Open Access Article
Open Access ArticleA new electrochemical DNA biosensor for determination of anti-cancer drug chlorambucil based on a polypyrrole/flower-like platinum/NiCo2O4/pencil graphite electrode
Hadi Mahmoudi-Moghaddam and
Zahra Garkani-Nejad
and
Zahra Garkani-Nejad *
*
Chemistry Department, Faculty of Science, Shahid Bahonar University of Kerman, Kerman, Iran. E-mail: z_garkani@uk.ac.ir; Tel: +98 3433257433
First published on 9th February 2022
Abstract
In the current study, DNA immobilization was performed on pencil graphite (PG) modified with a polypyrrole (PPy) and flower-like Pt/NiCo2O4 (FL-Pt/NiCo2O4) nanocomposite, as a new sensitive electrode to detect chlorambucil (CHB). Energy dispersive X-ray (EDX) analysis, X-ray diffraction (XRD) and scanning electron microscopy (SEM) techniques were employed to characterize the synthesized FL-Pt/NiCo2O4 and PPy/FL-Pt/NiCo2O4 nanocomposites. Moreover, differential pulse voltammetry (DPV) was selected to assess the guanine and adenine electrochemical responses on the DNA sensor. The CHB determination was performed using the maximum currents towards adenine and guanine in the acetate buffer solution (ABS). According to the results, ds-DNA/PPy/FL-Pt/NiCo2O4/PGE was able to detect the different concentrations of CHB in the range between 0.018 and 200 μM, with a detection limit of (LOD) of 4.0 nM. The new biosensor was also exploited for CHB determination in real samples (serum, urine and drug), the results of which revealed excellent recoveries (97.5% to 103.8%). Furthermore, the interaction between ds-DNA and CHB was studied using electrochemistry, spectrophotometry and docking whose outputs confirmed their effective interaction.
1. Introduction
4-{4-[Bis(2-chloroethyl) amino] phenyl} butanoic acid, chlorambucil (Scheme 1), is known as one of the basic agents to treat lymph node and blood cancers, including giant follicular lymphoma, leukemia, lymphosarcoma, chronic lymphocytic leukemia, lymphoma, Hodgkin's disease and rheumatoid arthritis. It has an adverse impact in suppressing bone marrow and reduces the ability of the body to generate specific blood cells. In addition, some severe skin responses have been reported for this anti-cancer drug.1,2 There are limited techniques for determination of CHB such as gas chromatography (GC) and high-performance liquid chromatography (HPLC).3,4 It should be noted that the mentioned techniques, in addition to advantages, are associated with some drawbacks, such as expensiveness, and also torturous protocols and sample preparations.5–7 Instead, individual analysis of these drugs can be performed in simple processes using electrochemical methods because of some unique properties such as rapid analysis, cost-effectiveness, portability and simplicity.8–16 Carbon paste, pencil graphite electrode, glassy carbon, screen-printed, etc. are the most regular electrodes for the electrochemical sensing of different analytes. The PGE is frequently employed as the electrochemical sensor in analytical chemistry due to its good reproducibility, lower toxicity and suitable stability for various electrochemical measurements.17–19Recently significant attention is aroused in the preparation of DNA sensors due to the specific properties including simple procedure, high speed and low cost in gene sequences detection, molecular diagnostic, virus detection and forensic applications.20–24 The important step for the preparation of the electrochemical DNA sensor is mainly dependent on the immobilization of ds-DNA on the surface of electrode.25–28 Nanomaterials have properties including high electrocatalytic activity, large surface area, biocompatibility and adsorption ability, which can enhance the signal intensity and sensor sensitivity. Therefore, a variety of materials such as conductive polymers, metal oxide, graphene and CNT have been employed to fabricate the electrochemical biosensors to improve their electrochemical characteristics.29–33
The electroanalytical potentials of electrochemical biosensors have been significantly improved due to nanomaterials' synergistic activities like the combination of conductive polymers (CPs) with binary metal oxides such as NiCo2O4. For example, polypyrrole is widely used as CPs in the fabrication of various biosensors, owing to its biocompatibility, high conductivity and easy deposition on the electrode surface.34,35
NiCo2O4 is a binary metal oxide with an inverse spinel structure containing nickel (Ni) ions with octahedral sites and cobalt (Co) ions distributed across the octahedral and tetrahedral sites. It has greater electrochemical potential and electronic conductivity when compared with Ni and Co oxides alone.36–38 In addition, metal oxide semiconductor doping can create an excess or deficiency of the count of electrons that can modulate the semiconductor conductivity. The noble metals such as Au, Ag, Pd and Pt can be used for doping the NiCo2O4 nanostructures to enhance active surface area and promote electronic properties, resulting in greater sensor response compared to NiCo2O4. Among these, Pt can improve the electrocatalytic properties of the NiCo2O4 nanostructures.39,40
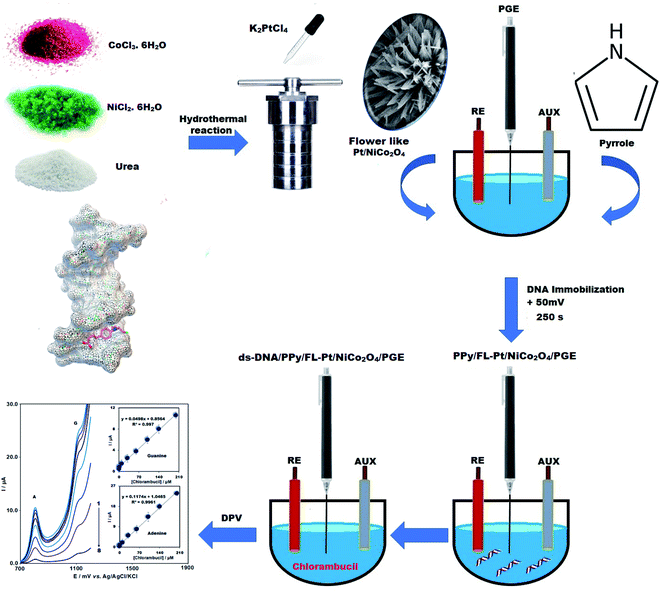
The present work, reports for the first time, a new strategy to fabricate a DNA biosensor for the CHB determination, along with the combination of polypyrrole, NiCo2O4, and Pt nanoparticles to form a new modifier deposited on a PGE. The ds-DNA/PPy/FL-Pt/NiCo2O4/PGE investigated in this study indicated that the integration of the ds-DNA, NiCo2O4, Pt and polypyrrole at the surface of PGE, is an effective method for the modification of electrode. Moreover, a greater active surface area and a larger enhancement of the conductivity were obtained using the modified electrode compared with the bare electrode. In addition, we fabricated a novel DNA biosensor based on ds-DNA/PPy/FL-Pt/NiCo2O4/PGE to detect CHB through the differential pulse voltammetry (DPV) method (Scheme 2). In addition, under optimized conditions, ds-DNA/PPy/FL-Pt/NiCo2O4/PGE could detect CHB in the real samples, successfully. Moreover, the CHB interaction with ds-DNA was investigated by different methods. The obtained results of DPV, docking and UV-Vis spectroscopy verified the effective ds-DNA–CHB interaction.
2. Experimental
2.1. Material and methods
We selected Sigma to purchase salmon sperm double-stranded DNA (ds-DNA), NiCl2·6H2O, pyrrole, CoCl3·6H2O, K2PtCl4 and urea. Graphite powder (particle size = <30 μm) was bought from Merck. High purity and analytical grade were considered for all chemicals. Based on the study procedure, general-purpose electrochemical system software was used to control experimental data using a personal computer connected to an Autolab potentiostat/galvanostat (PGSTAT 302N, Eco Chemie: The Netherlands). Reference, working and counter electrodes were Ag/AgCl/KCl (3.0 M) electrode, ds-DNA/FL-Pt/NiCo2O4/PGE, and Pt wire, respectively. The scanning electron microscopy (SEM, TESCAN Mira3 XMU) was employed to analyze the morphology of the nanocomposite. Philips analytical PC-APD X-ray diffractometer equipped with graphite monochromatic Cu and Kα radiation (α1, λ1 = 1.54056 Å, α2, λ2 = 1.54439 Å) was applied to take X-ray powder diffraction (XRD) spectra. A 2695 model LC (Waters) coupled with a Quattro Micro API Triple Quadrupole LC-MS was used for the determination of CHB.2.2. Preparation of flower-like Pt/NiCo2O4 nanocomposite (FL-Pt/NiCo2O4)
In this study, the flower-like Pt/NiCo2O4 nanocomposite was synthesized using a simple hydrothermal route. So, 0.62 g urea, 0.7931 g CoCl3·6H2O and 0.39615 g NiCl2·6H2O were dissolved in double-distilled water and 1 mL of K2PtCl4 (1000 mg L−1) was added to the solution. The solution was transferred to a Teflon-autoclave and was heated at 120 °C overnight. After cooling of autoclave at 25 °C, the product was collected and washed three times with ethanol and water. Finally, the obtained nanocomposite was dried in a vacuum oven at a temperature of 60 °C for 8 h.2.3. Fabrication of PPy/FL-Pt/NiCo2O4/PGE
The pyrrole electropolymerization was a substantial step in the fabrication of biosensors to obtain a uniform PPy surface coverage. Before the electropolymerization, to improve the electrochemical activity and the surface area of PGE, pretreatment of PGE was done by applying +1.40 V (vs. Ag/AgCl) in 0.50 M ABS (pH = 4.8) containing 20 mM NaCl for 60 seconds. The pyrrole electropolymerization in PBS (20 mM NaCl and pH = 7) was performed in presence of 30 mg L−1 FL-Pt/NiCo2O4, 100 mM NaClO4 and 0.15 M pyrrole via cyclic voltammetry (4 scans) using a potential range of 0 to +0.80 V and a scan rate of 30 mV s−1.2.4. Immobilization of ds-DNA
The potential of +0.5 V (vs. Ag/AgCl) was applied for 250 seconds to the PPy/FL-Pt/NiCo2O4/PGE for fabrication of the ds-DNA biosensor in 0.5 M ABS (pH 4.8) containing 0.02 M NaCl and 17 mg L−1 fish sperm ds-DNA.2.5. Real sample preparation
Based on the study protocol, 10 tablets of CHB (with each tablet containing 2.0 mg of CHB) were powdered; 20.0 mg of which was then dissolved in water (50 mL) under sonication. Next, certain volumes of obtained diluted solution were transferred into a 25 mL volumetric flask and reached to final volume using buffer solution. Additionally, serum and urine specimens were taken from a laboratory in Kerman, Iran, subsequently centrifuged at 6000 rpm for five minutes at an ambient temperature, and diluted using PBS (pH = 7.0). Finally, the test solutions were prepared by spiking the filtrate solution using a distinct CHB concentration.2.6. Docking study
The 1BNA and 1Z3F as DNA crystal structures were downloaded from RCSB Protein Data Bank (http://www.rcsb.org). For molecular docking of CHB, AutoDock 4.2 (ADT) software was used. The variables of the genetic algorithm were set at 150 population sizes and 100 runs. The results of docking were analyzed by Python molecular viewer and Discovery Studio 2020.3. Results and discussion
3.1. Characterization of the nanocomposite and modified electrode
The XRD pattern of FL-Pt/NiCo2O4 nanocomposite is shown in Fig. 1a. Diffraction peaks at 65.2°, 59.4°, 55.6°, 44.5°, 36.6°, 31.2° and 18.9° were correlated to (440), (511), (422), (400), (311), (220) and (111) planes, respectively that could be indexed to the cubic phase of NiCo2O4 (JCPDS card no. 20-0781).41 In the XRD pattern of FL-Pt/NiCo2O4 nanocomposite, we did not detect additional diffraction peaks that confirmed the high purity of obtained nanocomposite. Moreover, sharp diffraction peaks in the spectrum indicated the nanocrystalline nature of the product. The recorded diffraction peaks in lower angles show doping of platinum ions into the crystalline lattice of NiCo2O4. In addition, the doping of Pt in nanocomposite has not changed the crystal lattice of NiCo2O4. The Debye–Scherrer equation was used to calculate mean the crystallite size of NPs.42 The average crystallite size was calculated 29 nm.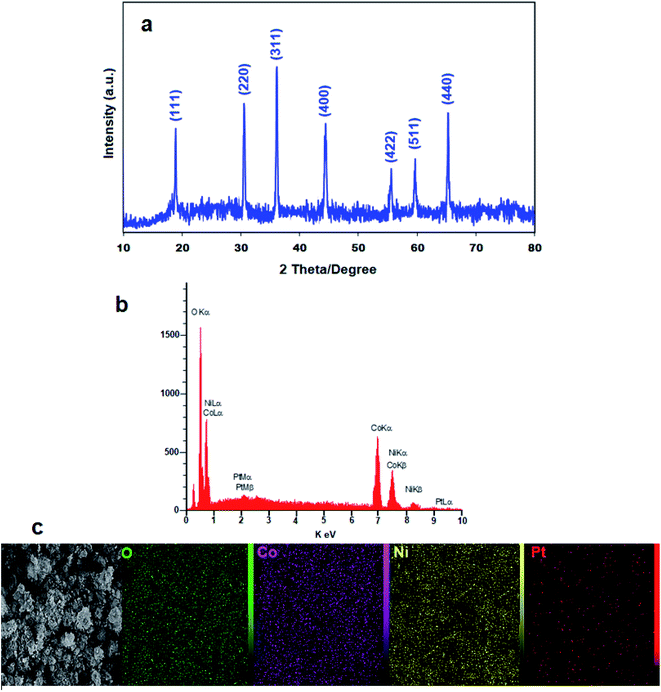 | ||
| Fig. 1 (a) XRD pattern, (b) EDX spectrum and (c) elemental mapping for flower-like Pt/NiCo2O4 nanocomposite. | ||
The EDX and elemental mapping were applied for further study of distribution and composition of FL-Pt/NiCo2O4 at the surface of nanocomposite, confirmed the presence of cobalt, nickel, platinum and oxygen in the products (Fig. 1b). Fig. 1c also displays the homogeneous distribution of Co, O, Ni and Pt in the nanocomposite.
Based on the SEM images in Fig. 2, the formation of flower-like nanostructure made up of point tipped nano-rods with the mean length of 400–500 nm and the mean diameter of 24–26 mm can be confirmed. The nano-flowers FL-Pt/NiCo2O4 possess formed clusters with uniform morphologies and without any aggregation are highly dispersed in a wide area of nanocomposite surface (Fig. 2a and b). The image of a flower in Fig. 2, indicates the similarity in the structure with the nanocomposite. Fig. 2c shows the branches consisting of symmetric point tipped nano-rods radially spread from the growth center.
 | ||
| Fig. 2 (a–c) SEM images of FL-Pt/NiCo2O4 nanocomposite in different scales, (d) SEM of PPy/PGE and (e) PPy/FL-Pt/NiCo2O4/PGE. | ||
Moreover, the SEM images were applied to evaluate the solid-state of PPy/FL-Pt/NiCo2O4/PGE the results of which revealed completely different morphology for PPy film prepared on PGE (Fig. 2d) compared to PPy/FL-Pt/NiCo2O4/PGE. The FL-Pt/NiCo2O4 nanoparticles (NPs) have been dispersed into the polymer matrix (Fig. 2e). A flower-like morphology was found for homogeneously dispersed FL-Pt/NiCo2O4 NPs deposited into the polymeric matrix.
3.2. Electrochemical behavior of modified electrodes
The CV voltammogram of PPy/FL-Pt/NiCo2O4/PGE was recorded in the ABS. As shown in the Fig. 3 (brown line), no peaks were found in the scanned range of potential. Therefore, a solution containing 5.0 mM [Fe(CN)6]3−/4− and 0.1 M KCl was used to investigate the electrochemical behavior of ds-DNA/PPy/FL-Pt/NiCo2O4/PGE (c), PPy/Pt/NiCo2O4/PGE (b) and PGE (a) (Fig. 3). The PPy/FL-Pt/NiCo2O4/PGE showed lower peak potential difference (ΔEp = 90 mV) compared to PGE (ΔEp = 213 mV) and also greater intensity in anodic and cathodic response, which means improved electron transfer kinetics due to synergistic effects of NiCo2O4, Pt and PPy. The ds-DNA immobilization on the ds-DNA/PPy/FL-Pt/NiCo2O4/PGE surface significantly reduced both the peak signals and enhanced peak-to-peak potential separation (ΔEp = 318 mV). The DNA phosphate backbone with a negative charge due to electrostatic repulsion inhibits the redox couple to reach the electrode surface, which reduces redox signals.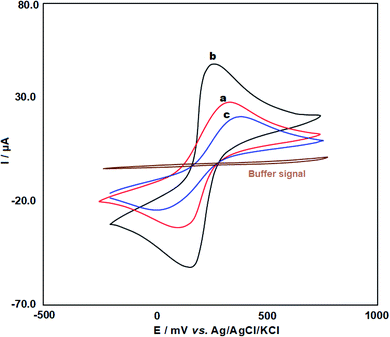 | ||
| Fig. 3 CV voltammograms for 5 mM [Fe(CN)6]3−/4− in 1.0 M KCl solution at the bare (a), PPy/FL-Pt/NiCo2O4/PGE (b) and ds-DNA/PPy/FL-Pt/NiCo2O4/PGE (c). | ||
The CV method was used to assess the real active surface area for various electrodes in 5 mM [Fe(CN)6]3−/4− solution, containing 0.1 mol L−1 KCl with the scan rate of 30–200 mV s−1. An elevation in the square root of scan rate linearly enhanced cathodic (Ipc) and anodic peak current (Ipa). The Randles–Sevcik equation (eqn (1))
| Ipa = ± (2.69 × 105)n3/2AD1/2Cv1/2 | (1) |
3.3. Optimization of ds-DNA immobilization and interaction between CHB and ds-DNA
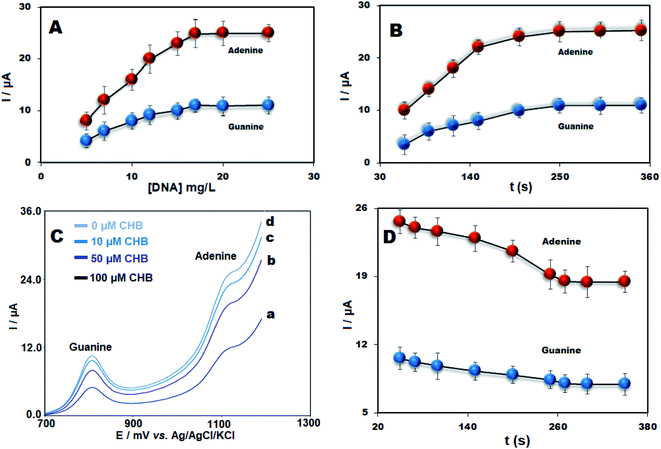
DNA biosensors were fabricated using the electrochemical method and variables like deposition time and concentration of ds-DNA on ds-DNA immobilization were investigated. The effect of ds-DNA concentration (5–25 mg L−1) was assessed on the peak currents of adenine and guanine. The signals of adenine and guanine oxidation were enhanced due to elevated concentration of ds-DNA up to 17 mg L−1 but after ds-DNA saturation at the surface of the electrode it remained constant (Fig. 4A). Accordingly, the ds-DNA concentration of 17 mg L−1 was selected as optimum content. The optimum immobilization time was also examined in the range between 60 and 350 seconds. The highest peak currents of DNA bases were recorded at the accumulation time of 250 seconds (Fig. 4B). Accordingly, 250 seconds as the optimal immobilization time was selected for ds-DNA to fabricate ds-DNA/PPy/FL-Pt/NiCo2O4/PGE.Moreover, the interaction between ds-DNA and CHB was investigated. The signals of adenine and guanine for ds-DNA/PPy/FL-Pt/NiCo2O4/PGE in the presence of 0.0, 10.0, 50.0 and 100.0 μM of CHB can be seen in Fig. 4C. There was a decrease in the signals of adenine and guanine oxidation with the addition of diverse CHB concentrations, which presented an effective interaction between CHB and ds-DNA and suggested this point as an analytical parameter to detect CHB concentration in the solution.
There is a need for the optimization of ds-DNA–CHB incubation time for sensitive determination of CHB. The short interaction time prevents proper biosensor interaction between DNA's bases and target drugs and in the long accumulation time, saturation can occur. The incubation of ds-DNA/PPy/FL-Pt/NiCo2O4/PGE in the presence of 50 mg L−1 of CHB for 50 to 350 seconds was performed. As shown in Fig. 4D the guanine and adenine oxidation currents significantly decreased at 200 s and then remained constant at t > 200 seconds. The results suggested the interaction time of 200 seconds is appropriate for subsequent experiments.
3.4. Determination of CHB
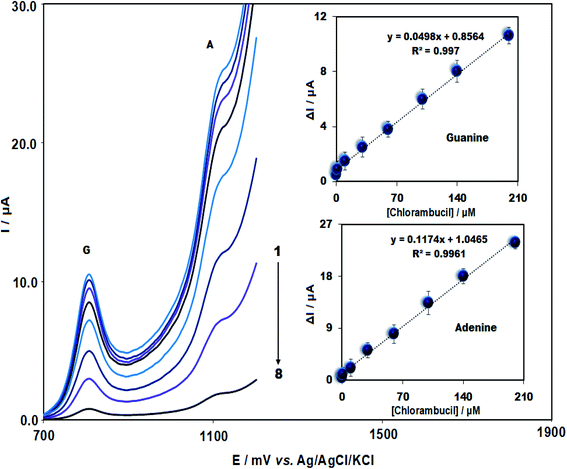
In this study, DPV was used for the determination of CHB. Fig. 5 depicts ΔI plot (difference in guanine or adenine current in the presence of CHB) as the CHB concentration function. It has been found that the plots were linear for CHB concentrations in the range between 0.018 and 200.0 μM (R2 = 0.997 and 0.996 for guanine and adenine, respectively) with a LOD of 4.0 nM.Table 1 compares the analytical potential of ds-DNA/PPy/FL-Pt/NiCo2O4/PGE to detect CHB with other electrochemical electrodes. The present sensor displays an acceptable and satisfactory linear range and limit of detection compared to other electrochemical sensors, which could be related to the synergistic impact of PPy, ds-DNA, NiCo2O4 and Pt.
| Electrode | Linear range (μM) | LOD (nM) | Ref. |
|---|---|---|---|
| a Glassy carbon electrode.b Poly(N-vinylcaprolactam) semi-interpenetrated conductive polypyrrole microgels.c One monomer molecularly imprinted polymers/reduced graphene oxide/ceramic electrode.d C60-molecularly imprinted polymer/ionic liquid-carbon ceramic electrode.e Double stranded DNA/polypyrrole/flower-like platinum/NiCo2O4/pencil graphite electrode. | |||
| MnO2@NiFe2O4/GCEa | 0.025–574.5 | 4.68 | 44 |
| PVCL/PPY semi-IPN MGsb | 0.02–420 | 1.98 | 45 |
| OMNiDIP/rGO/CEc | 0.00052–0.094 | 0.134 | 46 |
| C60-MIP/IL-CCEd | 0.0048–0.81 | 1.18 | 47 |
| ds-DNA/PPy/FL Pt/NiCo2O4/PGEe | 0.018–200.0 | 4.0 | This study |
3.5. Interference investigations
Several foreign compounds in the presence of CHB were evaluated to assess the modified electrode selectivity. Thus, the CHB concentration of 10.0 μM and the tolerance limit of ±5% were chosen in this research (mole ratio of foreign samples creating the relative error of ±5% on the determination of CHB). No interference was detected for the 100-fold concentration of sucrose, tyrosine, ascorbic acid lysine, methionine, uric acid, dopamine, norepinephrine, phenylalanine and alanine and also the 200-fold concentration of K+, Cl− and Zn2+ with the adenine and guanine oxidation signals (Fig. 6). This suggests ignoring the impact of other materials as well as the high selectivity of the biosensor to detect CHB.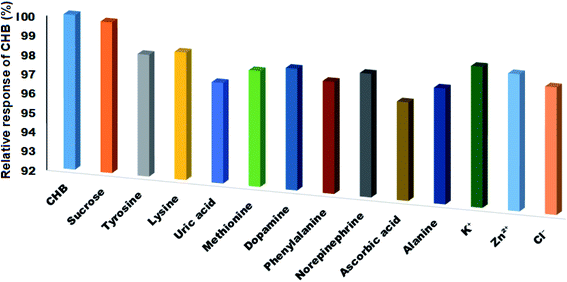 | ||
| Fig. 6 The influence of different interfering compounds on the signal of ds-DNA/PPy FL-Pt/NiCo2O4/PGE in the presence of 10.0 μM of CHB. | ||
3.6. Study of reproducibility, repeatability and stability of the modified electrode
The developed sensor was examined for reproducibility, repeatability and stability under optimized conditions. For assessing the sensor stability, ds-DNA/PPy/FL-Pt/NiCo2O4/PGE was maintained at an ambient temperature (25 °C) for seven days and one month to assess the changes in the current response. According to the results, the signals of adenine and guanine in the presence of 50 μM CHB after 7 and 30 days were approximately 98% and 94% of their initial responses, respectively. For evaluating the sensor repeatability, five successive measurements were performed in a 50 μM CHB solution using DPV, as well as the calculated RSD values were 2.37% for guanine and 2.87 for adenine that means excellent repeatability. Finally, for assessing the sensor reproducibility, the responses of five modified electrodes were recorded independently. The sensor reproducibility was verified based on RSDs% of 3.62 and 3.23 for the guanine and adenine signals.3.7. Analytical application
The applicability of the introduced sensor for electrochemical detection of CHB was tested in the real samples (serum, urine and drug samples). The CHB detection was performed in the prepared samples using the DPV technique in accordance with the standard addition method. The outcomes are shown in Table 2, highlighting acceptable RSD% (<3.8) and recovery of the spiked specimens (97.5% to 103.8%). Moreover, determination of CHB in real samples was performed using LC/MS and DPV methods and results were compared by the paired t-test (Table 3). The obtained p values were greater than 0.05 and obtained t were lower than t-table (95% CI), representing no statistically significant difference between the two methods.| Samples | Spiked (μM) | Found (μM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Chlorambucil tablet | 0.0 | 5.0 | — | 2.9 |
| 4.0 | 9.3 | 103.3 | 2.1 | |
| 8.0 | 13.5 | 103.8 | 2.4 | |
| 12.0 | 16.6 | 97.6 | 1.9 | |
| Urine | 0.0 | — | — | — |
| 5.0 | 4.9 | 98.0 | 3.1 | |
| 10.0 | 10.3 | 103.0 | 2.0 | |
| 15.0 | 15.4 | 102.6 | 2.5 | |
| Serum | 0.0 | — | — | — |
| 10.0 | 10.3 | 103.0 | 2.3 | |
| 20.0 | 19.5 | 97.5 | 2.9 | |
| 30.0 | 29.5 | 98.3 | 2.0 |
| Sample | Spiked (μM) | Found (μM) | tExp. | p value | |
|---|---|---|---|---|---|
| DPV | LC/MS | ||||
| Tablet | 0.0 | 5.04 ± 0.14 | 5.13 ± 0.19 | 0.6605 | 0.5450 |
| 12.0 | 16.61 ± 0.31 | 16.92 ± 0.23 | 1.3910 | 0.2366 | |
| Serum | 10.0 | 10.33 ± 0.23 | 10.21 ± 0.33 | 0.5167 | 0.6326 |
| 30.0 | 29.48 ± 0.59 | 30.23 ± 0.46 | 1.7364 | 0.1575 | |
3.8. Spectrophotometric analysis for the ds-DNA–CHB interaction
UV-Vis spectrophotometry as a routine method has been employed to assess enzyme catalysis, ligand-binding reactions, and conformational transitions in proteins and DNA. The study of the interaction between DNA and drug is related to changes in absorption spectra and wavelength, and also hypochromic/hyperchromic impacts on DNA or drug. Fig. 7 represents two peaks at 255 and 310 nm for the CHB in the UV-Vis spectrum which the peak at 255 is more significant than the second peak. The λmax of chlorambucil and ds-DNA was very close in the absorption spectra. Therefore, a fixed concentration of ds-DNA was poured in the cell of blank and baseline correction was performed. After adding different concentrations of ds-DNA, a gradual decrease was observed in the intensity of absorption bands at 255 nm, with no significant shift. Thus, alterations made in the absorption intensity for CHB are attributable to the generation of the CHB–ds DNA complex.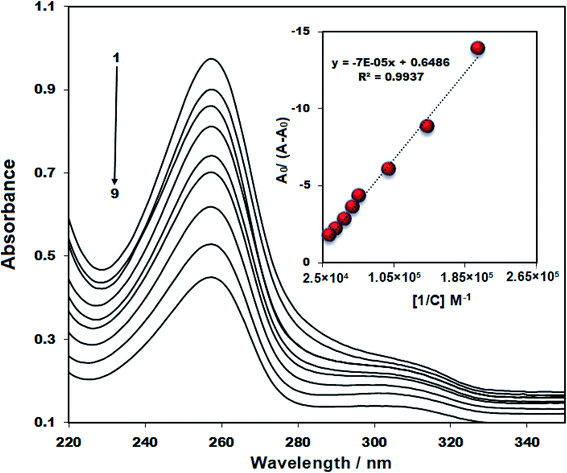 | ||
| Fig. 7 The UV-vis spectrum of 15 μM CHB in the presence of a different concentration of ds-DNA (0–20 μM; no: 1–9); inset: plot of A0/A − A0 versus 1/[DNA]. | ||
Based on the deviations of CHB absorbance at a wavelength of 255 nm within titration via ds-DNA, the binding constant (Kb) of ds-DNA/CHB complex was calculated in accordance with eqn (2);
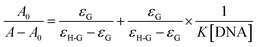 | (2) |
3.9. Docking study
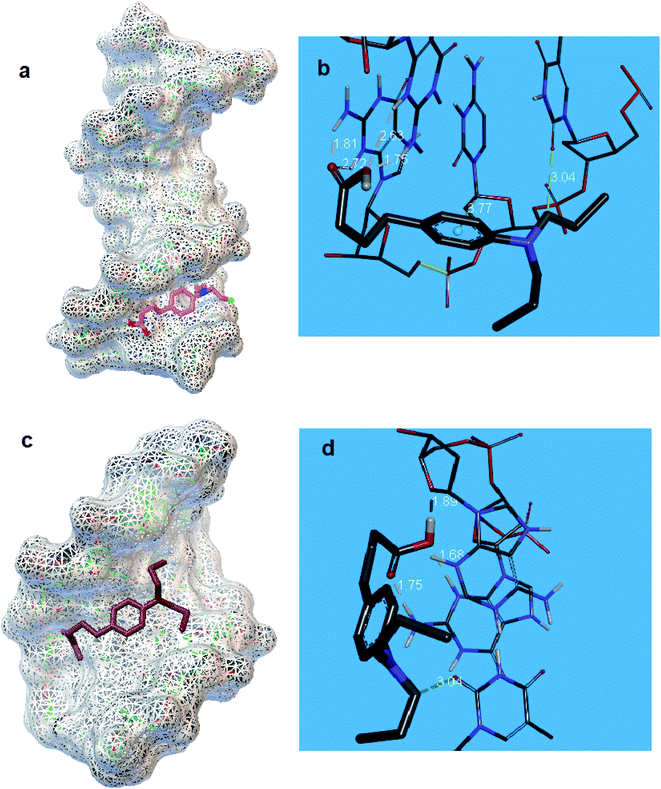
The 1BNA as canonical DNA was employed for the running of docking in the minor groove of DNA (Fig. 8a). It has been found that a π–σ interaction and five hydrogen bonds significantly contribute to the interaction of CHB and DNA bases (Fig. 8b). Notably, hydrogen bonds are as follows: hydrogen (H) 3 and 21 of guanine 44 (DG 44) would interact with oxygen (O) 15 from CHB (bond length: N–H⋯O 2.63 and 1.75 Å, bond angle: 124.35° and 157.78° respectively). Also, H 3 and 21 of guanine 222 (DG 222) interacted with O 16 of CHB (bond length of N–H⋯O: 2.72 and 1.81 Å, bond angle: 122.55° and 152.44° respectively). Moreover, C8–H of the CHB displayed an effective interaction with O2 of thymine 220 (DT 220) (bond length of C–H⋯O: 3.04 Å). Fig. 8b also shows one π–σ interaction between the aromatic ring of CHB molecule and C1′ of cytosine 221 (DC 221) (bond length: 3.77 Å). The analysis of docking confirmed that in the minor groove of DNA the effective interaction of CHB with the DNA bases can be occur.In the next stage, the interaction between CHB and DNA was also studied using 1Z3F of DNA (Fig. 8c). Based on the outputs of docking (Fig. 8d), stabilization of CHB in the major groove of ds-DNA can be done via five hydrogen bonds. Interactions are presented here. H 38 of CHB interacted with O′ 4 from adenine 33 (DA33) (bond length of O–H⋯O: 1.89 Å, bond angle: 140.66°). H 21 of guanine 22 established a hydrogen bond with O 19 of CHB (bond length of N–H⋯O: 1.75 Å, bond angle: 136.42°). In addition, H 3 of guanine 22 interacted with O 18 of CHB (bond length of N–H⋯O 1.68 Å, bond angle: 175.18°). Finally, C9–H showed a hydrogen bond with O 2 of thymine 44 (DT 44) (bond length of C–H⋯O: 3.04 Å, bond angle: 131.98°).
The docking showed that for CHB–DNA interaction, binding energy was −4.5 and −6.0 kcal mol−1 for major and minor groove binding, respectively. Based on the results obtained from the interaction of CHB in the DNA minor and major groove, the possibility of interaction of CHB in the minor groove is higher than the major groove of DNA.
4. Conclusion
The purpose of the present study was to detect CHB using ds-DNA/PPy/FL-Pt/NiCo2O4/PGE. Due to the larger surface area and suitable conductivity of PPy and FL-Pt/NiCo2O4 nanostructure, higher electrocatalytic activity and peak currents of guanine and adenine oxidation in the presence of CHB were reported for the proposed electrode. A wide linear range (0.018–200 μM) and a low limit of detection (4.0 nM) were obtained for the new DNA biosensor in the determination of CHB under the optimized conditions. The great stability, a lower limit of detection, cost-effectiveness and excellent anti-interference capability are advantages attributable to the present method. Accordingly, our biosensor can be an appropriate candidate for successful electrochemical analysis of CHB in real samples.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the Shahid Bahonar University of Kerman.References
- P. Hillmen, T. Robak and A. Janssens, Lancet, 2016, 387, 1376 Search PubMed.
- G. Dighiero, K. Maloum, B. Desablens, B. Cazin, M. Navarro, R. Leblay, M. Leporrier, J. Jaubert, G. Lepeu, B. Dreyfus, J.-L. Binet, P. Travade, F.-L. Turpin, G. Tertian and A. Bichoffe, N. Engl. J. Med., 1998, 338, 1506–1514 CrossRef CAS PubMed.
- X. Y. Wang, Q. Zhang, Q. Lin, Y. Zhang and Z. R. Zhang, J. Pharm. Biomed. Anal., 2014, 99, 74–78 CrossRef CAS PubMed.
- K. Lipska, A. Gumieniczek, R. Pietraś and A. A. Filip, Molecules, 2021, 26, 2903 CrossRef CAS PubMed.
- X. Chen, N. Li, Y. Rong, Y. Hou, Y. Huang and W. Liang, RSC Adv., 2021, 11, 28052–28060 RSC.
- C. Guo, C. Wang, H. Sun, D. Dai and H. Gao, RSC Adv., 2021, 11, 29590–29597 RSC.
- M. T. Maghsoodlou, N. Hazeri, S. M. H. Khorasani, H. M. Moghaddam, M. Nassiri and J. Salehzadeh, Phosphorus, Sulfur Silicon Relat. Elem., 2009, 184, 1713–1721 CrossRef CAS.
- B. Maleki, M. Baghayeri, M. Ghanei-Motlagh, F. Mohammadi Zonoz, A. Amiri, F. Hajizadeh, A. R. Hosseinifar and E. Esmaeilnezhad, Measurement, 2019, 140, 81–88 CrossRef.
- M. Brycht, A. Łukawska, M. Frühbauerová, K. Pravcová, R. Metelka, S. Skrzypek and M. Sýs, Food Chem., 2021, 338, 127975 CrossRef CAS PubMed.
- M. Brycht, O. Vajdle, K. Sipa, J. Robak, K. Rudnicki, J. Piechocka, A. Tasić, S. Skrzypek and V. Guzsvány, Ionics, 2018, 24, 923–934 CrossRef CAS.
- A. Farahi, H. Hammani, A. Kajai, S. Lahrich, M. Bakasse and M. A. El Mhammedi, Int. J. Environ. Anal. Chem., 2020, 100, 295–310 CrossRef CAS.
- S. Gürsoy, N. Dükar, Y. T. Yaman, S. Abaci and F. Kuralay, Anal. Chim. Acta, 2019, 1072, 15–24 CrossRef PubMed.
- G. Muthusankar, M. Sethupathi, S. M. Chen, R. K. Devi, R. Vinoth, G. Gopu, N. Anandhan and N. Sengottuvelan, Composites, Part B, 2019, 174, 106973 CrossRef CAS.
- N. Shahabadi and M. Razlansari, J. Biomol. Struct. Dyn., 2020, 1–15 CrossRef PubMed.
- M. Malakootian, H. Abolghasemi and H. Mahmoudi-Moghaddam, J. Electroanal. Chem., 2020, 876, 114474 CrossRef CAS.
- H. M. Moghaddam, M. Malakootian and B. Electrochemistry, Anal. Bioanal. Electrochem., 2018, 10, 520–530 CAS.
- T. Gan, C. Hu, Z. Chen and S. Hu, Electrochim. Acta, 2011, 56, 4512–4517 CrossRef CAS.
- T. Gan, C. Hu, Z. Chen and S. Hu, Talanta, 2011, 85, 310–316 CrossRef CAS PubMed.
- T. Gan, C. Hu and S. Hu, Anal. Methods, 2014, 6, 9220–9227 RSC.
- F. Jalali, Z. Hassanvand and A. Barati, J. Anal. Chem., 2020, 75, 544–552 CrossRef.
- M. Noszczyńska and Z. Piotrowska-Seget, Chemosphere, 2018, 201, 214–223 CrossRef PubMed.
- S. Ghosh, P. Kundu, B. K. Paul and N. Chattopadhyay, RSC Adv., 2014, 4, 63549–63558 RSC.
- H. Heidari and B. Limouei-Khosrowshahi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1114–1115, 24–30 CrossRef CAS PubMed.
- H. A. Javar, Z. Garkani-Nejad, G. Dehghannoudeh and H. Mahmoudi-Moghaddam, Anal. Chim. Acta, 2020, 1133, 48–57 CrossRef PubMed.
- X. Yang, M. Feng, J. Xia, F. Zhang and Z. Wang, J. Electroanal. Chem., 2020, 878, 114669 CrossRef CAS.
- W. Xu, T. Jin, Y. Dai and C. C. Liu, Biosens. Bioelectron., 2020, 155, 112100 CrossRef CAS PubMed.
- X. Wang, C. Xu, Y. Wang, W. Li and Z. Chen, Sens. Actuators, B, 2021, 343, 130151 CrossRef CAS.
- S. Han, W. Liu, M. Zheng and R. Wang, Anal. Chem., 2020, 92, 4780–4787 CrossRef CAS PubMed.
- L. Abad-Gil, M. J. Gismera, M. T. Sevilla and J. R. Procopio, Microchim. Acta, 2020, 187, 1–10 CrossRef PubMed.
- Z. Wang, H. Zhao, Q. Gao, K. Chen and M. Lan, Biosens. Bioelectron., 2021, 184, 113236 CrossRef CAS PubMed.
- K. Murtada and V. Moreno, J. Electroanal. Chem., 2020, 861, 113988 CrossRef CAS.
- M. Malakootian, S. Hamzeh and H. Mahmoudi-Moghaddam, Electroanalysis, 2021, 33, 38–45 CrossRef CAS.
- M. Amiri and H. Mahmoudi-Moghaddam, Microchem. J., 2021, 160, 105663 CrossRef CAS.
- G. Emir, Y. Dilgin, A. Ramanaviciene and A. Ramanavicius, Microchem. J., 2021, 161, 105751 CrossRef CAS.
- C. Tan, J. Zhao, P. Sun, W. Zheng and G. Cui, New J. Chem., 2020, 44, 4916–4926 RSC.
- C. Chen, H. Su, L. N. Lu, Y. S. Hong, Y. Chen, K. Xiao, T. Ouyang, Y. Qin and Z. Q. Liu, Chem. Eng. J., 2021, 408, 127814 CrossRef CAS.
- M. Amiri, H. Akbari Javar and H. Mahmoudi-Moghaddam, Electroanalysis, 2021, 33, 1205–1214 CrossRef CAS.
- J. Jin, J. Ding, X. Wang, C. Hong, H. Wu, M. Sun, X. Cao, C. Lu and A. Liu, RSC Adv., 2021, 11, 16161–16172 RSC.
- H. Cheng, W. Weng, H. Xie, J. Liu, G. Luo, S. Huang, W. Sun and G. Li, Microchem. J., 2020, 154, 104602 CrossRef CAS.
- N. Dimcheva, Curr. Opin. Electrochem., 2020, 19, 35–41 CrossRef CAS.
- G. Zhang, B. Y. Xia, X. Wang and X. W. Lou, Adv. Mater., 2014, 26, 2408–2412 CrossRef CAS PubMed.
- U. Holzwarth and N. Gibson, Nat. Nanotechnol., 2011, 6, 534 CrossRef CAS PubMed.
- I. Žutić, J. Fabian and S. Das Sarma, Rev. Mod. Phys., 2004, 76, 323–410 CrossRef.
- K. Sakthivel, A. Muthumariappan, S. M. Chen, Y. L. Li, T. W. Chen and M. A. Ali, Mater. Sci. Eng., C, 2019, 103, 109724 CrossRef CAS PubMed.
- B. Mutharani, P. Ranganathan, S. M. Chen and H. C. Tsai, Electrochim. Acta, 2021, 374, 137866 CrossRef CAS.
- S. Fatma, B. B. Prasad, K. Singh, R. Singh and S. Jaiswal, Sens. Actuators, B, 2019, 281, 139–149 CrossRef CAS.
- B. B. Prasad, R. Singh and A. Kumar, Biosens. Bioelectron., 2017, 94, 115–123 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |