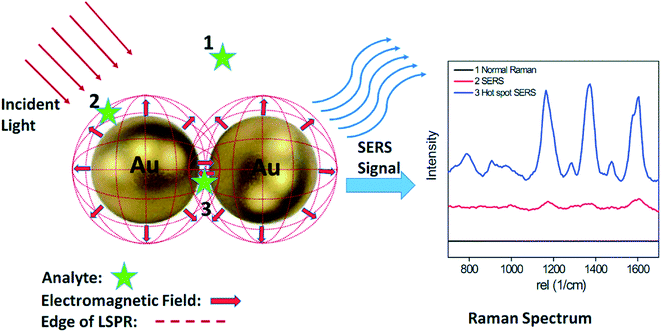 | ||
| Fig. 6 Schematic of SERS phenomenon for an organic analyte on AuNPs. Analyte molecules located within the dotted red circles (position 2 and 3) show clear Raman signals, while analytes located outside the circles (position 1) exhibit no detectable Raman signal. Commonly, significant Raman enhancements mainly occur within gaps smaller than 10 nm. These localized areas are often referred to as ‘hot-spots’ (position 3). Analyte molecules located within the hot spot exhibit a much stronger Raman signal than those located on AuNP surface (position 2). Reproduced from ref. 107 with permission from The Royal Society of Chemistry, Copyright 2015. | ||
The analyte or target element of a SERS-based sensor might be adsorbed directly to the surface of AuNPs or binds indirectly through a recognition element. This element can be an antibody, an aptamer, a small molecule or a polymer. Using AuNPs as SERS substrates, molecules containing thiol or cyanide group can be introduced onto the NP surface directly due to the high affinity between their functional groups and Au.108,109 Employing unfunctionalized AuNPs is simple to prepare. However, the lack of specificity may lead to the low selectivity of the sensors, especially for samples of complex matrices. More importantly, there are only few functional groups and ions that can attach to the surface of AuNPs. Thus, functionalization of AuNPs is a solution to avoid these two disadvantages because it allows specific binding of target elements on AuNPs. However, the limitation of this approach is that the functional molecule (or receptor) can be extremely large, for example, antibody, which makes the distance between the target element and the substrate too far to experience the signal enhancement.110 Thus, it has been a demand of investigating smaller molecules to functionalize AuNPs. Van Duyne's group has designed bisboronic acid receptors with two arms of boronic acid forming covalent bonds with glucose and one arm of thiol being immobilized on AuNP surface. In addition, the highly polarizable aromatic structure of the molecule acted as a Raman reporter to enhance SERS signals. The detection of glucose in physically relevant condition range allowed the diagnosis of both hypoglycemia and hyperglycemia.111 Besides, Raman signal of a target element can be amplified when it is adsorbed on hot-spot regions.107 Hence, in addition to directing the binding of analyte onto NPs, researchers also pay attention in creating hot-spots via material fabrication. Anisotropic AuNPs, such as nanostars, nanorods, have been synthesized and employed as SERS materials, leading to significant enhancement in SERS signal.112,113 Besides, hot-spots could be also created between isotropic AuNPs by self-assembly114 or polymer assisted assembly115 on the substrates, or even the targets themselves.116
In a strategy of hot-spots modulation, hot-spots are created or destroyed by the target elements themselves, consequently, SERS-based sensors are generated via either “turn-on” or “turn-off” mechanisms. “Turn-on” approach focuses on decreasing the distance between two adjacent AuNPs to create hot-spots to increase SERS signals. In contrast, in “turn-off” approach, the target elements disrupt the connections between AuNPs, elongate their interparticle distance to destroy the hot-spots, thus, decrease SERS signals. In fact, these mechanisms are similar to aggregation and anti-aggregation mechanisms in colorimetric sensing. An example for the “turn-on” method is the detection of Hg2+ ions based on the assembly of AuNPs functionalized with single stranded DNA into chains reported by Xu et al. in 2015. Thanks to the T-Hg2+-T bond between two thymine base pairs, double stranded DNA was form between NPs to decrease the interparticle distance. As a result, SERS signals are significantly enhanced, even at low concentration of Hg2+. Hence, the method was extremely sensitive with an LOD of 0.45 pg mL−1.117 On the other hand, heparin, an anticoagulant, was detected via “turn-off” method. Zhang et al. induced aggregation state of AuNPs using the interaction between 4-mercaptobenzoic on the surface of AuNPs and protamine. However, the higher affinity to protamine helped heparin to bind to this protein, resulting in anti-aggregation of the NPs and a decrease in SERS signals.118 To achieve high signals for SERS sensing, it is important to maintain the close distance between the substrates and the reporter. A similar strategy of “turn-on” and “turn-off” using the modulation of this distance can be also generated using the mechanisms mentioned above.119 However, applying either hot-spots between AuNPs or distance between AuNPs and reporter, “turn-on” approach is commonly preferred. The reason is that at low concentration levels of target elements, the decrease in SERS signals obtained by “turn-off” method is not as noticeable as the presence or not of the signal in “turn-on” approach.
Since SERS is a vibrational spectroscopy, once the structure or conformation of the recognition elements on the surface is changed, the spectral variations in some bands of SERS responds responses can be detected. In a 2019 study, Popp and coworkers reported the detection of Cu2+ in white wine using this principle. The interaction of Cu2+ and the dipicolylamine-based ligand bound to the surface of AuNPs caused the changes in SERS spectra associated with pyridine breathing mode.120 This chemo-optical sensing method is a new approach that takes advantage of the chemical sensitivity and specificity of SERS signals. However, it is not all of the chemical and conformational changes that can be detected in SERS responses. In general, changes of the groups linked directly to aromatic ring tend to have greater influence on the polarizability of the probe, therefore the spectral variations are also more noticeable.104 In addition to the active metal ions that can change the chemical/conformation of the ligands bound to the surface of NPs, large elements have also been detected using this mechanism. For example, the interaction between influenza-H1 antigens and their antibodies, which were linked to 4-ATP, led to the relaxation of the stretched aromatic linker on the surface of AuNPs. This conformational change resulted in significant up-shifts in the peaks corresponding to the C–S stretching and C–C breathing modes.121 This sensing mechanism has overcome the disadvantages of using large antibodies in SERS sensors as the long distance between target elements and AuNPs did not limit the SERS responses. This mechanism has paved the way to the development of novel methods to detect biological elements using SERS-based sensors.122,123
2.3. Advantages and disadvantages of AuNPs-based optical nanosensors
Taking advantage of the optical, chemical and electromagnetic properties of the AuNPs, combining with the knowledge about desired analytes, researchers can design and develop optical nanosensors with various of advantages. However, there are still several disadvantages with which researchers have to cope when working on these types of sensors. Although some advantages and disadvantages have been mentioned previously, in this section, we would like to summarize their pros and cons, in comparison to several other AuNPs-based nanosensors.Optical AuNPs-based sensors are simple and rapid.14,124 Especially, the results obtained for colorimetric sensors can be observed by bare-eye.14,125 However, they have to deal with a risk of false positive/negative results due to aggregation of AuNPs, especially in complex sample matrix.126,127 Moreover, the inadvertent aggregation of AuNPs may also reduce the sensibility and accuracy of SERS and FRET sensors.128,129 These risks in precision and accuracy can be avoided by functionalizing AuNPs with a recognition element specific for the desired analyte. Functionalization definitely improves the selectivity of optical AuNPs-based sensors, however, sometimes, it scarifies the stability of AuNPs.130 Nevertheless, the introduction of fine-tuning techniques resulted in more stable modified-AuNPs.131 Another drawback of functionalization of AuNPs in optical nanosensors is that large recognition element, such as polymer, protein, antibody, etc., elongate the distance between the surface of the particle and the analyte (in a SERS sensor) or the acceptor (in a FRET sensor), which prevents the analyte from experiencing SERS effect or prevents the fluorescence of the acceptor from being quenched. However, thanks to great quenching ability of AuNPs, the distance between them and the acceptor this distance to be extended up to 70–100 nm,89,91,94 thus, it is the problem of only the SERS sensors, and the solution is avoiding the large recognition elements despite their high specificity. Besides, the manufacture of multi-target optical sensors requires functionalizing AuNPs with multiple recognition probes. It can increase the complexity and the cost of sensor fabrication procedures, howbeit, to the best of our knowledge, researchers have to accept this drawback to design these optical sensors.132 In term of instrument, expect for colorimetric sensors, LSPR, FRET and SERS sensors all require specific spectroscopies for measurements.133 However, being developed for a long period of time, the spectroscopies for these techniques have become common and popular,134,135 which can be found in many laboratories all around the world, thus, it is convenient for researchers to generate these optical sensors.
Also using AuNPs as the main substrate for analyte detection, some other nanosensors can help researchers overcome the disadvantages of the optical sensors. Electrochemical sensors are promising for multi-target sensing as they can determine different analytes at the same time.136 However, they require noticeable electroactivity of the analytes, which limits the range of target molecules.137 Furthermore, electrode fouling may be a problem for sensing repeatability.138 Another novel technique based on plasmonic property of AuNPs is chaotic sensor, in which the evolution of absorption spectra of AuNPs was recorded under the excitation of a laser source and analyzed by electronic signals using a chaotic circuit.139 In another word, it is an opto-electric sensor, which senses the changes in conductivity arising from the absorption of the samples, thus, its sensibility is impressive. This sensor does not require any expensive spectroscopy. However, it is necessary to design a complicated electric modulator.139 Thus, it needs specialized human resources to design and operate the sensing system. In another approach, AuNPs can also act as an assisting material in terahertz (THz) sensors. THz waves are potential for detecting analytes at trace levels, especially biological elements whose absorption peaks fall in THz region. For example, L-histidine shows a strong absorption band at ∼0.78 THz.140 Unfortunately, AuNPs lack plasmonic property within this region.140 However, it has been reported that the assist of AuNPs could enhance the performance of metamaterials for THz sensors, leading to impressive LODs of the sensors.140,141 Avidin, Staphylococcus aureus (S. aureus), and microRNAs were detected using this kind of sensors at the concentration down to fM, pM, and even aM level, respectively.140–142 In spite of that excellent sensibility, THz sensors still exhibit several drawbacks. Compared to optical sensors, material fabrication for THz sensors is more complicated and expensive.140–143 Furthermore, being a novel technique, THz spectroscopy is not as popular.143 Moreover, due to several reported limitations, THz need to be further studied and modified before being employed for real samples.144 Therefore, the application of THz sensors is still limited.
Each of the AuNPs-based sensors has its advantages and disadvantages that researchers can consider developing the appropriate sensors based on their desired analytes and the conditions of their laboratories. With the convenience and simplicity of material fabrication and sensing system operation, as well as the popularity of the spectroscopies, AuNPs-based optical sensors still attract the interest of researchers with a huge number of sensors developed for food and health safety monitoring in recent years which will be presented in Tables 1 & 2 in the following sections.
3. AuNPs-based optical nanosensors for food and health safety monitoring
3.1. Food safety monitoring
The rising attention about food quality has triggered many studies on monitoring food safety. In general, they focus on the detection of the contaminants (i.e., heavy metals, pesticides, herbicides, farm animal drugs, etc.) and food additives (i.e., preservatives, food dyes). This part will present some examples of recent advances in AuNPs-based sensing to detect these targets (Fig. 7). Detailed information about limits of detection (LOD), linear ranges and real sample matrices is presented in Table 1. | ||
| Fig. 7 AuNPs-based optical sensors for food safety monitoring. Inset pictures: colorimetric sensor: reprinted from ref. 72 Copyright 2021, with permission from Elsevier. Fluorescence sensor: reproduced from ref. 145 with permission from The Royal Society of Chemistry, Copyright 2018. SERS sensor: reproduced from ref. 107 with permission from The Royal Society of Chemistry, Copyright 2015. | ||
In general, most of sensors using AuNPs as probes for detecting heavy metals operate based on the interaction between the ligands on the NP surface. There are several widely-used ligands that can be chosen to be suitable for the desired target. For example, the high specificity of thymine–Hg2+–thymine (T–Hg2+–T) affinity have been applied for many Hg2+ sensing systems as in the presence of Hg2+, T–T mismatches are formed. For example, Amiri et al. have introduced a FRET aptasensor for Hg2+ using carbon dots as donors and AuNPs as acceptors.145 The carbon dots were labeled with T-rich single stranded DNA, while the AuNPs were functionalized with a complementary DNA sequence (cDNA). When being mixed together, the AuNPs quenched the fluorescence of carbon dots. However, in the presence of Hg2+, the cDNA was replaced due to the formation of T–Hg2+–T on the T-rich single stranded DNA. As a result, fluorescence recovery occurred. The FRET sensor achieved an impressive LOD of 7.5 × 10−13 M of Hg2+.145 A similar principle has been employed to design an indirect colorimetric sensor, in which T-rich DNA sequence were used as a capturing probe for Hg2+. In the presence of Hg2+ ions, it bound to T-rich oligonucleotide to form a T–Hg2+–T complex. The formation of this complex prevented the adsorption of oligonucleotide on the surface of AuNPs. The AuNPs then were available as a nanozyme to oxidize TMB, resulting in a much deeper blue color of the solution compared to the previously coated AuNPs.147 Besides, the interaction between DNAzyme and Pb2+ also attract researchers' concern for anti-aggregation approach.148 In the presence of Pb2+, DNAzyme would cleavage the target sequence. Therefore, this phenomenon was employed to separate AuNPs and graphene quantum dots in a 2018 study, resulting in fluorescence recovery (Fig. 8A).149 The sensor had an LOD of 16.7 nM. However, compared to the utilization of T-rich oligonucleotide, using DNAzyme is more complicated and less convenient. Therefore, this approach is not widely used.
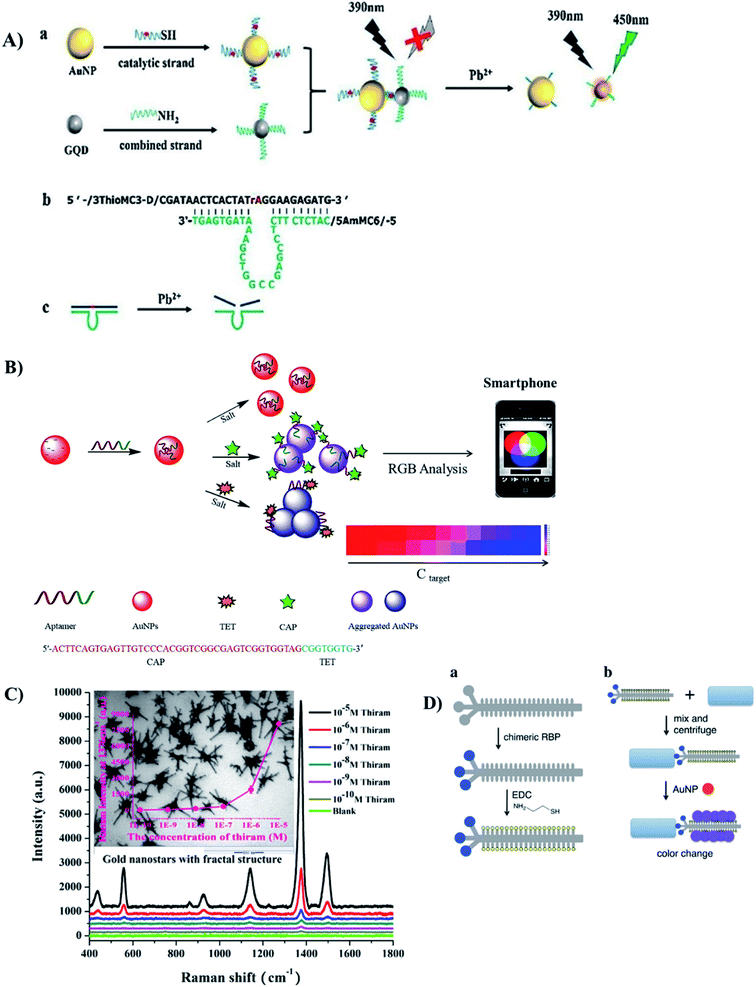 | ||
| Fig. 8 Different kinds of AuNPs-based optical sensors for detection of food contaminants. (A) FRET-based sensor for Pb2+ ions with the use of DNAzyme. Reprinted from ref. 149 Copyright 2018, with permission from Elsevier. (B) Colorimetric (de-protection) aptasensor for 2 kinds of antibiotics TET and CAP. Reprinted from ref. 132 Copyright 2018, with permission from Elsevier. (C) SERS-based sensor using Au nanostars to detect thiram. Reprinted from ref. 112 Copyright 2018, with permission from Elsevier. (D) Colorimetric (crosslinking) sensor for bacteria with the use of thiolated chimeric phage. Reprinted with permission from ref. 169 Copyright (2019) American Chemical Society. | ||
In addition to the interaction with oligonucleotide, heavy metals can also be captured by peptides that are rich in reactive groups. Glutathione (GSH) is an example. Each GSH possesses 8 binding sites, including 1 sulfhydryl, 1 amino group, 2 carboxyls and 4 carbonyl and amide donors.150 Therefore, heavy metal ions exhibit high affinity to this peptide. The thiol group allows GSH to be easily introduced onto the surface of AuNPs while other reactive groups are responsible for capturing heavy metal ions, causing the aggregation of AuNPs. Pb2+ and As3+ have been detected via this sensing principle.151–153 Incubated with GSH before the addition of AuNPs, Cd2+ exhibited such a high affinity to the peptide that AuNPs were prevented from binding to GSH, which led to the NP aggregation. The LOD of the method was calculated to be 4.3 pM.150 Similarly, other molecules containing reactive sites, either natural or synthesized ones, can become promising ligands to detect heavy metals in AuNPs-based assays.154
Another approach for the detection of pesticides is SERS-based sensors. Different kinds of organophosphorus pesticides and fungicides, such as thiram and carbendazim, have been detected using this method.112,159,160 In general, these residues can adsorb directly on the surface without any modifications. Therefore, to enhance the SERS signal, researchers have been focusing on the fabrication of the AuNPs. For example, Au nanostars (Fig. 8C), snowflake-like AuNPs, and spherical AuNPs deposited on a “sticky” tape have been employed as substrates for the detection of thiram and several organophosphorus pesticides.112,159–161
In general, there has been only few studies mentioning the detection of β-agonists using optical sensing systems. Most of them were based on the crossing-linking principle to cause aggregation of AuNPs. However, using different linking elements including melamine and aptamer has improved the selectivity of the detection,162,163 allowing them to perform in real complex samples. Recently, Han et al. reported on a ractopamine sensing system based on the growth of AuNPs in combination with a competitive immunoassay.164 In this system, ractopamine in the samples had to compete with ractopamine immobilized in the ELISA to bind to the antibodies. Then, biotinylated second antibodies were added, followed by the introduction of streptavidin label catalase, which could convert H2O2 to H2O and O2. In the presence of ractopamine, there was less catalase immobilized in the ELISA well, so Au3+ ions in the well were reduced by H2O2 rapidly, resulting the formation of AuNPs with the red color. In contrast, more catalase consumed a large amount of H2O2. The lack of H2O2 slowed down the kinetics of AuNP fabrication and induced the formation of aggregation with blue color.164
Antibiotics are also widely used in farming in a massive amount. The risk of antibiotic resistance caused by consuming agro-products containing antibiotics cannot be ignored. Therefore, for the last 10 years, many studies have been reported on the detection of antibiotics in food and animal samples. Most of them are colorimetric and fluorescent sensors, however, the detecting approaches are quite diverse. Using aptamers as capturing probes, ampicillin and kanamycin were detected via aggregation and anti-aggregation, respectively.165,166 In another approach, kanamycin, neomycin, streptomycin, and members of tetracycline family were employed to trigger the growth of AuNPs. The color of AuNPs confirmed their presence and concentrations in samples. Although these methods did not exhibit significant difference among different antibiotics,78,167 it is worth noting that most of those systems could be employed to detect antibiotics in milk and animal tissues. Recently, researchers have expressed a great interest in developing multi-residue sensors. In a 2020 study, Wu et al. described a colorimetric aptasensor for multiplex antibiotics, including chloramphenicol (CAP) and tetracycline (TET).132 A multifunctional aptamer was designed and adsorbed onto the AuNP surface. In the presence of one antibiotic, the specifically recognized fragment of the aptamer bound to that antibiotic and dissociated from the AuNP surface, while the non-specific one still attached and controlled the aggregation of AuNPs. Different antibiotics caused different states of AuNPs aggregation that could be distinguished by the colors of the solutions (Fig. 8B). Therefore, one sensing system was successful for the separate detection of two kinds of antibiotics with low LODs of 32.9 nM and 7.0 nM for TET and CAP, respectively.132 Similarly, Xiang's group reported an even more complicated system including two kinds of AuNPs labeled with 2 distinct aptamers to detect 5 types of aminoglycoside antibiotics.168 Due to different binding sites of those types of antibiotics, the obtained signal was distinguished.168 Therefore, 5 types of antibiotics could be detected separately using one sensing system. However, those two sensing systems were not possible to detect two or more antibiotics at once.
Melamine is a trimer of cyanamide which has been widely used in manufacture of polymer resins, fertilizers, kitchenware, etc. However, due to its high nitrogen contents, it has been added illegally into animal feeds and dairy products to inflate the protein contents.170 Consuming melamine containing food and drinks may cause serious damages to human kidneys and reproductive systems.170,171 Thus, it is important to develop reliable sensing methods to detect melamine. Due to the structure containing 3 amino groups, it is easily bound to 2–3 AuNPs at once, through the interactions with their coating agents such as citrate,171 thymine,172 and 1,4-dithiothreitol (DTT).173 Therefore, the detection of melamine is usually based on the crosslinking aggregation approach.171–175 Gao et al. even combine crosslinking and de-protection strategies to develop a colorimetric assay for the detection of melanine (Fig. 9A).174 To improve the sensitivity of the method, several novel strategies have been proposed. Zhang et al. took advantage of AuNP aggregation to generate a “turn-off” FRET sensor using conjugated polymer NPs as donors.176 In the absence of melamine, fluorescence signal was quenched by AuNPs. The introduction of melamine led to aggregation of the AuNPs, resulting in the fluorescence recovery. The method achieved a low LOD of 1.7 nM.176 Chen et al. focused on widening the dynamic detection range.170 The authors found that after introducing melamine to AuNP solution, AuNPs quickly aggregated and formed large aggregations. The precipitation of these aggregations led to the rapid evanishment of solution color, which reduced the accuracy of the colorimetric signals. Therefore, they introduced polyethylene glycol onto selected area of the AuNP surface, thereby stabilizing the AuNPs. The binding of melamine still occurred at the citrate-site of the AuNPs and induced a purple-to-blue color change. The linear range of the method covered from 1.05 nM to 1 mM.170
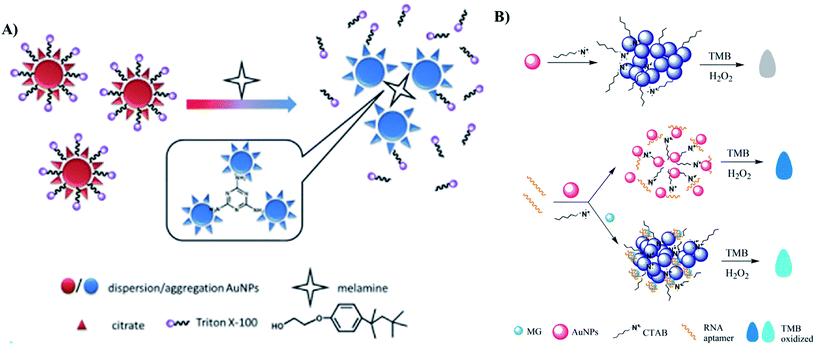 | ||
| Fig. 9 Different kinds of AuNPs-based optical sensors for detection of food additives. (A) Colorimetric sensor for detection of melanine based on two strategies: crosslinking and de-protection. Reprinted from ref. 174 Copyright 2018, with permission from Elsevier. (B) Colorimetric aptasensor for detecting Malachite Green based on the inhibition of the peroxidase-like activity of gold nanoparticle by CTAB. Reprinted by permission from Springer Nature: ref. 183 Copyright 2019. | ||
Preservatives have been playing an important role in the food industry by improving the duration of food products and allowing them to be stored and transported to consumers. However, illegal food preservatives are extremely toxic to human. For example, high intake nitrite and formaldehyde might lead to the risk of gastric cancer and leukemia.177,178 In general, there are only few studies on optical sensors to detect food preservatives, in comparison to electrochemical ones. Martínes-Aquino et al. employed resorcinol functionalized AuNPs for colorimetric detection of formaldehyde with an LOD of 0.05 ppm, but the method has not been applied in real samples.179 Nie et al. detected formaldehyde using non-functionalized AuNPs in a SERS sensor with an LOD of 1 × 10−4 μg mL−1, which exhibited satisfying performance in real samples of duck blood and rice flour.180 To detect thiabendazole, an antifungal agent and also a food preservative that is not approved in the EU, Alsammarraie et al. developed a SERS sensor, in which the utilization of Au nanorods as substrates has enhanced SERS signal.181 The sensor achieved LODs of 149, 216 and 179 μg L−1 in lemon, carrot and mango juice, respectively.181
Food dyes provide attractive colors to food and drinking products. However, due to their toxicity, many kinds of food dyes have been prohibited, including Sudan dyes, Malachite Green, Allura Red, Sunset Yellow, etc. Malachite Green (MG) is not only a dye but also an antimicrobial in aquaculture. Therefore, MG can be detected in fish and other seafood products. Due to its affinity to RNA aptamer, Jia et al. employed aptamer labeled AuNPs to detect MG using an anti-aggregation strategy.182 In the presence of MG, RNA aptamer would bind to MG instead of protecting AuNPs. As a result, AuNPs were aggregated at a high concentration of NaCl and the solution color turned from red to blue. The LOD of the sensor was as low as 15.95 nM.182 Also taking advantage of that affinity, Zhao et al. developed an indirect colorimetric sensor using CTAB as inhibitors for peroxidase-like activity of AuNPs.183 The system had 3 states of color. In the absence of RNA aptamer and MG, CTAB inhibited the enzymatic activity of AuNPs, so TMB could not be oxidized and the solution was nearly colorless (the concentration of AuNPs was low). In the addition of RNA aptamer, AuNPs were stabilized and protected, which consequently reduced the adsorption of CTAB on AuNPs, leading to the oxidization of TMB and a blue color in the solution. In the presence of MG, some of the aptamers bound to MG instead of AuNPs, reducing the level of protection, resulting in a light blue color in the solution. The LOD of the method was 1.8 nM (ref. 183) (Fig. 9B).
Biotoxins occur naturally in the environment, which are secreted by organisms to protect themselves from being eaten by others. For instance, mycotoxins are produced by the ones from fungus kingdom while saxitoxin is the most harmful shellfish toxin. To avoid food poisoning, it is important to determine these toxins in potential food before consuming. Therefore, several studies have been carried out to sense biotoxins using AuNPs-based optical sensors. Mycotoxins, such as fumonisin B1 and T-2, have been detected using colorimetric sensors via crosslinking and non-crosslinking approaches, respectively.197,198 Cao et al. employed cysteine-modified AuNPs as detecting probes to sense saxitoxin as the toxin could bind to cysteine via electrostatic interactions and hydrogen bonds. As a result, saxitoxin was detected with an LOD as low as 1 × 10−7 M.199
3.2. Health safety monitoring
For disease diagnostics, AuNPs-based sensors aim to detect the target molecules related to a specific disease. They can be molecules and peptides (e.g., glucose for diabetes, dopamine for schizophrenia disorder, glutathione for reperfusion injury, etc.), protein and nucleic acid biomarkers for non-infectious diseases (e.g., cancers, liver diseases, heart diseases, etc.), or infectious pathogens (e.g., bacteria, viruses, etc.) and their antibodies for infectious diseases in human blood, serum, saliva, urine, etc. Although it has been reported that appropriate nanomaterials for biomedical sensing should have the absorbance adjusted to the near-infrared region by changing their sizes or morphology. Thus, the ideal materials for disease diagnosis are large nanospheres, nanorods, nanocage, etc.75 However, it is the criteria for the substrates for in vivo sensing, i.e., bioimaging.75,223 In this review, we focus on in vitro disease diagnosis. Thus, there is no large difference in the nature of the samples in food and health safety monitoring sensors. Therefore, similar to the sensors described in the previous section, in this section, the selection of particle shape in each kind of AuNPs-based optical sensors for in vitro disease diagnosis is still obeyed what we mentioned in Section 2.2. Herein, we will introduce some AuNPs-based sensing systems for both non-infectious and infectious disease diagnostics, focusing on diabetes, cancers and pathogenic bacteria and viruses, especially SARS-CoV-2 (Fig. 10). The information about other sensors for other diseases is presented in Table 2.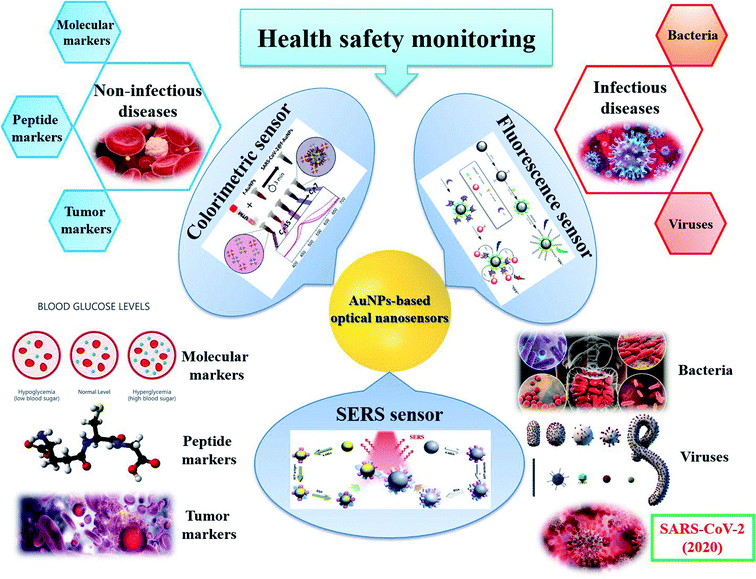 | ||
| Fig. 10 AuNPs-based optical sensors for health safety monitoring. Inset pictures: colorimetric sensor: reprinted with permission from ref. 224 Copyright (2020) American Chemical Society. Fluorescence sensor: reprinted by permission from Springer Nature: ref. 225 Copyright 2019. SERS sensor: reprinted by permission from Springer Nature: ref. 118 Copyright 2018. | ||
3.2.1.1. Diabetes. The glucose level in human blood, serum, saliva as well as urine is an indication of health status. Both high and low glycemic levels are harmful for human health. According to World Health Organization, a level below 3.0 mM (54 mg dL−1) of plasma glucose after a period of fasting can be considered as hypoglycemia.226 In contrast, plasma glucose maintaining at a high concentration of above 7.0 mM (126 mg dL−1) may present hyperglycemia.227 Persistently high glucose level can be the result of the inability to produce insulin of the pancreas or the inefficiency of utilizing insulin for uptaking glucose into the cells.228 This is one of the characterized symptoms for diabetes. Therefore, monitoring the glucose level is important to diagnose as well as manage diabetes. In this review, we focus on glucose as a typical example of the molecules that have been detected using many different sensing principles.
Glucose sensors using AuNPs as the detecting probes can be enzymatic or non-enzymatic. Glucose oxidase (GOx) is an enzyme for converting glucose to gluconolactone that releases H2O2 as a by-product.229 Thus, the level of glucose can be determined by the presence of this by-product. In 2020, Yang et al. designed a tapered fiber structure decorated with AuNPs and GOx to sense glucose.230 AuNPs integrated on surface of the tapered fiber were functionalized with GOx. In the addition of glucose solution, GOx decomposed the glucose and produced H2O2 that changed the refractive index (RI) of the medium surrounding the AuNPs. As mentioned in the previous part, the RI change Δn always leads to the shift of LSPR band. The absorbance peak shifted toward the longer wavelength region along with the increase in glucose level. This sensor achieved an LOD as low as 322 μM.230 This sensor also introduced the idea of designing a reusable sensor, which would reduce the cost of the system. However, the limitation of this sensor is that the RI can be affected by various factors. Intrinsic variabilities in complex biological samples can influence the accuracy of the method. This might be the reason why the sensing system was not employed to detect glucose in real samples.
Another strategy is taking the advantage of the reducing activity of H2O2 to accelerate AuNP etching reaction in the presence of halogenic ions. In a recent study, Donati et al. use Br− as the halogenic ion in a sensing system for glucose.77 Due to the enzymatic reaction between glucose and GOx in the AuNP solutions, H2O2 was produce. This by-product converted Br− ions into Br2 molecules, which quickly etched the multibranched AuNPs at their tips and reshaped them into spherical AuNPs. Interestingly, in real samples of saliva, the salivary thiols (i.e., cysteine, glutathione and others) acted as an organic shield that protected and stabilized the NP surface. Therefore, the AuNPs were not aggregated, and the blue-to-red color change of the solution was the result of the transformation of AuNP shapes. At a higher concentration of glucose, a pink color was observed in the reaction solution, indicating a hyperglycemia event (Fig. 11A). The LOD of the sensor calculated after analyzing saliva samples from different donors was 1.4 mg mL−1. Although this LOD is not in line with the best performing glucose sensors mentioned in this review, the system is promising to intrinsic biological variabilities or real clinic samples.77
 | ||
| Fig. 11 Several AuNPs-based optical sensors for detection of diabetes and cancers using different kinds of biomarkers. (A) Glucose (molecular biomarker). Colorimetric sensor based on AuNP growth. Reproduced with permission from ref. 77 Copyright 2020 Donati, Pomili, Boselli and Pompa. (B) p53 gene (DNA biomarker). FRET-based sensor with signal amplification strand displacement amplification of the target gene. Reprinted by permission from Springer Nature: ref. 241 Copyright 2019. (C) CA-125 (protein biomarker) FRET-based sensor with signal amplification by PCR. Reprinted from ref. 261 Copyright 2018, with permission from Elsevier. | ||
The by-product H2O2 can also acts as the analyte in an indirect sensor for glucose. Candem and coworkers designed a sensing system based on 3-mercaptophenylboronic acid (3-MPBA) modified (AuNPs).231 In the presence of H2O2, 3-MPBA is oxidized into 3-hydroxythiophenol (3-HTP). The change in the structure of boronate probe was presented by the change among two SERS spectra of the nanoprobe before and after the H2O2 treatment. For the real samples, GOx and nanoprobes were added into glucose-containing urine solutions, resulting in a change in SERS spectra indicating conversion of the boronate into the phenol as expected. A similar result was obtained in serum samples.231
Due to the specificity of the enzyme to its substrate, the enzymatic approach provides high selectivity to the AuNPs-based sensors. However, the presence of enzyme requires specific storage and reaction conditions. Thus, we also concern non-enzymatic strategies. Based on the formation of covalent bonds between glucose and boronic acids, Na et al. have employed 3-aminobenzeneboronic acid as the recognition element in a sensing assay using AuNPs and graphene oxide quantum dots (GOQDs).232 GOQDs displayed orange fluorescence, however, this fluorescence was quenched by AuNPs. In the presence of glucose and 3-aminobenzeneboronic acid, they bound together to form a cationic species that could cause aggregation of anionic AuNPs. As a result, the fluoresce signal of GOQD was recovered. This method resulted in an LOD of 0.65 μM,232 lower than that of a similar sensing system using phenylboronic acid, a boronic acid derivative in colorimetric approach.233 Besides, the complex of boronic acid and glucose is also useful to reduce the distance between AuNPs and create “hot spots” for SERS sensing.111
In another approach, glucose can act as a reducing and capping agent for AuNP synthesis. Hence, Pinheiro et al. have designed a colorimetric sensor with the direct detection of glucose without any intermediate product.234 The authors stated that low concentrations of glucose resulted in large AuNPs while high concentrations of glucose led to the formation of smaller AuNPs. The difference in sizes of AuNPs was claimed based on the LSPR shifts and the color changes of the reaction solutions. The method gave an LOD of 0.65 mM,234 which is not as low as some others that we have discussed above, but it is still appropriate in physiological range. More importantly, this study has introduced a fast and easily-prepared AuNPs-based sensor without NP modification.
In addition to glucose, glycated hemoglobin is another glycemic indicator, especially to measure the average glucose concentration in the blood of diabetic patients. In comparison to the glucose level, glycated hemoglobin level is more stable due to the 4 months lifespan of the hemoglobin.235 Therefore, Kwak et al. developed a sensor for detection of glycated hemoglobin in an enzymatic approach.236 Glycated hemoglobin consists of a glucose molecule binding to N-terminal valine residue of the hemoglobin β-chains. H2O2 was generated due to oxidative cleavage of fructosyl valine in glycated hemoglobin in the presence of fructosyl amino acid oxidase. Subsequently, H2O2 reduced Au3+ ions into Au atoms and formed AuNPs. The characteristic red color of AuNP solutions and their absorption peak at 520 nm represented for the presence of glycated hemoglobin. Based on the intensity of absorption peak, they calculated the ratio of glycated hemoglobin to total hemoglobin. The sensor exhibited an LOD of 0.124% of total hemoglobin and a linear range of 4.6% to 13.5% of total hemoglobin. Because normal level of glycated hemoglobin levels is within the range of 4–6%, this method was promising in vitro diagnosis for diabetes.236
3.2.1.2. Cancers. Cancers can be detected through the presence of biomarkers in blood, tissues and other body fluids.237 Cancer biomarkers can be proteins (either secreted or cell surface proteins), carbohydrates or nucleic acids (i.e., DNA, microRNA, etc.).238 Besides, in some studies, extracellular vesicles and even cancer cells were chosen to be biomarkers for cancer diagnosis.239,240 Most of AuNPs-based cancer sensors focus on detecting protein and nucleic acid biomarkers due to the well-understood knowledge of the specific interactions such as antigen–antibody, complementary nucleotide sequences, etc. The discovery of numerous cancer markers has enabled the detection of cancers, for example, p53 (tumor suppressor gene/protein),241,242 BRCA,243 HER2 (ref. 244) and Erα (breast cancer),245 AFP (liver cancer),246 PSA (prostate cancer),247 etc.
Nucleic acid biomarkers. To sense nucleic acid cancer biomarkers, the main strategy is based on the interaction between complementary nucleotide sequences. However, because the number of each gene copies in one cell is limited to 2, the concentration of DNA in the samples is consequently low, thus these sensors usually require a step of amplification.248,249 It can be either the amplification of target, which is carried out before the detection step, or the amplification of signal, which takes place after the detection step. As the most widely-used method to amplify DNA molecules is polymerase chain reaction (PCR), a colorimetric sensor for PSA3 has been designed by combining PCR technique and AuNP functionalization. In this 2019 study, Htoo et al. designed primers for PSA3 amplifying using PCR technique, in which the forward primer was thiolated.247 In the presence of the target gene (PSA3), PRC products were produced as thiolated double-stranded PSA3 sequences, which could bind to AuNPs due to the affinity between their thiol groups and the NP metallic surface. Thanks to the stabilization of the DNA ligands, AuNPs did not aggregate at high-salt condition (red color). In contrast, without the presence of the target gene, no PCR product was formed to stabilize AuNPs, thus, AuNPs aggregated at a high concentration of NaCl (blue color). The red or blue color of AuNP solution represents the presence or absence of PSA3 biomarker in the urine samples, respectively.247 Instead of thiolated primers, in another study, Trau and coworkers utilized biotin-contained primers to amplify 3 DNA sequences containing 3 important melanoma mutations.250 These amplicons could be conjugated onto streptavidin coated magnetic bead via biotin-streptavidin binding. Meanwhile, AuNPs were functionalized with 3 kinds of DNA tags complementary to 3 different barcodes on the forward primers for recognition. Moreover, each kind of AuNPs was modified with one Raman reporter. As a result, the presence of the targeted mutation was detected through the characteristic peaks of the appropriate reporter. Thanks to DNA amplification step, this multi-target SERS system achieved an impressive LOD of 0.1% mutation (10 copies).250
Despite of its advantages, DNA amplification still have several drawbacks including the risk of erroneously amplifying contamination and time-consuming, thus, researchers have developed procedures in which the response signal was amplified instead. For example, instead of amplifying the number of target DNA molecules, Yu and coworkers employed a strategy of “reusing” them.241 They introduced a sensor for p53 cancer gene using strand displacement amplification on functionalized AuNPs. FAM-labeled hairpin probes were bound on the surfaced of AuNPs, therefore, their fluorescence was quenched by AuNPs via FRET principle. In the presence of p53 gene, it hybridized with the hairpin probe due to the complementary sequence. The hybridization opened the hairpin, which not only led to fluorescence recovery, but also allowed the primer to bind to the opened hairpin and trigger the polymerization/displacement reactions. Once being replaced, p53 gene was release, then bound to another hairpin to start another round of reaction (Fig. 11B). The sequences of reactions have amplified the fluorescence signal, which allowed p53 gene to be quantified at a concentration as low as 1.6 pM.241
Another approach of signal amplifìcation is focusing on material fabrication. In a 2020 study, Bai et al. took advantage of the catalytic activity of AuNPs as they can trigger the reduction of 4-nitrophenol, resulting in a color change from yellow to colorless.243 They designed a capture probe and a signal probe, each of which contains a DNA sequence that is complementary to the target BRCA1 mutated gene. Capture probes were bound to the surface of the wafer while the signal ones were attached to 0D AuNPs. In the presence of the targets, three DNA sequences form a sandwich structure, and the AuNPs with signal probes were immobilized on the surface of the wafer. The colorimetric reaction catalyzed by these AuNPs provided signals for detection of BRCA1 mutation. Then AuNPs were grafted on the surface of 2D materials including GO and Bi2Se3 nanosheets, resulting in 4- and 10-fold amplification in signal, respectively. Thus, by improving the material property, the method reached an impressive LOD of 10−18 M and a linear range of 10−18 to 10−12 M.243
To simplify sensing procedures, researchers are also interested in fabrication of amplification-free sensors for DNA biomarkers. Yu et al. reported on a SERS sensor, in which the target sequence was sandwiched between 2 probe DNA-immobilized on the surface of 2 kinds of NPs: AuNPs and magnetic beads.251 AuNPs were modified with a Raman reporter. In fact, the structure of this sensing system is relatively similar to the one developed by Trau and coworkers that we mentioned above.250 However, Yu et al. did not amplify the target gene. Instead, their strategy was enhancing the SERS signal by utilizing hollow AuNPs, which could localize electromagnetic fields through their pinholes. Additionally, the targeted sequence was short (45 nucleotide), so the gaps between two particles were also small enough to create “hot-spots”. As a result, the method achieved an LOD of 2.7 fM.251 Despite the impressive result, it was only an experimental model, in which the target sequence was a PCA3 mimic gene. In real samples, it is difficult to isolate a gene sequence from genome without PCR. This technique might be more appropriate for other kinds of nucleic acid, which are short and single stranded. MicroRNA is a promising candidate.
A dual (colorimetric and FRET) sensor for microRNA-155 was developed by Borghei et al.252 This microRNA was reported to be over-expressed in many human cancers,253 so amplification was not necessary. They utilized DNA-modified silver nanoclusters (DNA-AgNCs) as donors and AuNPs as acceptors. In the absence of microRNA, the single-stranded DNA sequences were adsorbed onto AuNPs. Consequently, AuNPs quenched the fluorescence emitted from DNA-AgNCs. At the same time, this adsorption also prevented salt-induced aggregation of the AuNPs. In contrast, upon addition of microRNA, the hybridization between microRNA and the DNA immobilized on the AgNCs separated the DNA-AgNCs from the AuNPs. As a result, fluorescence recovery occurred. Also, AuNPs easily aggregated at high concentration of salt, reflected by a red to purple color change. Although this method exhibited an acceptable LODs of 0.6 nM and 0.4 pM for colorimetric and FRET sensor, respectively, it was reported in the article that those LODs were still higher than amplification step-containing methods.252 Hence, amplification is definitely important for the sensitivity of sensors for nucleic acid biomarkers.
Due to the highly specific interaction between complementary sequences, the selectivity of cancer sensors using nucleic acid targets is undeniable. In all of the studies mentioned above, an obvious difference was observed in the signal of the target genes in comparison with their mutant sequences, even with 1 or 2 mis-matched bases.241,243,247 However, the necessity of amplification can be a limitation to generate a fast and simple procedure of detecting cancer.
Protein biomarkers. Compared to nucleic acid, protein is a much richer source of biomarkers because many kinds of protein are over-expressed in cancer tissues.254 However, researchers may encounter the problems of selectivity as the mutations of a few nucleotides only cause a little change in the protein conformation.255 It requires full understandings of the interactions between these biomarkers and other biological molecules to generate sensitive and selective sensing systems for cancers.
The well-understood immune-interaction of antigen–antibody might be a simple and specific principle to design sensors for protein cancer biomarkers. In a 2015 study, Chen et al. reported on a gold nanorods-based LSPR sensor for cancer antigen 15-3 (CA15-3) using gold nanorods modified with CA15-3 antibody. The presence of CA15-3 led to increase of intensity of absorption spectra, red-shifts of the absorption peaks and the broadening of the peaks, corresponding to the assembly of the gold nanorods.256 Similarly, the mucin protein, MUC4, which is overexpressed in pancreatic cancer, was also recognized thanks to antibody-antigen binding in a SERS sensor.257 However, due to the large size of the antibody immobilized on AuNPs, the hot-spots for SERS signal enhancement were not created due to the assembly of AuNPs through binding with the same antigen. Instead, they designed a template stripped gold (TSG) substrate to immobilized antibodies. Thus, MUC4 antigens were captured between the substrate and the AuNPs in a sandwich model. The AuNPs were modified with a Raman reporter. Therefore, the signal of the reporter represented for the presence or absence of MUC4. In addition, the concentration of antibody immobilized on the substrate could be controlled to ensure the distance among those antibodies as well as interparticle distance among the AuNPs after the recognition step were close enough to create SERS hot-spots.257 Another example of the SERS sensor using this interaction is a sensor for AFP developed by Zhang et al. in 2018.258 This system consisted of both AuNPs and AgNPs, in which AgNPs were functionalized with AFP antibodies while AuNPs acted as the capturing probe for the attachment of AFP antigens in the sample. Due to the antibody–antigen interaction, the distance between the nanoparticles was reduced, leading to formation of “hot spots” that enhanced the SERS signal. As expected, the method exhibited high specificity. However, in complex real samples, it expressed poor sensitivity compared to other methods.258 This can be explained by the high biocompatibility of AuNPs, which allows the attachment of many other molecules rather than AFP antigens.
Recently, based on the interactions between protein molecules and DNA sequences, AuNPs have been functionalized with aptamers and employed as sensing probes for colorimetric sensors. Protein biomarkers such as PSA and p53 was detected using AuNPs-based aptasensors via cross-linking strategy.242,259 Meanwhile, de-protection strategy was employed for detection of others such as HER2,244 EAα245 and carcinoma embryonic antigen (CEA).260 In cross-linking models, the aptamers had to be designed as thiolated DNA242 or containing poly A sequences259 so that they could be attached to AuNPs. In the presence of target biomarker, aptamers-modified AuNPs aggregated due to the specific interaction between their coating agents and the target protein. As a result, a red to purple color change could be observed with bare eye.242,259 On the other hand, aptamer acted as a protecting agent to prevent salt-induced aggregation of AuNPs by absorbing onto the metallic surface in de-protection models. In the addition of target protein, the aptamer would bind to the target due to a higher affinity, and the AuNPs aggregated easily at high concentration of salt, which also resulted in a red to purple color change. This method is simple and fast with the incubation time of only 5–10 minutes.244,245,260 Moreover, it can avoid the use of antibodies, which are required strict conditions of storage and utilization. More importantly, the selectivity of aptamer is undeniable. For example, in the study on p53, the mutant p53 could be distinguished from the wild-type.242 The lack of a functional tetramerization domain in mutated p53 prevented it from inducing aggregation of aptamer-functionalized AuNPs. In contrast, the AuNPs aggregated in the presence of wild-type p53 as the aptamer contained recognition element for the binding of p53.
In another attempt to detect AFP, Zhou et al. combined both antibody–antigen and protein–DNA interactions in a FRET sensing system, where the aptamer labeled luminescent CdTe quantum dots (QDs) were the donors and AFP antibody grafted AuNPs were the acceptors.246 In the presence of AFP antigen, its affinity to antibody and aptamer reduced the distance between the two kinds of NPs. As a result, AuNPs quenched the fluorescence of QDs. This “turn-off” sensor exhibited a linear range of 0.5–45 ng mL−1, with an LOD of 400 pg mL−1. Moreover, it also showed a satisfying performance in human serum samples.246 It is worth noting that in this study, the author used 2 sites of recognition instead of one, which increased the specificity of the sensing method.
The combination of different strategies may result in interesting results. In a 2021 study, Shen et al. were successful to design and develop a FRET sensor for cancer antigen 125 (CA-125) at trace amounts by combining protein-aptamer interaction and DNA amplification.261 They designed 2 partly complementary sequences, named Oligo 1 and Oligo 2. Oligo 1 contained an aptamer for CA-125. In the absence of CA-125, the partially hybridized sequences were extended with Klenow fragment (exo) polymerase to obtain a new double stranded DNA molecule. This new DNA was employed as the template for PCR-amplification. Both forward and reverse primers contained a spacer, resulting in the formation of single-stranded DNA at two sides of the PCR product, which was complementary with Oligo 3 and Oligo 4 immobilized on the surface of AuNPs. Moreover, each forward primer also contained a fluorescein (FAM). After hybridization, the fluorescence of FAM was quenched by gold nanoparticles due to FRET effect, resulting in a decrease in fluorescence signal. In contrast, in the addition of CA-125, the antigen bound to Oligo 1 and prevented the hybridization between Oligo 1 and Oligo 2. It also terminated all the following process. Thanks to its conformation (Fig. 11C), the forward primer did not hybridize to the DNA probes on the AuNPs directly, thus, their fluorescence intensity remained. Despite its complexity, the method achieved an impressive LOD of 1.5 fg mL−1.261 Moreover, it has revealed the unlimited potential of combining different strategies and techniques in fabrication of novel systems.
Other biomarkers. In addition to nucleic acid and protein biomarkers, other tumor-related elements have been studied as new markers to detect cancers, including extracellular vesicles (EVs) secreted from cancer cells262 and cancer cells themselves.128 Among them, extracellular vesicles are more widely-used because they are the richer source. The advantages of using these large biomarker is that they contain many different membrane proteins, which can be employed as targets for detection.263 In 2020, Wang and coworkers introduced a sandwich-type SERS sensor for EVs tumor.264 The system included two kinds of aptamer targeting two different sites of the EVs that were used to label AuNPs and agarose beads, respectively. Even in complex samples, EVs could selectively bind to agarose beads, followed by the attachment of AuNPs to form sandwich complexes. The enhancement in SERS signal was observed due to this formation. The LOD of the sensor was as low as 2.44 pg μL−1.264 Similarly, Oliveira-Rodríguez et al. also employed two tetraspanins on the membrane of exosome as detecting target in their colorimetric sensor.263 The utilization of two binding sites on one target can increase the selectivity of the sensing systems.
On the other hand, cancer cells were the biomarkers in several studies. Butler et al. developed a near infrared SERS sensor based on 150 nm AuNPs to detect MCF-7 breast cancer cells.128 A significant enhancement of Raman signal was observed in the presence of MCF-7 cells. However, without specific functionalization, this sensing system were not effective in protein-rich samples, such as serum, with a low signal-to-noise ratio.128 Also in an effort of sensing MCF-7 cells, Li et al. introduced a colorimetric sensor, in which the cancer cells were recognized by Mucin 1 protein (MUC-1) aptamer-modified gold nanorods.265 In the presence of the cancer cells, the gold nanorods bound onto their membrane, which can be observed by UV-vis spectroscopy. Thanks to the specific binding of the aptamer and MUC-1 protein, the sensors could distinguish the target MCF-7 and untumorigenic cells HepG-2.265
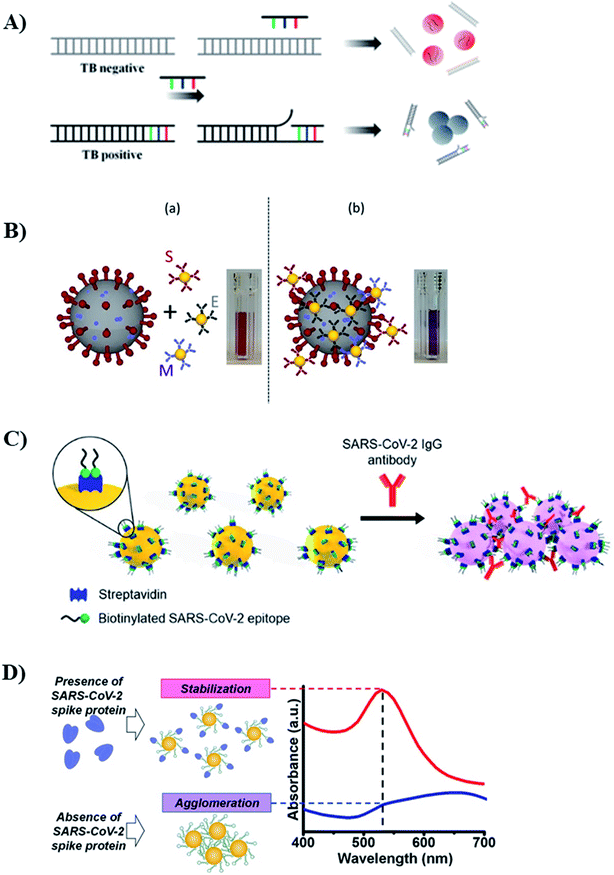 | ||
| Fig. 12 Several AuNPs-based colorimetric sensors for infectious pathogens designed for detecting different kinds of target. (A) Genome. Colorimetric (de-protection) sensor for detection of Tuberculosis. Reprinted with permission from ref. 268 Copyright (2017) American Chemical Society. (B) Whole virus particle. Colorimetric (crosslinking) sensor for detection of SARS-CoV-2. Reprinted with permission from ref. 224 Copyright (2020) American Chemical Society. (C) Antibody. Colorimetric (crosslinking) sensor for detection of SARS-CoV-2. Reprinted with permission from ref. 277. Copyright (2021) American Chemical Society. (D) Spike protein. Colorimetric (anti-aggregation) sensor for detection of SARS-CoV-2. Reprinted from ref. 279 Copyright 2021, with permission from Elsevier. | ||
Similarly, for virus detection, it is possible to detect the whole virus or its genome. Many kinds of influenza viruses were detected as whole virus particles,270–272 in which AuNPs were functionalized with recognition elements to sense the viruses. The most frequently-used capturing probe is antibody so that AuNPs would assemble on the surface of the virus. Thus, the presence of viruses could be determined due to a red to purple color change caused by that assembly.271,272 In another approach, Ahmed et al. reported that the organization of AuNPs on the surface of virus had enhanced the peroxidase-mimic enzymatic activity of positively-charged AuNPs.270 Compared to AuNPs only, AuNPs-decorated influenza viruses exhibited stronger catalytic activity when catalyzing the oxidation of TMB by H2O2, resulting in a deeper blue color in solution, reflecting in higher absorption intensity at 665 nm. It was the main principle to determine the presence of influenza virus H1N1 and H3N2.270 Instead of using antibody, Li and coworkers employed AuNPs modified with glycan containing terminal sialic acid (SA).273 The structure of this terminal SA played a decisive role for the binding of the functionalized AuNPs to the viruses. The binding of SA receptors to the viruses resulted in the aggregation of the AuNPs on the virus surface, and subsequently the blue color of the solution. In contrast, in case the SA receptor was not specific to the virus, the AuNPs would not bind to the viruses and the solution would remain the red color. Using a set of AuNPs with 7 different structures of SA receptors, the authors differentiated 14 influenza virus strains, including the most major subtypes in human and avian populations.273 Besides, an example for virus detection through its genome is a colorimetric assay to detect MERS-CoV developed by Kim et al.274 They designed a pair of thiolated probes at 5′ or 3′ end, which were complementary to MERS-CoV sequence. In the presence of the target DNA sequence, they reorganized into a double stranded DNA, which was assembled on the surface of AuNPs and prevented them from aggregation at high-salt condition. In contrast, the absence of target sequence led to the aggregation of AuNPs. The difference in color could be observed using bare eye or UV-Vis spectroscopy. The assay had a low LOD of 1 pmol μL−1.274
Jiao and coworkers have avoided the threat of being infected while working with viruses by choosing a different target, IgM antibody against SARS-CoV produced in the human body in the presence of the virus.86 They fabricated a testing strip, where SARS-CoV-2 nucleoprotein (SARS-CoV-2 NP) was coated on a membrane to capture the target antibody. Antihuman IgM was attached to AuNPs. In the presence of the antibody, it was immobilized between the nucleoprotein and AuNPs, which introduced a line in characteristic red color on the strip.86 Through this study, the authors have reported a fast testing of SARS-CoV-2 using serum samples rather than oral swabs. The limitation of this approach was that it required a relatively high concentration of antibody in human serum to form a visible red line on the testing strip. Therefore, SARS-CoV-2 could not be detected in early stage of infection using this method. In an effort to fabricate a more sensitive sensor for SARS-CoV-2 antibody, Lew et al. designed a colorimetric sensor to detect SARS-CoV-2 IgGs in patients' plasma.277 The IgG binding affinity of four immunodominant linear B-cell epitopes, located on the spike (S) and nucleocapsid (N) proteins, were characterized to find the two most sensitive ones, which were subsequently biotinylated and conjugated to streptavidin-coated AuNPs. In the presence of SARS-CoV-2 IgGs, these epitopes played the role of recognizing target antibodies, triggering to the aggregation of AuNPs (Fig. 12C). Moreover, co-immobilization of two epitopes led to an improvement in the sensitivity of the method, in comparison to single-epitope modified AuNPs. The assay achieved an LOD of 3.2 nM. Hence, it could be potentially used to detect IgG for convalescent COVID-19 patients. Using for 35 clinic samples, including Covid-19 patients' samples, the novel method successfully identified SARS-CoV-2 infection with 100% specificity and 83% sensitivity.277
In addition to the method using antigen–antibody interaction, recently, detecting SARS-CoV-2 using aptamers has attracted the interest of researchers.278,279 In a recent study, Aithal et al. employed aptamer-functionalized AuNPs to target S protein of the virus in a colorimetric sensor.279 In an anti-aggregation approach, the binding of S protein to the aptamer prevented to the aggregation of the AuNPs at high-salt condition (Fig. 12D). The sensor was well-performed in the presence of isolated S protein as well as heat-inactivated SARS-CoV-2 viruses. The nanoprobes could detect 3540 genome copies/μL inactivated SARS-CoV-2 virus. However, it should be further validated with real patients' samples.279
3.3. Potential of multi-metallic gold-based optical sensors for food and health safety monitoring
Recently, the introduction of multi-metallic nanomaterials, in general, and multi-metallic gold-based nanoparticles, in particular, has opened a potential of developing a new generation of optical sensors. To achieve the best plasmonic effect, noble metals such as silver (Ag), platinum (Pt), copper (Cu), etc. have been selected to combine with Au in the manufacture of multi-metallic nanostructures.289,290 It has been reported that these nanomaterials exhibited stronger LSPR bands than mono-metallic ones.289,291 Moreover, the position of LSPR band in UV-Vis spectrum of a multi-metallic material can be controlled by adjusting the ratio and the arrangement of its components.289–291 Besides, in another study, Bai et al. calculated electromagnetic field distributions of AuNPs and several multi-metallic NPs using Lorenz-Mie theory.292 The results revealed significant electromagnetic enhancement of alloy and core–shell multi-metallic structures (i.e., Ag–Au NPs and Au@Ag NP, respectively), and especially alloy core–shell structure (i.e., Au@Ag–Au NPs), in comparison with mono-metallic AuNPs.292 Therefore, multi-metallic gold-based NPs are promising to develop colorimetric (or LSPR) and SERS nanosensors. However, there has been no report on the property of quenching fluorescence of these NPs, especially in comparison to AuNPs. As a results, studies on multi-metallic gold-based NPs as substrates for optical sensors for food and health safety monitoring have focused on colorimetric and SERS sensors. For instance, the strong LSPR band of bimetallic Ag–Au NPs allowed George et al. to develop ultrasensitive colorimetric sensors for Mn2+ and ciprofloxacin, an antibiotic banned for animal food.293 Mn2+ and ciprofloxacin were detected at low concentrations down to 18.40 and 10.26 nM, respectively.293 In another approach, core–shell Au@Ag nanorods were employed to detect AFP, a protein marker of hepatocellular cancer.294 By controlling the thickness of Ag shell and the aspect ratio of Au core, the authors adjusted the color of nanorod solution to achieve the appropriate color range for bare-eyed observation.294 Besides, the noticeable electromagnetic enhancement of core–shell structures, especially at the internal gap between the core and the shell, has attracted the attention of researchers in the effort of designing and developing effective SERS substrates.292,295 For example, Au@Ag NPs was designed and optimized to detect pesticides, including tricyclazone and thiram in pear extracts, resulting in impressive LODs of 0.005 and 0.003 ppm, respectively.296 In addition, Cu@Au NPs exhibited a great potential to be appropriate SERS substrates for various biomolecules, including acetaminophen, dopamine, cholesterol, etc.290 Furthermore, the complex core–shell alloy structure of Au@Ag–Au NPs showed the best performance compared with core–shell (Au@Ag NPs), alloy (Ag–Au NPs) and mono-metallic structures (AuNPs) in SERS sensing system for cardiac troponin I, a biomarker for cardiac injury.292Despite the great potential, the application of multi-metallic gold-based NPs for development of optical nanosensors for food and health safety is still limited with only a few studies, and very few of them were carried out in real samples.296 Compared to mono-metallic AuNPs, complicated procedures of multi-metallic NP manufacture297 might have been prevented them from being widely-employed in sensing systems. Readers who are interested in the manufacture of multi-metallic NPs may find useful information in the established review by Srinoi et al.297 Nevertheless, the superior optical properties of multi-metallic gold-based NPs are undeniable, therefore, they still have space to grow as substrates for optical sensors for food and health safety monitoring in the future.
4. Trends and challenges of gold nanoparticles-based optical sensing systems for food and health safety monitoring
4.1. Trends
The huge amount of information in the tables has provided a picture of recent developments of AuNPs-based sensing systems for food safety control and in vitro disease diagnosis. Fig. 13 exhibits the most concerned detecting targets in both fields with the presence of long-term and emerging ones. In general, the development of AuNPs-based optical sensors have always caught up with urgent needs of the society. For example, virus outbreaks have triggered the investigation of their detecting sensors. High toxicity, even at low concentrations, of pesticides298 and melamine have led to the demand of monitoring these food contaminant/additive, especially when melamine contamination is usually related to enfant milk.299 Diabetes and cancers are also concerning as the number of patients suffering and dying of these disease keeps rising every year.300,301 Herein, we present several trends of developing AuNPs-based optical sensors in the recent 7 years. | ||
| Fig. 13 The most concerned detecting targets of AuNPs-based optical nanosensors for food and health safety (2015 – February 2022). | ||
With a large number of reported studies, obviously, AuNPs-based optical nanosensors are widely studied to monitor food safety. Since 2015, many food contaminants and additives have been detected using colorimetric, FRET- and SERS-based sensors. Starting with analytes possessing simple chemical structure such as heavy metals (e.g., Hg2+, Pb2+) and melamine, researchers have design various sensing systems based on their well-understood interactions with DNA,201 DNAzyme205 and peptides207 (for heavy metals) as well as citrate,219 methanobactin217 and DTT,173 etc. (for melamine). Thus, heavy metals and melamine were the targets for many reported optical nanosensors. Moreover, recent studies about melamine detection also concern the improvements in technologies, including newly-investigated ligands218 and the combination of known ligands170 to evaluate the performance of the optical sensors. They not only introduced effective sensing tools for the most popular contaminants and additive in food but also provided the concepts for the following studies. The enlarged knowledge about other food contaminants and their specific interactions to other compounds allow the fabrication of other sensing systems in the later period. We can clearly observe an upward trend in determination of toxins as many analytes in this category have been detected using different kinds of optical sensors and in various forms of real samples in recent 3 years.95,199,214 This trend may continue to grow in the following years. Detection of pesticides and herbicides is also a long-term concern of researchers. In fact, before 2016, pesticides and herbicides were determined using AuNPs-based optical sensors.156,302 However, it was not until 2017 that we observed a wave of research on pesticide and herbicide detection as many kinds of targets have been detected. The developments in nanotechnology have led to the significant improvements in the sensitivity of the sensors. For instance, SERS sensors for pesticide detection designed by Huang et al. in 2020159 resulted in impressive LODs which were 10 times lower than those of the studies reported in 2018 (ref. 112) and 2016,161 thanks to the fabrication of snowflake-like AuNPs and the utilization of specific ligands. Recently, many states and countries has participated in numerous Free Trade Agreements, which allows the trading of goods, including various kinds of food, all over the world. Therefore, cost- and time-effective sensing tools to monitor food safety and quality have been in high demand. It opens an opportunity for AuNPs-based optical sensors to be further investigated and applied in this large area in the near future.
Besides, the AuNPs have maintained their position as one of the most appropriate substrates to develop optical nanosensors for in vitro disease diagnosis during the observed period (i.e., 2015–now) thanks to their excellent biocompatibility.22,39,88 The non-infectious diseases such as diabetes and cancers have been always long-term concerns of researchers as many kinds of optical sensors have been generated to monitor the glucose level or determine varied tumor markers. Because diabetes and cancers are still unsolved problems of modern mankind,303,304 these studies may be continued in several years. On the other hand, the trends of investigating optical sensors for detection of infectious diseases usually follow their outbreaks. For examples, influenza viruses, which have been the perpetrators for the outbreaks of flu, including bird flu,273 swine flu,270 and seasonal flu,271 etc., have attract the interest of researchers for several years. The turning point came in December 2019 with the outbreak of a new corona virus (i.e., SARS-CoV-2), which then developed into COVID-19 pandemic, one of the most serious health crisis in the human history.305,306 Consequently, we are witnessing an explosion of researches on detecting COVID-19, including AuNPs-based optical sensors. Until now (February 2022), the world is still under the threat of the pandemic with the appearance of the new variants, and the virus shows no sign of stop. Therefore, the studies on SARS-CoV-2 will be continued, and the AuNPs-based optical sensing systems for rapid COVID-19 detection is still on the road to optimization.
Following the recent advances, we can see that antibody is losing their place as the most ideal recognition element. Although antibody provides selective targeting, more and more groups are choosing aptamer, which is more stable and smaller in size compared to antibodies. However, it cannot be denied that biological molecules are not as stable as chemical ones. Their affinity to analytes would reduce after a long time of storage, especially in hot weather. Researchers have started solving this challenge by finding alternative such as synthetic organic compounds,111 antibiotics190,195 and chimeric phages.169,196
Another trend is the utilization of smart phone in recording and analyzing signals, especially with colorimetric sensors, in RGB color model.132,158,221 Because almost everyone today possesses a smart phone, it is a step closer to the application of sensor in real life.
4.2. Challenges
Along with the developments of knowledge and new technology, researchers have focused on various AuNPs-based sensing systems for food and health safety monitoring. However, we are still facing several challenges to improve and optimize the performance of these sensing systems, including their sensibility, reliability and practicality.Besides, due to the complexity of real samples, each of which might contain more than one desired target, optical sensors have been designed and developed towards multi-target directions. However, among numerous examples that we have mentioned above, there is only one sensor that can detect more than one molecule at once. It is the sensing systems for 3 kinds of pesticides by Zhang et al. in 2020,158 in which 3 different fluorescence signals from 3 kinds of fluorophores were recorded, analyzed in RGB model to calculate the contents of each kind of pesticides. However, a fluorescence-based sensor always requires some instruments and tools to record and analyze the signals. Colorimetric sensor is a more user-friendly approach. Therefore, several groups have investigated multi-target colorimetric sensor. Wu et al. employed multi-functional aptamer labeled AuNPs132 while Yan et al. proposed a pattern recognition methods using two kinds of AuNPs functionalized with two different aptamers.168 However, due to the “red-to-blue” trap, these sensors are not accurate in complex samples contained two or more targets.
5. Conclusions and future prospects
Gold nanoparticles are excellent transducers for optical nanosensors including LSPR, colorimetric, FRET-based and SERS-based sensors for detection of food contaminants and diseases. Examples on advanced sensing systems have been summarized to draw a general picture of the recent developments of AuNPs-based optical nanosensors. In-laboratory works have shown impressive achievements in detecting many kinds of analytes. However, on the road of optimizing their sensing performance, there are several challenges. In the future, by solving all those challenges and taking advantage of the developments of every field of science and technology, sensor-developers may fabricate a new generation of AuNPs-based optical sensors, which is smarter, achieves significant improvements in sensing performance and accomplishes the goal of practical applications. To generate smart sensors, researchers need to combine the latest technologies including Internet of Things (IoT), Artificial Intelligence (AI) and machine learning to their sensing systems. These tools are able to collect, manage and analyze a large amount of data. Therefore, the data recorded by the sensors may be rapidly collated to the data bank, followed by the fast determination of the sensing system. Besides, the sensing performance of the sensors will be improved in every aspect. Knowledge about the interactions between analytes and recognition elements will be fulfilled, so more highly-specific elements (either natural or synthetic ones) at appropriate sizes will be employed for the detection of desired targets. Obviously, stable and durable ligands are highly desirable to enhance to stability of the systems. In addition, further studies on multi-metallic gold-based nanomaterials and their applications of optical sensing may be a direction to elevate the sensibility of the sensors. Finally, the new sensors will be more practicable. Standard but irreducible preparation procedures will be investigated in order to minimize the influence of environmental factors on the sensing performance, but still be user-friendly. Besides, the success of glucose meter has established the idea of developing hand-held spectrometers for LSPR, fluorescence and SERS measurements. The manufacture of these devices in combination with the development of professional analysis tools in smartphones might make AuNPs-based optical nanosensors become accessible for everyone. Furthermore, multi-target sensors will be the new direction to monitor complex samples, especially food products. Beside the red-to-blue range, sensor-developer may add more color ranges by combining indirect colorimetric sensing to the systems. In another approach, taking advantage of the different colors of fluorophores may be the key for the detection of several targets at once using fluorescence sensors. In term of material fabrication, AuNPs should be synthesized using robotic systems. It will ensure the homogeneity among different synthesis batches. Moreover, it will not only reduce the requirement of specialized human resources, but also allow highly-yielded manufacture. As a result, AuNP fabrication will be more cost- and time-effective, which is essential for the goal of high practicability. After several decades of research, the practical applications of optical sensors based on AuNPs are still limited. Hence, greater efforts should be made towards their practical utilization in the future.Author contributions
N. H. Anh: conceptualization, methodology, formal analysis, writing-original draft; M. Q. Doan: conceptualization, validation, investigation, writing-original draft; N. X.Dinh: conceptualization, writing-review & editing; T. Q. Huy: writing-review & editing, formal analysis; D. Q. Tri: funding acquisition, visualization, writing-review & editing; L. T. N. Loan: methodology, writing-review & editing; B. V. Hao: formal analysis, writing-review & editing; L. A. Tuan: conceptualization, methodology, supervision, project administration, writing-review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 103.02-2018.48. The authors would like to thank application-driven research program on Smart Sensing Technologies between ITIMS-HUST, IMS-VAST and MSE-NANO at the Phenikaa University.References
- W. Xing, H. Zhang, K. G. Scheckel and L. Li, Environ. Monit. Assess., 2015, 188, 23 CrossRef PubMed
.
- C. Xiu and K. K. Klein, Food Pol., 2010, 35, 463–470 CrossRef
.
- A. European Food Safety, H. Reich and G. A. Triacchini, EFSA J., 2018, 16, e05164 Search PubMed
.
- F. Federspiel and M. Ali, BMC Public Health, 2018, 18, 1338 CrossRef PubMed
.
- Q. Song, Y.-J. Zheng, Y. Xue, W.-G. Sheng and M.-R. Zhao, Neurocomputing, 2017, 226, 16–22 CrossRef
.
- A. N. Tchana, P. F. Moundipa and F. M. Tchouanguep, Int. J. Environ. Res. Public Health, 2010, 7, 178–188 CrossRef CAS PubMed
.
- T. Ghazi, T. Arumugam, A. Foolchand and A. A. Chuturgoon, Cells, 2020, 9, 2004 CrossRef CAS PubMed
.
- T. Borahan, T. Unutkan, A. Şahin and S. Bakırdere, J. Sep. Sci., 2019, 42, 678–683 CrossRef CAS
.
- D. T. Usmanov, S. Akhunov, U. Khasanov, V. M. Rotshteyn and B. Kasimov, Eur. J. Mass Spectrom., 2019, 26, 153–157 CrossRef
.
- H. Nakagawa, R. Hashimoto, Y. Matsuo, Y. Sago, K. Yokoyama and H. Takahashi, Curr. Microbiol., 2020, 77, 3057–3064 CrossRef CAS
.
- V. C. Fernandes, M. Freitas, J. P. G. Pacheco, J. M. Oliveira, V. F. Domingues and C. Delerue-Matos, J. Chromatogr. A, 2018, 1566, 1–12 CrossRef CAS PubMed
.
- A. Agarwal, R. Agrawal, D. V. Gunasekaran, D. Raje, B. Gupta, K. Aggarwal, S. L. Murthy, M. Westcott, S. P. Chee, P. McCluskey, H. S. Ling, S. Teoh, L. Cimino, J. Biswas, S. Narain, M. Agarwal, P. Mahendradas, M. Khairallah, N. Jones, I. Tugal-Tutkun, K. Babu, S. Basu, E. Carreño, R. Lee, H. Al-Dhibi, B. Bodaghi, A. Invernizzi, D. A. Goldstein, C. P. Herbort, T. Barisani-Asenbauer, J. J. González-López, S. Androudi, R. Bansal, B. Moharana, S. Mahajan, S. Esposti, A. Tasiopoulou, S. Nadarajah, M. Agarwal, S. Abraham, R. Vala, R. Singh, A. Sharma, K. Sharma, M. Zierhut, O. M. Kon, E. Cunningham, Q. D. Nguyen, C. Pavesio and V. Gupta, Ocul. Immunol. Inflamm., 2019, 27, 465–473 CrossRef CAS PubMed
.
- Y. Li, Z. Wang, L. Sun, L. Liu, C. Xu and H. Kuang, TrAC, Trends Anal. Chem., 2019, 113, 74–83 CrossRef CAS
.
- G. Liu, M. Lu, X. Huang, T. Li and D. Xu, Sensors, 2018, 18, 4166 CrossRef PubMed
.
- D. O'Hare, in Body Sensor Networks, ed. G.-Z. Yang, Springer London, London, 2014, DOI:10.1007/978-1-4471-6374-9_2, pp. 55–115
.
- S. Mohammadi, A. Salimi, S. Hamd-Ghadareh, F. Fathi and F. Soleimani, Anal. Biochem., 2018, 557, 18–26 CrossRef CAS PubMed
.
- Q. Liu, L. Li, Y. Zhao and Z. Chen, Mikrochim. Acta, 2018, 185, 301 CrossRef PubMed
.
- S. Dalirirad and A. J. Steckl, Sens. Actuators, B, 2019, 283, 79–86 CrossRef CAS
.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed
.
- N. Elahi, M. Kamali and M. H. Baghersad, Talanta, 2018, 184, 537–556 CrossRef CAS PubMed
.
- P. Zhao, N. Li and D. Astruc, Coord. Chem. Rev., 2013, 257, 638–665 CrossRef CAS
.
- M. Azharuddin, G. H. Zhu, D. Das, E. Ozgur, L. Uzun, A. P. F. Turner and H. K. Patra, Chem. Commun., 2019, 55, 6964–6996 RSC
.
- C. C. Chang, C. P. Chen, T. H. Wu, C. H. Yang, C. W. Lin and C. Y. Chen, Nanomaterials, 2019, 9, 861 CrossRef CAS PubMed
.
- P. Jiang, Y. Wang, L. Zhao, C. Ji, D. Chen and L. Nie, Nanomaterials, 2018, 8, 977 CrossRef PubMed
.
- E. Petryayeva and U. J. Krull, Anal. Chim. Acta, 2011, 706, 8–24 CrossRef CAS PubMed
.
- Y. Zhang, W. Chu, A. D. Foroushani, H. Wang, D. Li, J. Liu, C. J. Barrow, X. Wang and W. Yang, Materials, 2014, 7, 5169–5201 CrossRef PubMed
.
- L. Li, M. Zhang and W. Chen, J. Food Drug Anal., 2020, 28, 642–654 CrossRef
.
- M. Cordeiro, F. Ferreira Carlos, P. Pedrosa, A. Lopez and P. V. Baptista, Diagnostics, 2016, 6, 43 CrossRef PubMed
.
- L. Tessaro, A. Aquino, A. P. A. d. Carvalho and C. A. Conte-Junior, Sens. Actuators Rep., 2021, 3, 100060 CrossRef
.
- B. Negahdari, M. Darvishi and A. A. Saeedi, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 455–461 CrossRef CAS PubMed
.
- V. Ramalingam, Adv. Colloid Interface Sci., 2019, 271, 101989 CrossRef CAS PubMed
.
- D. T. Nguyen, D.-J. Kim and K.-S. Kim, Micron, 2011, 42, 207–227 CrossRef CAS PubMed
.
- P. Slepička, N. Slepičková Kasálková, J. Siegel, Z. Kolská and V. Švorčík, Materials, 2019, 13, 1 CrossRef PubMed
.
- T. Schmutzler, T. Schindler, T. Zech, S. Lages, M. Thoma, M.-S. Appavou, W. Peukert, E. Spiecker and T. Unruh, ACS Appl. Nano Mater., 2019, 2, 3206–3219 CrossRef CAS
.
- J.-E. Park, M. Atobe and T. Fuchigami, Ultrason. Sonochem., 2006, 13, 237–241 CrossRef CAS PubMed
.
- Y. Ren, C. Xu, M. Wu, M. Niu and Y. Fang, Colloids Surf., A, 2011, 380, 222–228 CrossRef CAS
.
- P. Priecel, H. Adekunle Salami, R. H. Padilla, Z. Zhong and J. A. Lopez-Sanchez, Chin. J. Catal., 2016, 37, 1619–1650 CrossRef CAS
.
- C. Kohout, C. Santi and L. Polito, Int. J. Mol. Sci., 2018, 19, 3385 CrossRef PubMed
.
- X. Hu, Y. Zhang, T. Ding, J. Liu and H. Zhao, Front. Bioeng. Biotechnol., 2020, 8, 990 CrossRef PubMed
.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC
.
- G. Frens, Nat. Phys. Sci., 1973, 241, 20–22 CrossRef CAS
.
- M. Brust, M. Walker, D. Bethell, D. J. Schiffrin and R. Whyman, J. Chem. Soc., Chem. Commun., 1994, 801–802, 10.1039/C39940000801
.
- M. Shah, V. Badwaik, Y. Kherde, H. K. Waghwani, T. Modi, Z. P. Aguilar, H. Rodgers, W. Hamilton, T. Marutharaj, C. Webb, M. B. Lawrenz and R. Dakshinamurthy, Front. Biosci., 2014, 19, 1320–1344 CrossRef PubMed
.
- R. Herizchi, E. Abasi, M. Milani and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2014, 44, 1–7 Search PubMed
.
- M. Grzelczak, J. Pérez-Juste, P. Mulvaney and L. M. Liz-Marzán, Chem. Soc. Rev., 2008, 37, 1783–1791 RSC
.
- S. E. Lohse and C. J. Murphy, Chem. Mater., 2013, 25, 1250–1261 CrossRef CAS
.
- C. Kan, C. Wang, J. Zhu and H. Li, J. Solid State Chem., 2010, 183, 858–865 CrossRef CAS
.
- M. A. Wall, S. Harmsen, S. Pal, L. Zhang, G. Arianna, J. R. Lombardi, C. M. Drain and M. F. Kircher, Adv. Mater., 2017, 29, 1605622 CrossRef PubMed
.
- J. Gubitosa, V. Rizzi, P. Fini, R. Del Sole, A. Lopedota, V. Laquintana, N. Denora, A. Agostiano and P. Cosma, Mater. Sci. Eng., C, 2020, 106, 110170 CrossRef CAS PubMed
.
- J. Yu, D. Xu, H. N. Guan, C. Wang, L. K. Huang and D. F. Chi, Mater. Lett., 2016, 166, 110–112 CrossRef CAS
.
- S. Palanisamy, S. K. Ramaraj, S.-M. Chen, T.-W. Chiu, V. Velusamy, T. C. K. Yang, T.-W. Chen and S. Selvam, J. Colloid Interface Sci., 2017, 496, 364–370 CrossRef CAS PubMed
.
- M. Annadhasan, J. Kasthuri and N. Rajendiran, RSC Adv., 2015, 5, 11458–11468 RSC
.
- F. Correard, K. Maximova, M.-A. Estève, C. Villard, M. Roy, A. Al-Kattan, M. Sentis, M. Gingras, A. V. Kabashin and D. Braguer, Int. J. Nanomed., 2014, 9, 5415–5430 CAS
.
- V. Amendola, S. Polizzi and M. Meneghetti, J. Phys. Chem. B, 2006, 110, 7232–7237 CrossRef CAS PubMed
.
- M. Vinod, R. S. Jayasree and K. G. Gopchandran, Curr. Appl. Phys., 2017, 17, 1430–1438 CrossRef
.
- M. Morita, T. Tachikawa, S. Seino, K. Tanaka and T. Majima, ACS Appl. Nano Mater., 2018, 1, 355–363 CrossRef CAS
.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103–1170 CrossRef CAS PubMed
.
- E. Colangelo, J. Comenge, D. Paramelle, M. Volk, Q. Chen and R. Lévy, Bioconjugate Chem., 2017, 28, 11–22 CrossRef CAS PubMed
.
- O. S. Muddineti, B. Ghosh and S. Biswas, Int. J. Pharm., 2015, 484, 252–267 CrossRef CAS PubMed
.
- Q. Shao and C. K. Hall, Langmuir, 2016, 32, 7888–7896 CrossRef CAS PubMed
.
- B. Sajjanar, B. Kakodia, D. Bisht, S. Saxena, A. K. Singh, V. Joshi, A. K. Tiwari and S. Kumar, J. Nanopart. Res., 2015, 17, 234 CrossRef
.
- S. Giorgi-Coll, M. J. Marín, O. Sule, P. J. Hutchinson and K. L. H. Carpenter, Mikrochim. Acta, 2019, 187, 13 CrossRef PubMed
.
- J. Cao, T. Sun and K. T. V. Grattan, Sens. Actuators, B, 2014, 195, 332–351 CrossRef CAS
.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef
.
- X. Lu, M. Rycenga, S. E. Skrabalak, B. Wiley and Y. Xia, Annu. Rev. Phys. Chem., 2009, 60, 167–192 CrossRef CAS PubMed
.
- Z. Zhang, Z. Chen, F. Cheng, Y. Zhang and L. Chen, Biosens. Bioelectron., 2017, 89, 932–936 CrossRef CAS PubMed
.
- Y. A. Attia, D. Buceta, F. G. Requejo, L. J. Giovanetti and M. A. López-Quintela, Nanoscale, 2015, 7, 11273–11279 RSC
.
- L. M. Liz-Marzán, Langmuir, 2006, 22, 32–41 CrossRef PubMed
.
- A. Bigdeli, F. Ghasemi, H. Golmohammadi, S. Abbasi-Moayed, M. A. F. Nejad, N. Fahimi-Kashani, S. Jafarinejad, M. Shahrajabian and M. R. Hormozi-Nezhad, Nanoscale, 2017, 9, 16546–16563 RSC
.
- S. Megarajan and A. Veerappan, Opt. Mater., 2020, 108, 110177 CrossRef CAS
.
- W. H. Wu, M. Li, Y. Wang, H. X. Ouyang, L. Wang, C. X. Li, Y. C. Cao, Q. H. Meng and J. X. Lu, Nanoscale Res. Lett., 2012, 7, 658 CrossRef PubMed
.
- L. Mao, Q. Wang, Y. Luo and Y. Gao, Talanta, 2021, 222, 121506 CrossRef CAS PubMed
.
- J. Zhang, X. Sun and J. Wu, Appl. Sci., 2019, 9, 489 CrossRef CAS
.
- L. Guo, X. Wu, L. Liu, H. Kuang and C. Xu, Small, 2018, 14, 1701782 CrossRef PubMed
.
- A. J. Mieszawska, W. J. M. Mulder, Z. A. Fayad and D. P. Cormode, Mol. Pharm., 2013, 10, 831–847 CrossRef CAS PubMed
.
- C.-C. Chang, C.-P. Chen, T.-H. Wu, C.-H. Yang, C.-W. Lin and C.-Y. Chen, Nanomaterials, 2019, 9, 861 CrossRef CAS PubMed
.
- P. Donati, T. Pomili, L. Boselli and P. P. Pompa, Front. Bioeng. Biotechnol., 2020, 8, 601216 CrossRef PubMed
.
- L. Shen, J. Chen, N. Li, P. He and Z. Li, Anal. Chim. Acta, 2014, 839, 83–90 CrossRef CAS PubMed
.
- C. Jiang, Z. Li, Y. Wu, W. Guo, J. Wang and Q. Jiang, Bull. Korean Chem. Soc., 2018, 39, 625–630 CrossRef CAS
.
- Y. Wu, Y. Chen, Y. Li, J. Huang, H. Yu and Z. Wang, Sens. Actuators, B, 2018, 270, 443–451 CrossRef CAS
.
- C. Jiang, J. Zhu, Z. Li, J. Luo, J. Wang and Y. Sun, RSC Adv., 2017, 7, 44463–44469 RSC
.
- V. Kumar, D. Bano, D. K. Singh, S. Mohan, V. K. Singh and S. H. Hasan, ACS Sustain. Chem. Eng., 2018, 6, 7662–7675 CrossRef CAS
.
- Y. Zhao, Y. Huang, H. Zhu, Q. Zhu and Y. Xia, J. Am. Chem. Soc., 2016, 138, 16645–16654 CrossRef CAS PubMed
.
- S. M. Taghdisi, N. M. Danesh, M. Ramezani, A. S. Emrani and K. Abnous, ACS Appl. Mater. Interfaces, 2018, 10, 12504–12509 CrossRef CAS PubMed
.
- E. Guler, T. Yilmaz Sengel, Z. P. Gumus, M. Arslan, H. Coskunol, S. Timur and Y. Yagci, Anal. Chem., 2017, 89, 9629–9632 CrossRef CAS PubMed
.
- C. Huang, T. Wen, F.-J. Shi, X.-Y. Zeng and Y.-J. Jiao, ACS Omega, 2020, 5, 12550–12556 CrossRef CAS PubMed
.
- C. Shende, C. Brouillette and S. Farquharson, Analyst, 2019, 144, 5449–5454 RSC
.
- M. Cordeiro, F. Ferreira Carlos, P. Pedrosa, A. Lopez and P. V. Baptista, J. Diagn., 2016, 6, 43 Search PubMed
.
- J. Shi, F. Tian, J. Lyu and M. Yang, J. Mater. Chem. B, 2015, 3, 6989–7005 RSC
.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS PubMed
.
- J. Xu, Y. Li, L. Wang, Y. Huang, D. Liu, R. Sun, J. Luo and C. Sun, Dyes Pigm., 2015, 123, 55–63 CrossRef CAS
.
- G. Chen, F. Song, X. Xiong and X. Peng, Ind. Eng. Chem. Res., 2013, 52, 11228–11245 CrossRef CAS
.
- S. Mayilo, M. A. Kloster, M. Wunderlich, A. Lutich, T. A. Klar, A. Nichtl, K. Kürzinger, F. D. Stefani and J. Feldmann, Nano Lett., 2009, 9, 4558–4563 CrossRef CAS PubMed
.
- P. C. Ray, A. Fortner and G. K. Darbha, J. Phys. Chem. B, 2006, 110, 20745–20748 CrossRef CAS PubMed
.
- C. Wang, Y. Li, C. Zhou and Q. Zhao, Mikrochim. Acta, 2019, 186, 728 CrossRef PubMed
.
- N.-T. Chen, S.-H. Cheng, C.-P. Liu, J. S. Souris, C.-T. Chen, C.-Y. Mou and L.-W. Lo, Int. J. Mol. Sci., 2012, 13, 16598–16623 CrossRef CAS PubMed
.
- W. Dong, R. Wang, X. Gong and C. Dong, Anal. Bioanal. Chem., 2019, 411, 6687–6695 CrossRef CAS PubMed
.
- Y. Wang, M. Lv, Z. Chen, Z. Deng, N. Liu, J. Fan and W. Zhang, Front. Chem., 2020, 8, 238 CrossRef CAS PubMed
.
- J. Shi, C. Chan, Y. Pang, W. Ye, F. Tian, J. Lyu, Y. Zhang and M. Yang, Biosens. Bioelectron., 2015, 67, 595–600 CrossRef CAS PubMed
.
- M. I. Halawa, J. Lai and G. Xu, Mater. Today
Nano, 2018, 3, 9–27 CrossRef
.
- C. Muehlethaler, M. Leona and J. R. Lombardi, Anal. Chem., 2016, 88, 152–169 CrossRef CAS PubMed
.
- M. G. Albrecht and J. A. Creighton, J. Am. Chem. Soc., 1977, 99, 5215–5217 CrossRef CAS
.
- D. L. Jeanmaire and R. P. Van Duyne, J. Electroanal. Chem. Interfacial Electrochem., 1977, 84, 1–20 CrossRef CAS
.
- X. Gu, M. J. Trujillo, J. E. Olson and J. P. Camden, Annu. Rev. Anal. Chem., 2018, 11, 147–169 CrossRef CAS PubMed
.
- S. Kumar, P. Kumar, A. Das and C. S. Pathak, 2020, DOI:10.5772/intechopen.92614.
- S. L. Kleinman, R. R. Frontiera, A.-I. Henry, J. A. Dieringer and R. P. Van Duyne, Phys. Chem. Chem. Phys., 2013, 15, 21–36 RSC
.
- H. Wei, S. M. Hossein Abtahi and P. J. Vikesland, Environ. Sci.: Nano, 2015, 2, 120–135 RSC
.
- Q. Xiao, H. Gao, C. Lu and Q. Yuan, TrAC, Trends Anal. Chem., 2012, 40, 64–76 CrossRef CAS
.
- D. Senapati, S. S. R. Dasary, A. K. Singh, T. Senapati, H. Yu and P. C. Ray, Chem. – Eur. J., 2011, 17, 8445–8451 CrossRef CAS PubMed
.
- C. Catala, B. Mir-Simon, X. Feng, C. Cardozo, N. Pazos-Perez, E. Pazos, S. Gómez-de Pedro, L. Guerrini, A. Soriano, J. Vila, F. Marco, E. Garcia-Rico and R. A. Alvarez-Puebla, Adv. Mater. Technol., 2016, 1, 1600163 CrossRef
.
- B. Sharma, P. Bugga, L. R. Madison, A.-I. Henry, M. G. Blaber, N. G. Greeneltch, N. Chiang, M. Mrksich, G. C. Schatz and R. P. Van Duyne, J. Am. Chem. Soc., 2016, 138, 13952–13959 CrossRef CAS PubMed
.
- J. Zhu, M.-J. Liu, J.-J. Li, X. Li and J.-W. Zhao, Spectrochim. Acta, Part A, 2018, 189, 586–593 CrossRef CAS PubMed
.
- Y. Ou, X. Wang, K. Lai, Y. Huang, B. A. Rasco and Y. Fan, J. Agric. Food Chem., 2018, 66, 2954–2961 CrossRef CAS PubMed
.
- N. Ha Anh, M. Quan Doan, N. Xuan Dinh, T. Quang Huy, D. Quang Tri and A.-T. Le, Appl. Surf. Sci., 2022, 584, 152555 CrossRef CAS
.
- M. Chen, W. Luo, Q. Liu, N. Hao, Y. Zhu, M. Liu, L. Wang, H. Yang and X. Chen, Anal. Chem., 2018, 90, 13647–13654 CrossRef CAS PubMed
.
- A. M. Paul, Z. Fan, S. S. Sinha, Y. Shi, L. Le, F. Bai and P. C. Ray, J. Phys. Chem. C, 2015, 119, 23669–23675 CrossRef CAS PubMed
.
- L. Xu, H. Yin, W. Ma, H. Kuang, L. Wang and C. Xu, Biosens. Bioelectron., 2015, 67, 472–476 CrossRef CAS PubMed
.
- C. Zhang, X. Liang, T. You, N. Yang, Y. Gao and P. Yin, Anal. Methods, 2017, 9, 2517–2522 RSC
.
- Y. Shi, H. Wang, X. Jiang, B. Sun, B. Song, Y. Su and Y. He, Anal. Chem., 2016, 88, 3723–3729 CrossRef CAS PubMed
.
- V. Dugandžić, S. Kupfer, M. Jahn, T. Henkel, K. Weber, D. Cialla-May and J. Popp, Sens. Actuators, B, 2019, 279, 230–237 CrossRef
.
- K. W. Kho, U. S. Dinish, A. Kumar and M. Olivo, ACS Nano, 2012, 6, 4892–4902 CrossRef CAS PubMed
.
- H. Ma, X. Sun, L. Chen, W. Cheng, X. X. Han, B. Zhao and C. He, Anal. Chem., 2017, 89, 8877–8883 CrossRef CAS PubMed
.
- C. Song, S. Guo, S. Jin, L. Chen and Y. M. Jung, Chemosensors, 2020, 8, 118 CrossRef CAS
.
- G. Zhang, Nanotechnol. Rev., 2013, 2, 269–288 CAS
.
- G. Liu, R. Zhang, X. Huang, L. Li, N. Liu, J. Wang and D. Xu, Sensors, 2018, 18, 1595 CrossRef PubMed
.
- S. H. Hwang, S. Jeong, H. J. Choi, H. Eun, M. G. Jo, W. Y. Kwon, S. Kim, Y. Kim, M. Lee and K. S. Park, J. Nanomater., 2019, 2019, 3676384 CrossRef
.
- M. S. Cordray, M. Amdahl and R. R. Richards-Kortum, Anal. Biochem., 2012, 431, 99–105 CrossRef CAS PubMed
.
- H. J. Butler, S. W. Fogarty, J. G. Kerns, P. L. Martin-Hirsch, N. J. Fullwood and F. L. Martin, Analyst, 2015, 140, 3090–3097 RSC
.
- M. Swierczewska, S. Lee and X. Chen, Phys. Chem. Chem. Phys., 2011, 13, 9929–9941 RSC
.
- J. Manson, D. Kumar, B. J. Meenan and D. Dixon, Gold Bull., 2011, 44, 99–105 CrossRef CAS
.
- H. Daima, P. R. Selvakannan, R. Shukla, S. Bhargava and V. Bansal, PLoS One, 2013, 8, e79676 CrossRef CAS PubMed
.
- Y.-Y. Wu, P. Huang and F.-Y. Wu, Food Chem., 2020, 304, 125377 CrossRef CAS PubMed
.
- J. B. Wiester, in Raman Spectroscopy in the Undergraduate Curriculum, American Chemical Society, 2018, vol. 1305, ch. 2, pp. 13–33 Search PubMed
.
- A. Saletnik, B. Saletnik and C. Puchalski, Molecules, 2021, 26, 1537 CrossRef CAS PubMed
.
- M. Nachtmann, J. Deuerling and M. Rädle, Micromachines, 2020, 11, 353 CrossRef PubMed
.
- G. Maduraiveeran and R. Ramaraj, J. Anal. Sci. Technol., 2017, 8, 14 CrossRef
.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Sensors, 2008, 8, 1400–1458 CrossRef CAS PubMed
.
- J. Barek, Chemosensors, 2021, 9, 12 CrossRef CAS
.
- H. Martines-Arano, B. E. García-Pérez, M. A. Vidales-Hurtado, M. Trejo-Valdez, L. H. Hernández-Gómez and C. Torres-Torres, Sensors, 2019, 19, 4728 CrossRef CAS PubMed
.
- W. Xu, L. Xie, J. Zhu, X. Xu, Z. Ye, C. Wang, Y. Ma and Y. Ying, ACS Photonics, 2016, 3, 2308–2314 CrossRef CAS
.
- K. Yang, J. Li, M. Lamy de la Chapelle, G. Huang, Y. Wang, J. Zhang, D. Xu, J. Yao, X. Yang and W. Fu, Biosens. Bioelectron., 2021, 175, 112874 CrossRef CAS PubMed
.
- K. Yang, W. Yu, G. Huang, J. Zhou, X. Yang and W. Fu, RSC Adv., 2020, 10, 26824–26833 RSC
.
- K. Smart, J. Du, L. Li, D. Wang, K. Leslie, F. Ji, X. D. Li and D. Z. Zeng, Sensors, 2016, 16, 579 CrossRef PubMed
.
- V. A. Trofimov and S. A. Varentsova, Sensors, 2016, 16, 502 CrossRef PubMed
.
- S. Amiri, R. Ahmadi, A. Salimi, A. Navaee, S. Hamd Qaddare and M. K. Amini, New J. Chem., 2018, 42, 16027–16035 RSC
.
- H. Chen, K. Zhou and G. Zhao, Trends Food Sci. Technol., 2018, 78, 83–94 CrossRef CAS
.
- Y. Qi, D. Song and Y. Chen, Sci. Total Environ., 2021, 766, 142579 CrossRef CAS PubMed
.
- Z. Huang, J. Chen, Z. Luo, X. Wang and Y. Duan, Anal. Chem., 2019, 91, 4806–4813 CrossRef CAS PubMed
.
- X. Niu, Y. Zhong, R. Chen, F. Wang, Y. Liu and D. Luo, Sens. Actuators, B, 2018, 255, 1577–1581 CrossRef CAS
.
- L. Li, B. Liu and Z. Chen, Mikrochim. Acta, 2018, 186, 37 CrossRef PubMed
.
- L. Beqa, A. K. Singh, S. A. Khan, D. Senapati, S. R. Arumugam and P. C. Ray, ACS Appl. Mater. Interfaces, 2011, 3, 668–673 CrossRef CAS PubMed
.
- L. Gong, B. Du, L. Pan, Q. Liu, K. Yang, W. Wang, H. Zhao, L. Wu and Y. He, Mikrochim. Acta, 2017, 184, 1185–1190 CrossRef CAS
.
- W. Chu, Y. Zhang, D. Li, C. J. Barrow, H. Wang and W. Yang, Biosens. Bioelectron., 2015, 67, 621–624 CrossRef CAS PubMed
.
- A. Anwar, A. Minhaz, N. A. Khan, K. Kalantari, A. B. M. Afifi and M. R. Shah, Sens. Actuators, B, 2018, 257, 875–881 CrossRef CAS
.
- L. Yang, X. Zhang, H. Li and L. Jiang, Anal. Methods, 2017, 9, 6139–6147 RSC
.
- W. Bai, C. Zhu, J. Liu, M. Yan, S. Yang and A. Chen, Environ. Toxicol. Chem., 2015, 34, 2244–2249 CrossRef CAS PubMed
.
- S. Wu, D. Li, J. Wang, Y. Zhao, S. Dong and X. Wang, Sens. Actuators, B, 2017, 238, 427–433 CrossRef CAS
.
- C. Zhang, Z. Jiang, M. Jin, P. Du, G. Chen, X. Cui, Y. Zhang, G. Qin, F. Yan, A. M. Abd El-Aty, A. Hacimüftüoğlu and J. Wang, Food Chem., 2020, 326, 126813 CrossRef CAS PubMed
.
- D. Huang, J. Zhao, M. Wang and S. Zhu, Food Control, 2020, 108, 106835 CrossRef CAS
.
- X. Chen, M. Lin, L. Sun, T. Xu, K. Lai, M. Huang and H. Lin, Food Chem., 2019, 293, 271–277 CrossRef CAS PubMed
.
- J. Chen, Y. Huang, P. Kannan, L. Zhang, Z. Lin, J. Zhang, T. Chen and L. Guo, Anal. Chem., 2016, 88, 2149–2155 CrossRef CAS PubMed
.
- Y. Zhou, P. Wang, X. Su, H. Zhao and Y. He, Talanta, 2013, 112, 20–25 CrossRef CAS PubMed
.
- P. Wang, X. Su, L. Shi and Y. Yuan, Mikrochim. Acta, 2016, 183, 2899–2905 CrossRef CAS
.
- S. Han, T. Zhou, B. Yin and P. He, Mikrochim. Acta, 2018, 185, 210 CrossRef PubMed
.
- K.-M. Song, E. Jeong, W. Jeon, M. Cho and C. Ban, Anal. Bioanal. Chem., 2012, 402, 2153–2161 CrossRef CAS PubMed
.
- N. Zhou, J. Zhang and Y. Tian, Anal. Methods, 2014, 6, 1569–1574 RSC
.
- X. Zhang, Y. Zhang, H. Zhao, Y. He, X. Li and Z. Yuan, Anal. Chim. Acta, 2013, 778, 63–69 CrossRef CAS PubMed
.
- S. Yan, X. Lai, G. Du and Y. Xiang, Anal. Chim. Acta, 2018, 1034, 153–160 CrossRef CAS PubMed
.
- H. Peng and I. A. Chen, ACS Nano, 2019, 13, 1244–1252 CAS
.
- X.-Y. Chen, W. Ha and Y.-P. Shi, Talanta, 2019, 194, 475–484 CrossRef CAS PubMed
.
- M. Zhou, J. Yin, X. Zhao, Y. Fu, X. Jin, X. Liu and T. Jiao, Colloids Surf., A, 2020, 603, 125293 CrossRef CAS
.
- J. Du, Z. Wang, X. Peng and J. Fan, Ind. Eng. Chem. Res., 2015, 54, 12011–12016 CrossRef CAS
.
- C. Xiao, X. Zhang, J. Liu, A. Yang, H. Zhao, X. Li, Y. He and Z. Yuan, Anal. Methods, 2015, 7, 924–929 RSC
.
- N. Gao, P. Huang and F. Wu, Spectrochim. Acta, Part A, 2018, 192, 174–180 CrossRef CAS PubMed
.
- J. Li, P. Huang and F. Wu, J. Nanopart. Res., 2016, 18, 156 CrossRef
.
- C.-j. Zhang, Z.-y. Gao, Q.-b. Wang, X. Zhang, J.-s. Yao, C.-d. Qiao and Q.-z. Liu, Polymers, 2018, 10, 873 CrossRef PubMed
.
- P. Song, L. Wu and W. Guan, Nutrients, 2015, 7, 9872–9895 CrossRef CAS PubMed
.
- S.-C. Kwon, I. Kim, J. Song and J. Park, Ann. Occup. Environ. Med., 2018, 30, 5 CrossRef PubMed
.
- C. Martínez-Aquino, A. M. Costero, S. Gil and P. Gaviña, Nanomaterials, 2019, 9, 302 CrossRef PubMed
.
- X. Nie, Z. Chen, Y. Tian, S. Chen, L. Qu and M. Fan, Food Chem., 2021, 340, 127930 CrossRef CAS PubMed
.
- F. K. Alsammarraie, M. Lin, A. Mustapha, H. Lin, X. Chen, Y. Chen, H. Wang and M. Huang, Food Chem., 2018, 259, 219–225 CrossRef CAS PubMed
.
- J. Jia, S. Yan, X. Lai, Y. Xu, T. Liu and Y. Xiang, Food Anal. Methods, 2018, 11, 1668–1676 CrossRef
.
- C. Zhao, C.-y. Hong, Z.-z. Lin, X.-m. Chen and Z.-Y. Huang, Mikrochim. Acta, 2019, 186, 322 CrossRef PubMed
.
- N. Bhardwaj, S. K. Bhardwaj, M. K. Nayak, J. Mehta, K.-H. Kim and A. Deep, TrAC, Trends Anal. Chem., 2017, 97, 120–135 CrossRef CAS
.
- Y. Zou, J. Liang, Z. She and H.-B. Kraatz, Talanta, 2019, 193, 15–22 CrossRef CAS PubMed
.
- K. Fu, Y. Zheng, J. Li, Y. Liu, B. Pang, X. Song, K. Xu, J. Wang and C. Zhao, J. Agric. Food Chem., 2018, 66, 9516–9521 CrossRef CAS PubMed
.
- B. Jin, S. Wang, M. Lin, Y. Jin, S. Zhang, X. Cui, Y. Gong, A. Li, F. Xu and T. J. Lu, Biosens. Bioelectron., 2017, 90, 525–533 CrossRef CAS PubMed
.
- R. Gupta, A. Kumar, S. Kumar, A. K. Pinnaka and N. K. Singhal, Sens. Actuators, B, 2021, 329, 129100 CrossRef CAS
.
- J. Yi, P. Wu, G. Li, W. Xiao, L. Li, Y. He, Y. He, P. Ding and C. Chen, Mikrochim. Acta, 2019, 186, 711 CrossRef PubMed
.
- X. Tao, Z. Liao, Y. Zhang, F. Fu, M. Hao, Y. Song and E. Song, Chin. Chem. Lett., 2021, 32, 791–795 CrossRef CAS
.
- J. Feng, Q. Shen, J. Wu, Z. Dai and Y. Wang, Food Control, 2019, 98, 333–341 CrossRef CAS
.
- J. Huang, J. Sun, A. R. Warden and X. Ding, Food Control, 2020, 108, 106885 CrossRef CAS
.
- M. Esmaeillou, G. Zarrini, M. Ahangarzadeh Rezaee, J. Shahbazi Mojarrad and A. Bahadori, Adv. Pharm. Bull., 2017, 7, 479–483 CrossRef CAS PubMed
.
- C. Wang, B. Gu, Q. Liu, Y. Pang, R. Xiao and S. Wang, Int. J. Nanomed., 2018, 13, 1159–1178 CrossRef CAS PubMed
.
- M. Yu, H. Wang, F. Fu, L. Li, J. Li, G. Li, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2017, 89, 4085–4090 CrossRef CAS PubMed
.
- H. Peng, R. E. Borg, A. B. N. Nguyen and I. A. Chen, ACS Sens., 2020, 5, 1491–1499 CrossRef CAS PubMed
.
- W. Zhang, Y. Wang, M. Nan, Y. Li, J. Yun, Y. Wang and Y. Bi, Food Chem., 2021, 348, 129128 CrossRef CAS PubMed
.
- T. Chotchuang, W. Cheewasedtham, T. J. Jayeoye and T. Rujiralai, Mikrochim. Acta, 2019, 186, 655 CrossRef PubMed
.
- C. Cao, P. Li, H. Liao, J. Wang, X. Tang and L. Yang, Anal. Bioanal. Chem., 2020, 412, 4609–4617 CrossRef CAS PubMed
.
- S. Liu, X. Leng, X. Wang, Q. Pei, X. Cui, Y. Wang and J. Huang, Mikrochim. Acta, 2017, 184, 1969–1976 CrossRef CAS
.
- Y. Liu, Q. Ouyang, H. Li, M. Chen, Z. Zhang and Q. Chen, J. Agric. Food Chem., 2018, 66, 6188–6195 CrossRef CAS PubMed
.
- W. Jin, P. Huang, Y. Chen, F. Wu and Y. Wan, J. Nanopart. Res., 2015, 17, 358 CrossRef
.
- M. H. Hu, W. H. Huang, L. L. Suo, L. H. Zhou, L. F. Ma and H. F. Zhu, Anal. Methods, 2017, 9, 5598–5603 RSC
.
- S. Thatai, P. Khurana, S. Prasad, S. K. Soni and D. Kumar, Microchem. J., 2016, 124, 104–110 CrossRef CAS
.
- W. Diao, G. Wang, L. Wang, L. Zhang, S. Ding, T. Takarada, M. Maeda and X. Liang, ACS Appl. Bio Mater., 2020, 3, 7003–7010 CrossRef CAS PubMed
.
- N. Ratnarathorn, O. Chailapakul and W. Dungchai, Talanta, 2015, 132, 613–618 CrossRef CAS PubMed
.
- B. Feng, R. Zhu, S. Xu, Y. Chen and J. Di, RSC Adv., 2018, 8, 4049–4056 RSC
.
- H. Yu, M. Wang, J. Cao, Y. She, Y. Zhu, J. Ye, A. M. Abd El-Aty, A. Hacımüftüoğlu, J. Wang and S. Lao, Food Chem., 2021, 364, 130326 CrossRef CAS PubMed
.
- D. Li, S. Wang, L. Wang, H. Zhang and J. hu, Anal. Bioanal. Chem., 2019, 411, 2645 CrossRef CAS PubMed
.
- G. Liu, R. Zhang, L. Li, X. Huang, T. Li, M. Lu, D. Xu and J. Wang, Nanomaterials, 2018, 8, 499 CrossRef PubMed
.
- S.-H. Hung, J.-Y. Lee, C.-C. Hu and T.-C. Chiu, Food Chem., 2018, 260, 61–65 CrossRef CAS PubMed
.
- H. Luo, X. Wang, Y. Huang, K. Lai, B. A. Rasco and Y. Fan, J. Sci. Food Agric., 2018, 98, 3892–3898 CrossRef CAS PubMed
.
- L. Zou, X. Li and Y. Lai, Microchem. J., 2021, 162, 105858 CrossRef CAS
.
- S. H. Jalalian, P. Lavaee, M. Ramezani, N. M. Danesh, M. Alibolandi, K. Abnous and S. M. Taghdisi, Spectrochim. Acta, Part A, 2021, 246, 119062 CrossRef CAS PubMed
.
- M. Ramezani, S. H. Jalalian, S. M. Taghdisi, K. Abnous and M. Alibolandi, in Biomedical Engineering Technologies: Volume 1, ed. M. R. Ossandon, H. Baker and A. Rasooly, Springer US, New York, NY, 2022, pp. 417–436, DOI:10.1007/978-1-0716-1803-5_21
.
- S. Sun and Y. Xie, Anal. Methods, 2021, 13, 1255–1260 RSC
.
- J.-y. Xin, L.-x. Zhang, D.-d. Chen, K. Lin, H.-c. Fan, Y. Wang and C.-g. Xia, Food Chem., 2015, 174, 473–479 CrossRef CAS PubMed
.
- X. Bao, J. Liu, Q. Zheng, L. Duan, Y. Zhang, J. Qian and T. Tu, Chin. Chem. Lett., 2021, 3023–3026 CrossRef CAS
.
- J. Zhu, H. Chang, J.-J. Li, X. Li and J.-W. Zhao, Sens. Actuators, B, 2017, 239, 906–915 CrossRef CAS
.
- P. Kumar, P. Kumar, S. Manhas and N. K. Navani, Sens. Actuators, B, 2016, 233, 157–161 CrossRef CAS
.
- K. Chaisiwamongkhol, S. Labaidae, S. Pon-in, S. Pinsrithong, T. Bunchuay and A. Phonchai, Microchem. J., 2020, 158, 105273 CrossRef CAS
.
- P. Kumar, P. Ramulu Lambadi and N. Kumar Navani, Biosens. Bioelectron., 2015, 72, 340–347 CrossRef CAS PubMed
.
- H. S. Ali, B. M. El-Haj, S. Saifullah and M. Kawish, in Metal Nanoparticles for Drug Delivery and Diagnostic Applications, ed. M. R. Shah, M. Imran and S. Ullah, Elsevier, 2020, pp. 43–58, DOI:10.1016/B978-0-12-816960-5.00004-5
.
- B. D. Ventura, M. Cennamo, A. Minopoli, R. Campanile, S. B. Censi, D. Terracciano, G. Portella and R. Velotta, ACS Sens., 2020, 5, 3043–3048 CrossRef CAS PubMed
.
- J. Peng, N. Zhou, Y. Zhong, Y. Su, L. Zhao and Y.-T. Chang, Mikrochim. Acta, 2019, 186, 618 CrossRef PubMed
.
- Division of Child Health and Development and Maternal Health and Safe Motherhood Program, Hypoglycaemia of the newborn: review of the literature, World Health Organization, Geneva, 1997, https://apps.who.int/iris/handle/10665/63362 Search PubMed
.
- WHO and IDF, Definition and diagnosis of diabetes mellitus and intermediate hyperglycemia, 2006 Search PubMed
.
- S. Kawahito, H. Kitahata and S. Oshita, World J. Gastroenterol., 2009, 15, 4137–4142 CrossRef CAS PubMed
.
- E.-H. Yoo and S.-Y. Lee, Sensors, 2010, 10, 4558–4576 CrossRef PubMed
.
- Q. Yang, X. Zhang, S. Kumar, R. Singh, B. Zhang, C. Bai and X. Pu, Plasmonics, 2020, 15, 841–848 CrossRef CAS
.
- X. Gu, H. Wang, Z. D. Schultz and J. P. Camden, Anal. Chem., 2016, 88, 7191–7197 CrossRef CAS PubMed
.
- W. Na, H. Liu, M. Wang and X. Su, Mikrochim. Acta, 2017, 184, 1463–1470 CrossRef CAS
.
- Y.-P. Li, L. Jiang, T. Zhang, M. Lin, D.-B. Tian and H. Huang, Chin. Chem. Lett., 2014, 25, 77–79 CrossRef CAS
.
- T. Pinheiro, J. Ferrão, A. C. Marques, M. J. Oliveira, N. M. Batra, P. M. F. J. Costa, M. P. Macedo, H. Águas, R. Martins and E. Fortunato, Nanomaterials, 2020, 10, 2027 CrossRef CAS PubMed
.
- F. B. Araujo, D. S. Barbosa, C. Y. Hsin, R. C. Maranhão and D. S. P. Abdalla, Atherosclerosis, 1995, 117, 61–71 CrossRef CAS PubMed
.
- J. Kwak, H. J. Park and S. S. Lee, Bull. Korean Chem. Soc., 2018, 39, 156–160 CrossRef CAS
.
- M. Nagpal, S. Singh, P. Singh, P. Chauhan and M. A. Zaidi, Natl. J. Maxillofac. Surg., 2016, 7, 17–20 CrossRef PubMed
.
- Y. Zhang, M. Li, X. Gao, Y. Chen and T. Liu, J. Hematol. Oncol., 2019, 12, 137 CrossRef PubMed
.
- N. L. Henry and D. F. Hayes, Mol. Oncol., 2012, 6, 140–146 CrossRef CAS PubMed
.
- R. E. Lane, D. Korbie, M. M. Hill and M. Trau, Clin. Transl. Med., 2018, 7, 14 CAS
.
- W. Zhao, H. Li, Y. Tang, M. Liu, S. Wang and R. Yu, Mikrochim. Acta, 2019, 186, 517 CrossRef PubMed
.
- E. Assah, W. Goh, X. T. Zheng, T. X. Lim, J. Li, D. Lane, F. Ghadessy and Y. N. Tan, Colloids Surf., B, 2018, 169, 214–221 CrossRef CAS PubMed
.
- Y. Bai, H. Li, J. Xu, Y. Huang, X. Zhang, J. Weng, Z. Li and L. Sun, Biosens. Bioelectron., 2020, 166, 112424 CrossRef CAS PubMed
.
- V. Ranganathan, S. Srinivasan, A. Singh and M. C. DeRosa, Anal. Biochem., 2020, 588, 113471 CrossRef CAS PubMed
.
- R. Ahirwar and P. Nahar, Anal. Bioanal. Chem., 2016, 408, 327–332 CrossRef CAS PubMed
.
- L. Zhou, F. Ji, T. Zhang, F. Wang, Y. Li, Z. Yu, X. Jin and B. Ruan, Talanta, 2019, 197, 444–450 CrossRef CAS PubMed
.
- K. P. P. Htoo, V. Yamkamon, S. Yainoy, T. Suksrichavalit, W. Viseshsindh and W. Eiamphungporn, Clin. Chim. Acta, 2019, 488, 40–49 CrossRef CAS PubMed
.
- A. Kumar, B. Mazinder Boruah and X.-J. Liang, J. Nanomater., 2011, 2011, 202187 Search PubMed
.
- B. P. Crulhas, C. R. Basso, G. R. Castro and V. A. Pedrosa, ECS J. Solid State Sci. Technol., 2021, 10, 047004 CrossRef CAS
.
- E. J. H. Wee, Y. Wang, S. C.-H. Tsao and M. Trau, Theranostics, 2016, 6, 1506–1513 CrossRef CAS PubMed
.
- J. Yu, J. Jeon, N. Choi, J. O. Lee, Y.-P. Kim and J. Choo, Sens. Actuators, B, 2017, 251, 302–309 CrossRef CAS
.
- Y.-S. Borghei, M. Hosseini, M. R. Ganjali and H. Ju, Mikrochim. Acta, 2018, 185, 286 CrossRef PubMed
.
- G. Higgs and F. Slack, J. Clin. Bioinf., 2013, 3, 17 CrossRef PubMed
.
- M. C. Boonstra, S. W. L. de Geus, H. A. J. M. Prevoo, L. J. A. C. Hawinkels, C. J. H. van de Velde, P. J. K. Kuppen, A. L. Vahrmeijer and C. F. M. Sier, Biomarkers Cancer, 2016, 8, 119–133 CAS
.
- R. Sasidharan and C. Chothia, Proc. Natl. Acad. Sci. U.S.A., 2007, 104, 10080–10085 CrossRef CAS PubMed
.
- S. Chen, Q. Zhao, L. Zhang, L. Wang, Y. Zeng and H. Huang, Sens. Actuators, B, 2015, 221, 1391–1397 CrossRef CAS
.
- A. V. Krasnoslobodtsev, M. P. Torres, S. Kaur, I. V. Vlassiouk, R. J. Lipert, M. Jain, S. K. Batra and Y. L. Lyubchenko, Nanomedicine, 2015, 11, 167–173 CrossRef CAS PubMed
.
- C. Zhang, Y. Gao, N. Yang, T. You, H. Chen and P. Yin, Mikrochim. Acta, 2018, 185, 90 CrossRef PubMed
.
- O. H. Shayesteh and R. Ghavami, Spectrochim. Acta, Part A, 2020, 226, 117644 CrossRef CAS PubMed
.
- C. Luo, W. Wen, F. Lin, X. Zhang, H. Gu and S. Wang, RSC Adv., 2015, 5, 10994–10999 RSC
.
- R. Shen, J. Zhang, W. Huang, S. Wu, G. Li, S. Zou and L. Ling, Anal. Chim. Acta, 2021, 1145, 87–94 CrossRef CAS PubMed
.
- K. Abhange, A. Makler, Y. Wen, N. Ramnauth, W. Mao, W. Asghar and Y. Wan, Bioact. Mater., 2021, 6, 3705–3743 CrossRef CAS PubMed
.
- M. Oliveira-Rodríguez, S. López-Cobo, H. T. Reyburn, A. Costa-García, S. López-Martín, M. Yáñez-Mó, E. Cernuda-Morollón, A. Paschen, M. Valés-Gómez and M. C. Blanco-López, J. Extracell. Vesicles, 2016, 5, 31803 CrossRef PubMed
.
- M. Hou, D. He, H. Bu, H. Wang, J. Huang, J. Gu, R. Wu, H.-W. Li, X. He and K. Wang, Analyst, 2020, 145, 6232–6236 RSC
.
- Y. Li, Y. Zhang, M. Zhao, Q. Zhou, L. Wang, H. Wang, X. Wang and L. Zhan, Chem. Commun., 2016, 52, 3959–3961 RSC
.
- W.-J. Shieh and S. R. Zaki, in Advanced Techniques in Diagnostic Microbiology, ed. Y.-W. Tang and C. W. Stratton, Springer US, Boston, MA, 2013, pp. 873–890, DOI:10.1007/978-1-4614-3970-7_45
.
- W. Sabiiti, K. Azam, E. Esmeraldo, N. Bhatt, A. Rachow and S. H. Gillespie, J. Clin. Microbiol., 2019, 57, e01778–01718 CrossRef CAS PubMed
.
- T.-T. Tsai, C.-Y. Huang, C.-A. Chen, S.-W. Shen, M.-C. Wang, C.-M. Cheng and C.-F. Chen, ACS Sens., 2017, 2, 1345–1354 CrossRef CAS PubMed
.
- N. A. Owens, A. Pinter and M. D. Porter, J. Raman Spectrosc., 2019, 50, 15–25 CrossRef CAS
.
- S. R. Ahmed, J. Kim, T. Suzuki, J. Lee and E. Y. Park, Biotechnol. Bioeng., 2016, 113, 2298–2303 CrossRef CAS PubMed
.
- S. G. Hwang, K. Ha, K. Guk, D. K. Lee, G. Eom, S. Song, T. Kang, H. Park, J. Jung and E.-K. Lim, Sci. Rep., 2018, 8, 12999 CrossRef PubMed
.
- Y. Liu, L. Zhang, W. Wei, H. Zhao, Z. Zhou, Y. Zhang and S. Liu, Analyst, 2015, 140, 3989–3995 RSC
.
- L. Zheng, J. Wei, X. Lv, Y. Bi, P. Wu, Z. Zhang, P. Wang, R. Liu, J. Jiang, H. Cong, J. Liang, W. Chen, H. Cao, W. Liu, G. F. Gao, Y. Du, X. Jiang and X. Li, Biosens. Bioelectron., 2017, 91, 46–52 CrossRef CAS PubMed
.
- H. Kim, M. Park, J. Hwang, J. H. Kim, D.-R. Chung, K.-s. Lee and M. Kang, ACS Sens., 2019, 4, 1306–1312 CrossRef CAS PubMed
.
- H. A. Alfassam, M. S. Nassar, M. M. Almusaynid, B. A. Khalifah, A. S. Alshahrani, F. A. Almughem, A. A. Alshehri, M. O. Alawad, S. Massadeh, M. Alaamery, I. M. Aldeailej, A. A. Alamri, A. Z. Binjomah and E. A. Tawfik, Pharmaceutics, 2021, 13, 06 CrossRef PubMed
.
- L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen, C. Zhao, J. Xu, W. Hu, J. Ji, H. Xu and G. L. Liu, Biosens. Bioelectron., 2021, 171, 112685 CrossRef CAS PubMed
.
- T. T. S. Lew, K. M. M. Aung, S. Y. Ow, S. N. Amrun, L. Sutarlie, L. F. P. Ng and X. Su, ACS Nano, 2021, 15, 12286–12297 CrossRef CAS PubMed
.
- Z. Chen, Q. Wu, J. Chen, X. Ni and J. Dai, Virol. Sin., 2020, 35, 351–354 CrossRef CAS PubMed
.
- S. Aithal, S. Mishriki, R. Gupta, R. P. Sahu, G. Botos, S. Tanvir, R. W. Hanson and I. K. Puri, Talanta, 2022, 236, 122841 CrossRef CAS PubMed
.
- P. A. Nair and K. Sreenivasan, Anal. Methods, 2016, 8, 2082–2087 RSC
.
- Y. Xiong, Y. Zhang, P. Rong, J. Yang, W. Wang and D. Liu, Nanoscale, 2015, 7, 15584–15588 RSC
.
- B. Brasiunas, A. Popov, A. Ramanavicius and A. Ramanaviciene, Food Chem., 2021, 351, 129238 CrossRef CAS PubMed
.
- M. L. Firdaus, E. Saputra, S. M. Ginting, S. Wyantuti, D. R. Eddy, L. Rahmidar and B. Yuliarto, Sens. Bio-Sens. Res, 2022, 35, 100472 CrossRef
.
- H.-H. Deng, G.-L. Hong, F.-L. Lin, A.-L. Liu, X.-H. Xia and W. Chen, Anal. Chim. Acta, 2016, 915, 74–80 CrossRef CAS PubMed
.
- N. Mohseni and M. Bahram, Spectrochim. Acta, Part A, 2018, 193, 451–457 CrossRef CAS PubMed
.
- M. Oliveira-Rodríguez, S. López-Cobo, H. T. Reyburn, A. Costa-García, S. López-Martín, M. Yáñez-Mó, E. Cernuda-Morollón, A. Paschen, M. Valés-Gómez and M. C. Blanco-López, J. Extracell. Vesicles, 2016, 5, 31803 CrossRef PubMed
.
- M. K. Hameed, J. B. M. Parambath, M. T. Gul, A. A. Khan, Y. Park, C. Han and A. A. Mohamed, Appl. Surf. Sci., 2022, 583, 152504 CrossRef CAS
.
- Y. Feng, G. Liu, M. La and L. Liu, Molecules, 2022, 27, 615 CrossRef CAS PubMed
.
- S. Kunwar, P. Pandey and J. Lee, ACS Omega, 2019, 4, 17340–17351 CrossRef CAS PubMed
.
- G. Cabello, K. C. Nwoko, J. F. Marco, M. Sánchez-Arenillas, A. M. Méndez-Torres, J. Feldmann, C. Yáñez and T. A. D. Smith, J. Alloys Compd., 2019, 791, 184–192 CrossRef CAS
.
- M. Sui, S. Kunwar, P. Pandey and J. Lee, Sci. Rep., 2019, 9, 16582 CrossRef PubMed
.
- T. Bai, M. Wang, M. Cao, J. Zhang, K. Zhang, P. Zhou, Z. Liu, Y. Liu, Z. Guo and X. Lu, Anal. Bioanal. Chem., 2018, 410, 2291–2303 CrossRef CAS PubMed
.
- J. M. George, R. N. Priyanka and B. Mathew, Microchem. J., 2020, 155, 104686 CrossRef CAS
.
- F. Zhang, J. Zhu, J.-J. Li and J.-W. Zhao, J. Mater. Chem. C, 2015, 3, 6035–6045 RSC
.
- B. Liu, H. Ni, D. Zhang, D. Wang, D. Fu, H. Chen, Z. Gu and X. Zhao, ACS Sens., 2017, 2, 1035–1043 CrossRef CAS PubMed
.
- N. Hussain, H. Pu and D.-W. Sun, Food Chem., 2021, 350, 129025 CrossRef CAS PubMed
.
- P. Srinoi, Y.-T. Chen, V. Vittur, M. D. Marquez and T. R. Lee, Appl. Sci., 2018, 8, 1106 CrossRef
.
- I. Ansari, M. M. El-Kady, C. Arora, M. Sundararajan, D. Maiti and A. Khan, in Global Climate Change, ed. S. Singh, P. Singh, S. Rangabhashiyam and K. K. Srivastava, Elsevier, 2021, pp. 361–391, DOI:10.1016/B978-0-12-822928-6.00017-4
.
- Q. Li, P. Song and J. Wen, Curr. Opin. Food Sci., 2019, 30, 79–84 CrossRef
.
- J. Reed, S. Bain and V. Kanamarlapudi, Diabetes, Metab. Syndr. Obes.: Targets Ther., 2021, 14, 3567–3602 CrossRef PubMed
.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed
.
- Q. Xu, S. Du, G.-d. Jin, H. Li and X. Y. Hu, Mikrochim. Acta, 2011, 173, 323–329 CrossRef CAS
.
- A. T. Kharroubi and H. M. Darwish, World J. Diabetes, 2015, 6, 850–867 CrossRef PubMed
.
- V. Schirrmacher, Int. J. Oncol., 2019, 54, 407–419 CAS
.
- M. Simeoni, T. Cavinato, D. Rodriguez and D. Gatfield, Commun. Biol., 2021, 4, 715 CrossRef CAS PubMed
.
- T.-A. T. Le, K. Vodden, J. Wu and G. Atiwesh, Int. J. Environ. Res. Public Health, 2021, 18, 559 CrossRef CAS PubMed
.
- Y.-C. Tyan, M.-H. Yang, S.-B. Jong, C.-K. Wang and J. Shiea, Anal. Bioanal. Chem., 2009, 395, 729–735 CrossRef CAS PubMed
.
- J. Khodaveisi, S. Dadfarnia, A. M. H. Shabani and D. Saberi, Sens. Actuators, B, 2017, 239, 1300–1306 CrossRef CAS
.
Footnote |
| † N. H. Anh and M. Q. Doan contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |