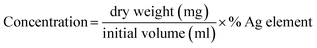Open Access Article
Open Access ArticleStreptomyces chiangmaiensis SSUT88A mediated green synthesis of silver nanoparticles: characterization and evaluation of antibacterial action against clinical drug-resistant strains†
A'liyatur Rosyidahab,
Oratai Weeranantanapanac,
Nuannoi Chudapongseac,
Wanwisa Limphiratd and
Nawarat Nantapong *ac
*ac
aSchool of Preclinical Sciences, Institute of Science, Suranaree University of Technology, Nakhon Ratchasima 30000, Thailand. E-mail: nawarat@sut.ac.th
bResearch Center for Biology, National Research and Innovation Agency (BRIN), West Java, 16911, Indonesia
cCenter of Excellence on Advanced Functional Materials, Suranaree University of Technology, Nakhon Ratchasima 30000, Thailand
dSynchrotron Light Research Institute (SLRI), Nakhon Ratchasima 30000, Thailand
First published on 2nd February 2022
Abstract
This study involved the characterization of AgNPs synthesized from soil isolate Streptomyces sp. SSUT88A and their antimicrobial activities. The strain SSUT88A revealed 98.8% similarity of the 16s rRNA gene to Streptomyces chiangmaiensis TA4-1T. The AgNPs were synthesized by mixing either intracellular or extracellular cell-free supernatant of strain SSUT88A with AgNO3. The synthesized AgNPs from intracellular cell-free supernatant and extracellular cell-free supernatant were designated as IS-AgNPs and ES-AgNPs, respectively. The IS-AgNPs showed maximum absorbance of UV-vis spectra at 418 nm, while ES-AgNPs revealed maximum absorbance at 422 nm. The TEM observation of synthesized AgNPs revealed a spherical shape with an average diameter of 13.57 nm for IS-AgNPs and 30.47 nm for ES-AgNPs. The XRD and XANES spectrum profile of both synthesized AgNPs exhibited similar spectrum energy, which corresponded to AgNPs. The IS-AgNPs revealed antimicrobial activity against clinical isolate drug-resistant bacteria (Acinetobacter baumannii, Escherichia coli 8465, Klebsiella pneumoniae 1617, and Pseudomonas aeruginosa N90PS), while ES-AgNPs had no antimicrobial activity. When compared to commercial AgNPs, IS-AgNPs exhibited antibacterial efficacy against all clinical isolate bacteria including A. baumannii, one of the most threatening multi-drug resistant strains, while commercial AgNPs did not. Thus, IS-AgNPs has potential to be further developed as an antimicrobial agent against drug-resistant bacteria.
1. Introduction
Metal nanoparticles has gained increasing attention during the last decade because of their unique characteristics such as antimicrobial, anticancer, physicochemical, optical, magnetic, and electronic properties.1–3 Different metal nanoparticles like copper, titanium, zinc, gold, magnesium, and silver have been synthesized. However, silver nanoparticles (AgNPs) are the most effective for their antimicrobial properties.4 The antimicrobial properties of silver have been recorded since 1000 BC. The first medical use of silver for blood purification, palpitations of the heart, and offensive breath was reported in 980 AD.2 Nowadays, silver has been widely used to treat superficial and deep dermal burn wounds in the form of silver sulfadiazine and silver nitrate.2,5–7 The silver in nanoscale has different properties from the bulk material due to its small size, large surface area, and strong toxicity to a wide range of microorganisms.8The AgNPs can be synthesized by using chemical, physical, and biological methods. However, toxic reducing agents such as 2-mercaptoethanol and sodium borohydride are used for the synthesis by chemical methods. These toxic substances negatively impact the environment. While the physical method produces a low yield of product and it is difficult to control the particle size.3,9–12 On the other hand, the biological method provides a possible option to synthesize AgNPs due to its simplicity, cost-effective, high-yield production capability, and eco-friendliness. AgNPs can be synthesized from plants, bacteria, fungi, and algae.9,13
Streptomycetes are Gram-positive filamentous soil bacteria. They have been recognized as a major source of antibiotics.14,15 It has been shown that the members in the genus Streptomyces, Streptomyces rochei MHM13, Streptomyces coelicolor, Streptomyces sp. LK3, Streptomyces griseorubens AU2, and Streptomyces viridodiastaticus SSHH-1, are used to synthesize of AgNPs.16–19 The AgNPs has been reported for their antimicrobial properties against Gram-positive and Gram-negative bacteria.20 Morones et al. reported the antimicrobial activity of AgNPs against Gram-negative bacteria, Escherichia coli, Vibrio cholera, Pseudomonas aeruginosa, and Salmonella typhi.21 The antimicrobial activity of AgNPs was also observed on multidrug-resistant bacteria.4,22,23 Several biological syntheses of AgNPs have been reported to be effective against multidrug-resistant strains of Acinetobacter baumannii, Staphylococcus aureus, and P. aeruginosa.24,25 Therefore, AgNPs provide a potential candidate as an inhibitor for multidrug-resistant microorganisms.
Because of their antimicrobial properties, the AgNPs has been widely used in personal care products, dressing for external wound treatment, surgical instruments, and ointment.26,27 Furthermore, their applications expand to the biological and biomedical fields as drug and gene delivery, biosensors, diagnostics, medical devices, and implantable materials.1,28 In this research, the AgNPs were synthesized by a green method using two different cell-free supernatants of S. chiangmaiensis SSUT88A isolated from soil sample at Sakaerat Environmental Research Station. The characterization of synthesized AgNPs and the antimicrobial activity against drug-resistant pathogens were investigated.
2. Experimental
2.1 Isolation of S. chiangmaiensis SSUT88A
Soil samples were collected from Sakaerat Environmental Research Station, Thailand. Soil Streptomyces were isolated using a serial dilution method and cultured on the actinomycete isolation agar (AIA, Himedia) medium. Selected colonies were transferred to an International Streptomyces Project 2 (ISP2) agar medium for purification.2.2 Identification of S. chiangmaiensis SSUT88A
Genomic DNA extraction was performed by a method used for fungi with some modifications.29 For PCR amplification, the universal 16 s rRNA gene primers, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1525R (5′-AAGGAGGTGATCCAGCC-3′) were used. The PCR products were purified using FavorPrep™ Gel/PCR Purification Mini Kit (Favorgen). The purified PCR products were used for DNA sequencing analysis. The obtained 16S rRNA gene sequence was analysed and compared to the 16s rRNA online gene database, EzBioCloud (http://www.ezbiocloud.net).2.3 Preparation of cell-free supernatant
The S. chiangmaiensis SSUT88A was inoculated in a 250 mL Erlenmeyer flask containing 100 mL ISP2 medium. A flask was incubated in a rotary shaker at 200 rpm, 37 °C for 4 days. After incubation, the culture was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min to separate bacterial cells and extracellular cell-free supernatant.16 To prepare intracellular cell-free supernatant, the bacterial cells were washed three times with sterile deionized (DI) water to remove traces of medium. The S. chiangmaiensis SSUT88A biomass was resuspended in an Erlenmeyer flask containing 100 mL sterile DI water and incubated at 200 rpm, 37 °C for 24 h. After incubation, the culture was centrifuged at 13
000 rpm for 5 min to separate bacterial cells and extracellular cell-free supernatant.16 To prepare intracellular cell-free supernatant, the bacterial cells were washed three times with sterile deionized (DI) water to remove traces of medium. The S. chiangmaiensis SSUT88A biomass was resuspended in an Erlenmeyer flask containing 100 mL sterile DI water and incubated at 200 rpm, 37 °C for 24 h. After incubation, the culture was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min to separate bacterial cells and intracellular cell-free supernatant.30
000 rpm for 5 min to separate bacterial cells and intracellular cell-free supernatant.30
2.4 Biosynthesis of AgNPs
Fifty-millilitre of Streptomyces cell-free supernatant was mixed with 50 mL of 1 mM silver nitrate (AgNO3) (POCH SA silver nitrate 99.9%), and the pH was adjusted to 7. The extracellular and intracellular cell-free supernatant mixtures were incubated at 37 °C in the dark under 200 rpm shaking condition for 2 and 5 days, respectively. The ISP2 medium, DI water, and AgNO3 were used as a control experiment.2.5 Characterization of AgNPs
2.6 Antimicrobial activity assay and determination of minimum inhibitory concentration (MIC)
The antibacterial activity of synthesized AgNPs was evaluated against clinical isolate drug-resistant pathogens, A. baumannii, E. coli 8465, K. pneumoniae 1617, and P. aeruginosa N90PS, by using an agar well diffusion assay. The concentration of synthesized AgNPs was calculated based on the percentage of Ag in the EDXRF results by following equation:The test pathogens were obtained from Suranaree University of Technology Hospital, Thailand. The 5 × 105 CFU mL−1 of mid-log phase test pathogens were seeded onto Mueller Hinton agar (MHA) plates. Then, a 6 mm hole was punched aseptically with a sterile cork borer. A volume of 100 μL of 0.2 mg mL−1 AgNPs was introduced into the well. Commercial AgNPs (no. 576832 Sigma-Aldrich) was used as an AgNPs standard. The plates were incubated at 37 °C. The diameter of the clear zone was recorded.31 The MIC of synthesized AgNPs was determined by the microdilution method based on the Clinical and Laboratory Standard Institute (CLSI) guideline. The experiments were performed in triplicate.
3. Result and discussion
3.1 Identification of Streptomyces sp. SSUT88A
The current study was focused on the synthesis of AgNPs by Streptomyces sp. SSUT88A using intracellular and extracellular cell-free supernatant. The Streptomyces sp. SSUT88A was isolated from a soil sample collected at Sakaerat Environmental Research Station, Thailand by using serial dilution and spread plate method on a streptomycetes selective medium, AIA agar, and incubated at 37 °C for 14 days.33 The AIA medium was developed primarily to increase the yield of an isolation of Streptomyces spp., subgroup of actinomycetes, from soil and water.33,34 The isolates were purified and maintained on ISP2 medium.35 From 64 isolates of Streptomyces spp., the percentage yield of Streptomyces strain SSUT88A was 1.5%. The colony morphology of strain SSUT88A was velvety with white substrate mycelium. The grey spore was produced from aerial mycelium after nine days of incubation. It produced yellowish diffusible pigment when cultivated in the Mueller Hinton agar (MHA), ISP2, and starch casein agar (SCA) media.A 16s rRNA gene analysis revealed 98.8% similarity to Streptomyces chiangmaiensis TA4-1T. The bacteria in the Streptomyces genera belong to the Streptomycetaceae family. They are abundant in soil habitat, especially in alkaline soil and soil rich in organic matter.36 The members of this genus have extensively been isolated and screened due to their ability to produce a variety of bioactive metabolites such as antibacterial, antifungal, antiviral, anticancer, as well as several industrial important enzymes.14,37–39 According to BacDive, the S. chiangmaiensis belongs to biosafety level 1 (BSL 1) risk group, which unlikely to causes any human and animal diseases.40 They pose no threat to individuals, communities, or the environment.41–43 Several species in the genus Streptomyces are widely employed by pharmaceutical companies as a source of many antibiotics.44–46 Therefore, S. chiangmaiensis SSUT88A (BSL1) can be used safely in the healthcare industry.
3.2 Biosynthesis of AgNPs
In the biological method of the AgNPs synthesis, the AgNPs can be synthesized using bacteria, fungi, algae, and plants.9 Bacteria are good for the study of AgNPs synthesize due to their abundance, fast growing microorganism, inexpensive to cultivate, easy to adapt in the extreme condition, and easy to manipulate. On the other hand, the growth condition of the bacteria such as incubation time, temperature, and oxygenation can be easy controlled.47 Streptomyces have been proved as nano factories for developing non-toxic and clean procedures to synthesize silver and gold nanoparticles.48 In this study, the AgNPs was synthesized using intracellular and extracellular cell-free supernatant of S. chiangmaiensis SSUT88A as reducing agents. The synthesis of AgNPs was performed by mixing either intracellular or extracellular cell-free supernatant with 1 mM AgNO3 and incubated at 37 °C under 200 rpm shaking condition for 2 and 5 days, respectively. The overall processes of AgNPs synthesis consist of reduction, capping, and stabilization. In the biological method of AgNPs synthesis, the biological molecules released by bacteria play an essential role in reducing Ag+ to Ag0 and stabilizing AgNPs.49–51 After incubation, the formation of AgNPs can be visually observed by the change of the mixture color from light yellow to brown (Fig. 1). The color change from light yellow to brown during the synthesis of AgNPs was due to the surface plasmon resonance (SPR). The appearance of colour change was a clear indicator of the reduction of Ag+ to Ag0.52 In this study, the S. chiangmaiensis SSUT88A was successfully used for the biosynthesis of AgNPs because not all Streptomyces can be used to synthesize AgNPs. The synthesized AgNPs from intracellular cell-free supernatant and extracellular cell-free supernatant were designated as IS-AgNPs and ES-AgNPs, respectively. To date, this is the first report of S. chiangmaiensis being isolated from soil sample and used for the green synthesis of AgNPs.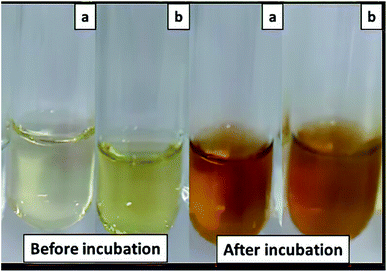 | ||
| Fig. 1 Visible observation of biosynthesis AgNPs by S. chiangmaiensis SSUT88A. (a) IS-AgNPs, and (b) ES-AgNPs. | ||
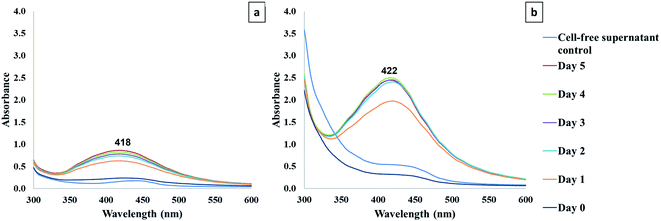
3.3 Characterization of AgNPs using UV-vis spectroscopy
The formation of AgNPs was monitored using the UV-vis spectroscopy technique from day 0 to day 5. The UV-visible spectra of synthesized AgNPs from cell-free supernatant at different points of reaction were recorded (Fig. 2). The results displayed the maximum absorption peaks corresponded to AgNPs at 418 nm and 422 nm for IS-AgNPs and ES-AgNPs, respectively. The absorbance of AgNPs ranges from 380 to 450 nm, which corresponds to the surface plasmon resonance of AgNPs.12,53 The λmax of the samples reflects an alteration of size, shape, and the scattering colour of AgNPs. As diameter increases, the plasmon resonance peak of AgNPs was broader and shifted to longer wavelength. On the other hand, the plasmon resonance of AgNPs shifted toward longer wavelength as the particle becomes more oblate. When the AgNPs aggregate and the conduction electrons near each particle surface become delocalized and shared among neighbouring particles, the surface plasmon resonance shifted to lower energies and scattering peaks shifted to longer wavelength.1,53–57 The absorbance of ES-AgNPs was higher than IS-AgNPs, indicated that the nanoparticle number of ES-AgNPs was higher than that of IS-AgNPs. The reduction of Ag+ could happen through the reducing agents that contain in the cell-free supernatants of S. chiangmaiensis SSUT88A. However, the exact mechanism of Ag+ reduction to Ag0 by cell-free supernatants remains unclear. Several researchers proposed that microbial enzymes could play a significant role as reducing agents such as NADH-dependent reductase.3,58–60In the current study, the synthesis period to obtain maximum yield of AgNPs was achieved at day 5 with intracellular cell-free supernatant, while that of extracellular cell-free supernatant was at day 2. Thus, it suggested that the synthesis process using extracellular cell-free supernatant was faster than that of intracellular cell-free supernatant. An optimum reaction time of synthesis was required to produce stable AgNPs. As the duration of reaction increases, more AgNPs are formed. However, the agglomeration of AgNPs can be formed when the reaction time exceeds the optimum time, resulting in the larger particle size.61
3.4 Analysis of hydrodynamic size and zeta potential
The hydrodynamic size distributions of IS-AgNPs and ES-AgNPs were determined using dynamic light scattering (DLS) technique (Table 1). For IS-AgNPs, DLS analysis showed nanoparticles with 77.03 nm average hydrodynamic size, while that of the ES-AgNPs was bigger, 82.44 nm. The stability of AgNPs is important since it can directly impact their biological activity.62 The stability of synthesized AgNPs was confirmed by measuring their zeta potential. The zeta potential value of the IS-AgNPs and ES-AgNPs were −32.0 and −27.9 mV, respectively, which indicated the presence of negative repulsion among the nanoparticles (Table 1). It has been known that stable nanoparticles exhibit more than +30 mV or less than −30 mV of zeta potential value.63–65 The negative or positive value of zeta potential is defined by capping or stabilizing agent used during the synthesis.66 In this study, the zeta potential values of IS-AgNPs and ES-AgNPs were negative. The negative zeta potential of microbial-based biosynthesis of AgNPs was recorded from the synthesis of AgNPs using Fusarium oxysporum and Aspergillus niger.22,67 The different zeta potential value of IS-AgNPs and ES-AgNPs might attribute to the difference of capping and stabilizing molecules presented in cell-free supernatants. The results of this study indicated that the ES-AgNPs were less stable than IS-AgNPs since the zeta potential value of ES-AgNPs was higher.| AgNPs sample | Parameter | |
|---|---|---|
| Hydrodynamic size (nm) | Zeta potential (mV) | |
| IS-AgNPs | 77.03 | −27.9 |
| ES-AgNPs | 82.44 | −32.0 |
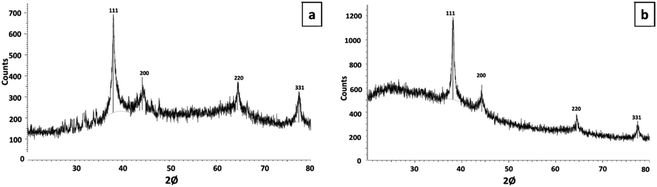
3.5 Analysis of XRD
The crystalline structure of synthesized AgNPs were confirmed by XRD (Fig. 3). Both of the synthesized AgNPs expressed the intense peaks at 38.11°, 44.27°, 64.42°, and 77.47°, which were corresponded to (111), (200), (220), and (311) planes of face-centered cubic (fcc) of metallic silver Standard Joint Committee for Powdered Diffraction Set (JCPDS files). The crystalline profile of AgNPs expressed four intense peaks that corresponded to the face-centred cubic structure of metallic silver.13,68–723.6 Analysis of XANES
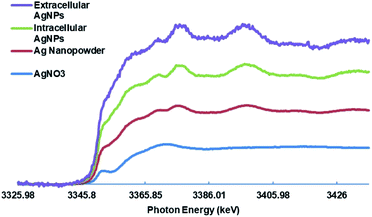
The XANES analysis was conducted to investigate the oxidation state of the synthesized AgNPs. XANES spectroscopy is a powerful tool for proving the oxidation state.73–75 AgNO3 and commercial AgNPs were used as a standard for Ag+ and Ag0, respectively. Different spectra energy of AgNO3 and commercial AgNPs obtained from the XANES measurement are shown in Fig. 4. This result proved that the silver in AgNO3 and commercial AgNPs were in a different oxidation state and spectrum. The spectra of IS-AgNPs and ES-AgNPs showed clear spectrum energy which were similar to commercial AgNPs. The result indicated that Ag that present in the synthesized AgNPs was completely reduced to nanoparticles during the synthesis.3.7 Analysis of FTIR
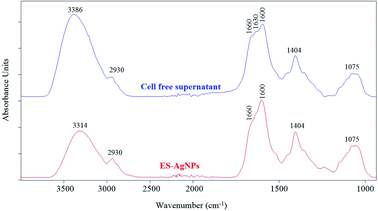
In the biological method, the biomolecules present in the cell-free supernatant of bacteria, such as protein, enzymes, inorganic complexes, and biosurfactants, can be served as reducing, capping, and stabilizing agents.11,95 The FTIR was carried out to determine the biomolecules responsible for reducing, capping, and stabilizing agents of AgNPs. The intracellular cell-free supernatant of S. chiangmaiensis SSUT88A showed a maximum peak at 3386 cm−1 correspond to the O–H stretching. The peaks at 1660, 1630, and 1600 cm−1 were a broad peak that correspond to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N stretching and N–H stretching. The C–N stretching peak was observed at 1404 cm−1. Also, the C–O–C stretching was observed at 1075 cm−1 (Fig. 5). In contrast, the IS-AgNPs showed shifted O–H stretching peak from 3386 to 3286 cm−1 and the peak at 2543 cm−1 which represented C–H stretching. The peaks at 1660 and 1630 cm −1 correspond to the C
O, C–N stretching and N–H stretching. The C–N stretching peak was observed at 1404 cm−1. Also, the C–O–C stretching was observed at 1075 cm−1 (Fig. 5). In contrast, the IS-AgNPs showed shifted O–H stretching peak from 3386 to 3286 cm−1 and the peak at 2543 cm−1 which represented C–H stretching. The peaks at 1660 and 1630 cm −1 correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of amide I, 1404 cm−1 correspond to the C–N stretching of amide III, 1047 cm −1 correspond to the C–O–C stretching, and 835 cm−1 correspond to the C–N stretching.
O stretching of amide I, 1404 cm−1 correspond to the C–N stretching of amide III, 1047 cm −1 correspond to the C–O–C stretching, and 835 cm−1 correspond to the C–N stretching.
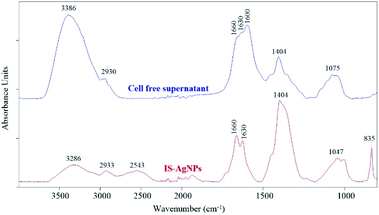 | ||
| Fig. 5 FTIR spectra of IS-AgNPs and intracellular cell-free supernatant of S. chiangmaiensis SSUT88A. | ||
The FTIR spectra of extracellular cell-free supernatant of S. chiangmaiensis SSUT88A and ES-AgNPs are shown in Fig. 6. In the extracellular cell-free supernatant, the absorption peaks at 3386 cm−1 corresponds to the O–H stretching, whereas the peaks at 1660, 1630, and 1600 cm−1 are C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N stretching and N–H stretching, respectively. The peaks at 1404 cm−1 of C–H, and 1075 cm−1 of C–O–C stretching were also observed. On the other hand, the main FTIR spectrum of ES-AgNPs remains the same except for the spectrum around 1600–1700 cm−1 which represents C
O, C–N stretching and N–H stretching, respectively. The peaks at 1404 cm−1 of C–H, and 1075 cm−1 of C–O–C stretching were also observed. On the other hand, the main FTIR spectrum of ES-AgNPs remains the same except for the spectrum around 1600–1700 cm−1 which represents C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and N–H stretching. In Based on the FTIR results (Fig. 5 and 6), the structural change of peaks around 1600–1700 cm−1 in the synthesized AgNPs indicated the presence of amide group that might be responsible for capping and stabilizing the AgNPs. Similar results were reported by Mohamedin et al., Abirami & Kanabiran, and Abd-Elnaby et al.16,19,31 The FTIR result suggests that amide groups of protein can bind to the metal, indicating that proteins play a role in the capping and stabilizing of AgNPs. The amide group of protein could form a layer covering the AgNPs to prevent agglomeration and stabilize the particles.16,31,76 Moreover, the presence of peaks at 2543 and 835 cm−1 corresponded to C–H stretching and C–N stretching of the IS-AgNPs might also function as capping and stabilizing agents.
O and N–H stretching. In Based on the FTIR results (Fig. 5 and 6), the structural change of peaks around 1600–1700 cm−1 in the synthesized AgNPs indicated the presence of amide group that might be responsible for capping and stabilizing the AgNPs. Similar results were reported by Mohamedin et al., Abirami & Kanabiran, and Abd-Elnaby et al.16,19,31 The FTIR result suggests that amide groups of protein can bind to the metal, indicating that proteins play a role in the capping and stabilizing of AgNPs. The amide group of protein could form a layer covering the AgNPs to prevent agglomeration and stabilize the particles.16,31,76 Moreover, the presence of peaks at 2543 and 835 cm−1 corresponded to C–H stretching and C–N stretching of the IS-AgNPs might also function as capping and stabilizing agents.
 | ||
| Fig. 6 FTIR spectra of ES-AgNPs and extracellular cell-free supernatant of S. chiangmaiensis SUT88A. | ||
3.8 Analysis of TEM and EDXRF
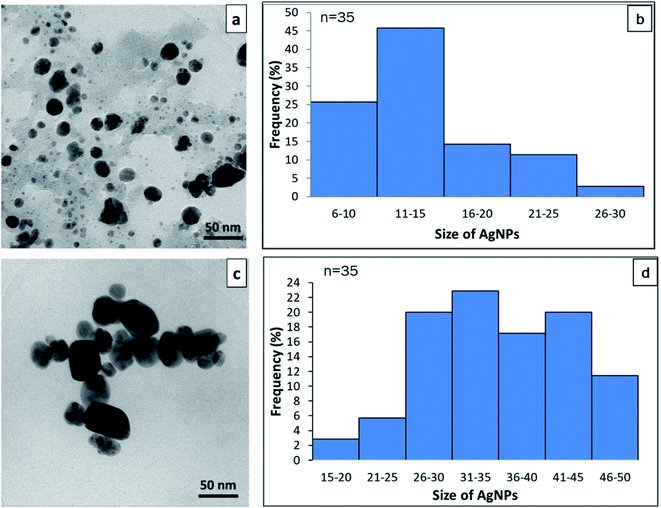
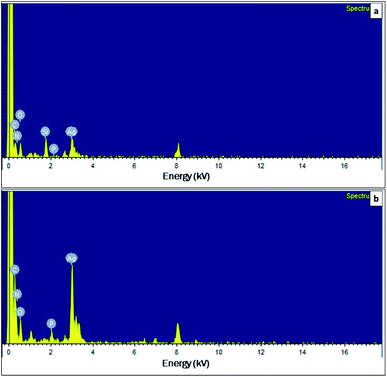
TEM was used to confirm the size and shape of synthesized AgNPs. The TEM image of synthesized AgNPs from cell-free supernatant of S. chiangmaiensis SSUT88A showed a spherical form. A spherical shape is a common form of biological synthesis AgNPs by microbes.19,77 A spherical shape of AgNPs has been reported to synthesize from S. ghanaensis VITHM1, S. rochei MHM13, and Streptomyces sp. LK3.16,17,31 Our results also indicated that different cell-free supernatant used for a synthesis produced different sizes of AgNPs. The size of the IS-AgNPs was varied, ranging from 6.1 to 23.75 nm, with an average size around 13.57 nm. While the size of the ES-AgNPs was larger than that of the IS-AgNPs. It was varied between 15.35 and 47.68 nm, with 30.47 nm average (Fig. 7). The size of ES-AgNPs observed under TEM was two times larger than IS-AgNPs, while the size measurement of synthesized AgNPs using DLS are larger than those from TEM (77.03 nm for IS-AgNPs and 82.44 nm for ES-AgNPs). TEM was considered as the gold standard technique for nanoparticle sizing. Typically, TEM measurements represent only the core particle and exclude the size of capping agents or other molecules associated with AgNPs that would increase the size. On the other hand, DLS measured the hydrodynamic diameter of the AgNPs, which include Ag core, capping agents, and other molecules that absorbed to the surface. The presence of aggregates in the suspensions also be measured and contribute to the higher size values. Therefore, the size obtained from TEM measurements often converges more accurately than DLS.78–80 The factors that could affect the size and shape of nanoparticles are the type and concentration of reducing agents as well as the temperature used during the synthesis.81,82 Therefore, the different sizes between IS-AgNPs and ES-AgNPs might cause by the reducing agents presented in the cell-free supernatant.The dispersive energy of XRF was used to determine the elemental composition of synthesized AgNPs. The result exhibited clear identification of an elemental Ag at 3 kV in both IS-AgNPs and ES-AgNPs (Fig. 8). According to the EDXRF, the amount of Ag in the IS-AgNPs and ES-AgNPs were 21.93% and 49.09%, respectively. The signal of carbon (C), nitrogen (N), oxygen (O), silicon (Si), and phosphorus (P) were able to detect from both IS-AgNPs and ES-AgNPs (Fig. 8). The EDXRF analysis also showed the presence of Ag spectrum at 3 kV, which indicated a signal for metallic silver.19 The presence of Ag in ES-AgNPs was two times higher than IS-AgNPs. This result was correlated to the UV-vis spectroscopy. Other components, such as carbon, nitrogen, sodium, and silica detected in the colloidal sample of AgNPs might be due to a signal of biological molecules in the cell-free supernatant.
3.9 Antimicrobial activity of AgNPs
The increasing phenomena of antibiotic-resistant bacteria happen due to the continuous use of antibiotics throughout the world. With the emergence of antibiotic-resistant bacterial infection, it is essential to look for a novel alternative antimicrobial agent.9,83 Currently, nanotechnology is used to control the consumption of antibiotics and combat antibiotic-resistant microorganisms.84 Nowadays, the AgNPs is gaining significant interest due to their unique properties. The AgNPs are known that they are relatively more toxic than bulk silver.85 The antimicrobial activities of synthesized AgNPs from cell-free supernatant of S. chiangmaiensis SSUT88A against clinical isolate drug-resistant bacteria are shown in Table 2. The antimicrobial activity was performed using an agar well diffusion method with the concentration of 0.2 mg mL−1. The inhibition zone of IS-AgNPs against A. baumannii, K. pneumoniae 1617, P. aeruginosa N90PS, and E. coli 8465 was 20.3 ± 1.7 mm, 12.6 ± 1.5 mm, 20.3 ± 0.5 mm, and 22 ± 1.7 mm, respectively. While ES-AgNPs and commercial AgNPs showed no clear zone, suggesting that there was no antimicrobial action against the tested pathogens. The MIC values of IS-AgNPs against clinical isolate drug-resistant bacteria were also investigated (Table 2). The MIC values of IS-AgNPs against A. baumannii, K. pneumoniae 1617, P. aeruginosa N90PS, and E. coli 8564 were 0.0068, 0.027, 0.013, and 0.027 mg mL−1, respectively. In the current study, the ES-AgNPs showed no antimicrobial activity against tested pathogen, while IS-AgNPs inhibit all tested drug-resistant bacteria (A. baumannii, K. pneumoniae 1617, P. aeruginosa N90PS, and E. coli 8465). The different capping and stabilizing agents of IS-AgNPs and ES-AgNPs might provide a different antimicrobial activity. Compared with the commercial AgNPs, IS-AgNPs had better antimicrobial activity with lower MIC against drug-resistant bacteria (Table 2). Because commercial AgNPs has greater particle sizes (∼100 nm) than IS-AgNPs which may impair their antibacterial effectiveness. As previously stated, the smaller particles have a higher bactericidal effect due to their increased surface area. Previously, commercial AgNPs was reported to have antimicrobial activity against methicillin-resistant Staphylococcus aureus (MRSA) with a MIC of 1.35 mg mL−1.86 However, in this study, IS-AgNPs exhibited antibacterial activity against clinical isolate drug-resistant strains, whereas commercial AgNPs did not show any antibacterial activity. It can therefore be concluded that IS-AgNPs is a promising candidate for controlling nosocomial infections caused by drug-resistant strains. Among all tested bacteria, A. baumannii was one of the major causes of nosocomial infections. In recent years, with the development of antimicrobial resistance, A. baumannii became resistant to numerous commonly used antibiotics, such as aminopenicillin, first- and second-generation cephalosporin. Therefore, A. baumannii become one of the most MDR organism threatening current antibiotics therapy.76,87–89 The WHO has assigned this bacterium as critical priority pathogen in which new antimicrobial agents are urgently needed.90 Thus, the AgNPs has more potential to be further developed as antimicrobial agent against A. baumannii. Previously, the antimicrobial activity of green synthesized AgNPs from Penicillium polonicum against MDR A. baumannii was reported by Neethu et al. with the MIC of 15.62 μg mL−1.91 Compared to the current study, the MIC of IS-AgNPs was two time lower than those AgNPs synthesized from P. polonicum. Therefore, the IS-AgNPs has more potential to be further developed as antimicrobial agent against A. baumannii. The antimicrobial action of AgNPs was to attach to the cell membrane, penetrate the bacteria, generate ROS, and modulate signal transduction pathways.9,11,18,92| Clinical isolate pathogen | Antimicrobial activity | |||||
|---|---|---|---|---|---|---|
| IS-AgNPs | ES-AgNPs | Commercial AgNPs | ||||
| Inhibition zone (mm) | MIC (mg mL−1) | Inhibition zone (mm) | MIC (mg mL−1) | Inhibition zone (mm) | MIC (mg mL−1) | |
| a — = no clear zone; measurements were performed in triplicate. | ||||||
| MDR A. baumannii | 20.3 ± 1.7 | 0.0068 | — | Not determined | — | ≥1.35 |
| K. pneumoniae 1617 | 12.6 ± 1.5 | 0.027 | — | Not determined | — | ≥1.35 |
| P. aeruginosa N90PS | 20.3 ± 0.5 | 0.013 | — | Not determined | — | ≥1.35 |
| E. coli 8465 | 21.0 ± 1.7 | 0.027 | — | Not determined | — | ≥1.35 |
The antimicrobial activity of AgNPs depends on the size, shape, coating or capping agent, stability, and type of bacteria.11 It has been showed that the smaller the nanoparticles, the higher the bactericidal effect. Since the small nanoparticles contain a large surface area for interaction with bacterial cells, thus they can easily adhere to the bacterial cell components.9,11,92
The stability of AgNPs is affected not only by the capping agents but also by storage time and conditions.93 In this study, the IS-AgNPs were stored at room temperature (30–35 °C) for one month and examined their antimicrobial activity. The antimicrobial activity of IS-AgNPs was not considerably affected after storage for one month at room temperature (Table S1†). On the other hand, IS-AgNPs was also tested for stability under 4 °C storage for five months by examining their antibacterial activity and UV-vis spectrum. The results showed that there was no significant change in antibacterial activity or UV-vis spectrum (Table S1 and Fig. S1†). As a result of these findings, IS-AgNPs can be stored at 30–35 °C for at least one month or at 4 °C for at least five months. Velgosova et al. demonstrated that AgNPs synthesized from green method exhibited mild particle agglomeration after 100 days of storage at room temperature but remained stable after six months at 5 °C. Therefore, they proposed that the green synthesized AgNPs should be kept at 5 °C in a dark condition.94 Thus, our data is in agreement with Velgosova et al. that the refrigerator is an ideal storage setting for AgNPs.
4. Conclusions
The soil isolated S. chiangmaiensis SSUT88A has been exploited for the synthesis of AgNPs. The ES-AgNPs showed a larger size compared to IS-AgNPs. The IS-AgNPs showed antimicrobial activity against drug-resistant bacteria A. baumannii, K. pneumoniae 1617, P. aeruginosa N90PS, and E. coli 8465, while ES-AgNPs and commercial AgNPs showed no antimicrobial activity against all tested pathogens. This study revealed that IS-AgNPs synthesized from S. chiangmaiensis SSUT88A was an effective antimicrobial agent against drug-resistant pathogens.Author contributions
AR: experiments design, investigation, data analysis, data curation, data presentation, manuscript preparation; NC: study consultation, manuscript editing; OW: study consultation, manuscript editing; WL: study consultation, data analysis (FTIR, XAS, XANES), manuscript reviewing; NN: conceptualization, experiments design, project administration, study validation, manuscript reviewing and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Suranaree University of Technology, Thailand Science Research and Innovation (TSRI), and National Science, Research, and Innovation Fund (NSRF) (NRIIS number 160363). The authors also thank Ms Supatcharee Siriwong for her assistance with FTIR analysis.References
- S. H. Lee and B. H. Jun, Int. J. Mol. Sci., 2019, 20, 1–24 Search PubMed.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS PubMed.
- M. Ovais, A. T. Khalil, M. Ayaz, I. Ahmad, S. K. Nethi and S. Mukherjee, Int. J. Mol. Sci., 2018, 19, 1–20 Search PubMed.
- E. D. Cavassin, L. F. de Figueiredo, J. P. Otoch, M. M. Seckler, R. A. de Oliveira, F. F. Franco, V. S. Marangoni, V. Zucolotto, A. S. Levin and S. F. Costa, J. Nanobiotechnology, 2015, 13, 1–16 CrossRef PubMed.
- J. W. Alexander, Surg. Infect., 2009, 10, 289–292 CrossRef PubMed.
- R. Li, J. Chen, T. C. Cesario, X. Wang, J. S. Yuan and P. M. Rentzepis, Proc. Natl. Acad. Sci, 2016, 113, 13612–13617 CrossRef CAS PubMed.
- M. J. Carter, K. Tingley-Kelley and R. A. Warriner 3rd, J. Am. Acad. Dermatol., 2010, 63, 668–679 CrossRef CAS PubMed.
- J. L. Elechiguerra, J. L. Burt, J. R. Morones, A. Camacho-Bragado, X. Gao, H. H. Lara and M. J. Yacaman, J. Nanobiotechnology, 2005, 3, 1–10 CrossRef PubMed.
- S. Gurunathan, J. W. Han, D. N. Kwon and J. H. Kim, Nanoscale Res. Lett., 2014, 9, 1–17 CrossRef PubMed.
- S. Iravani, H. Korbekandi, S. V. Mirmohammadi and B. Zolfaghari, Results Pharma Sci., 2014, 9, 385–406 CrossRef CAS PubMed.
- K. S. Siddiqi, A. Husen and R. A. K. Rao, J. Nanobiotechnology, 2018a, 16, 1–28 Search PubMed.
- X. F. Zhang, Z. G. Liu, W. Shen and S. Gurunathan, Int. J. Mol. Sci., 2016, 17, 1–34 Search PubMed.
- K. Anandalakshmi, J. Venugobal and V. Ramasamy, Appl. Nanosci., 2015, 6, 399–408 CrossRef.
- E. A. Barka, P. Vatsa, L. Sanchez, N. Gaveau-Vaillant, C. Jacquard, J. P. Meier-Kolthoff, H. P. Klenk, C. Clement, Y. Ouhdouch and G. P. van Wezel, Microbiol. Mol. Biol. Rev., 2016, 80, 1–43 CrossRef PubMed.
- A. Raja and P. Prabakarana, Am. J. Drug Discovery Dev., 2011, 1, 75–84 CrossRef.
- H. M. Abd-Elnaby, G. M. Abo-Elala, U. M. Abdel-Raouf and M. M. Hamed, Egypt. J. Aquat. Res., 2016, 42, 301–312 CrossRef.
- L. Karthik, G. Kumar, A. V. Kirthi, A. A. Rahuman and K. V. Bhaskara Rao, Bioprocess Biosyst. Eng., 2014, 37, 261–267 CrossRef CAS PubMed.
- D. Manikprabhu and K. Lingappa, J. Pharmacol. Res., 2013, 6, 255–260 CAS.
- A. Mohamedin, N. E.-A. El-Naggar, S. Shawqi Hamza and A. A. Sherief, Curr. Nanosci., 2015, 11, 640–654 CrossRef CAS.
- H. Jiang, S. Manolache, A. C. L. Wong and F. S. Denes, J. Appl. Polym. Sci., 2004, 93, 1411–1422 CrossRef CAS.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramirez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed.
- S. Scandorieiro, L. C. de Camargo, C. A. Lancheros, S. F. Yamada-Ogatta, C. V. Nakamura, A. G. de Oliveira, C. G. Andrade, N. Duran, G. Nakazato and R. K. Kobayashi, Front. Microbiol., 2016, 7, 1–14 Search PubMed.
- S. K. Santos, A. M. Barbosa, L. Pereira da Costa, M. S. Pinheiro, M. B. Oliveira and F. Ferreira Padilha, Molecules, 2016, 21, 1–7 CrossRef.
- P. Wintachai, S. Paosen, C. T. Yupanqui and S. P. Voravuthikunchai, Microb. Pathog., 2019, 126, 245–257 CrossRef CAS PubMed.
- Y. G. Yuan, Q. L. Peng and S. Gurunathan, Int. J. Mol. Sci., 2017, 18, 1–22 CrossRef PubMed.
- S. D. Bansod, M. S. Bawaskar, A. K. Gade and M. K. Rai, IET Nanobiotechnol., 2015, 9, 165–171 CrossRef PubMed.
- D. M. Eby, H. R. Luckarift and G. R. Johnson, ACS Appl. Mater. Interfaces, 2009, 1, 1553–1560 CrossRef CAS PubMed.
- M. Ahamed, M. S. Alsalhi and M. K. Siddiqui, Clin. Chim. Acta, 2010, 411, 1841–1848 CrossRef CAS.
- M. R. Green and J. Sambrook, Cold Spring Harb. Protoc., 2018, 6, 484–485 Search PubMed.
- S. Krishnakumar and V. D. M. Bai, Int. J. TechnoChem Res., 2015, 1, 112–118 CAS.
- M. Abirami and K. Kannabiran, Front. Chem. Sci. Eng., 2016, 10, 542–551 CrossRef CAS.
- B. Ravel and M. Newville, J. Synchrotron Radiat., 2005, 12, 537–541 CrossRef CAS PubMed.
- J. E. L. Daquioag and G. M. Penuliar, Int. J. Microbiol., 2021, 2021, 6699430 Search PubMed.
- Z. A. Z. Abidin, N. A. Malek, Z. Zainuddin and A. J. K. Chowdhury, Front. Life Sci., 2015, 9, 24–31 CrossRef.
- E. B. Shirling and D. Gottlieb, Int. J. Syst. Bacteriol., 1966, 16, 313–340 CrossRef.
- S. S. Purohit, A. K. Saluja and H. N. Kakrani, Pharmaceutical Microbiology, Agrobios, India, 2008 Search PubMed.
- D. P. Labeda, M. Goodfellow, R. Brown, A. C. Ward, B. Lanoot, M. Vanncanneyt, J. Swings, S. B. Kim, Z. Liu, J. Chun, T. Tamura, A. Oguchi, T. Kikuchi, H. Kikuchi, T. Nishii, K. Tsuji, Y. Yamaguchi, A. Tase, M. Takahashi, T. Sakane, K. I. Suzuki and K. Hatano, Antonie van Leeuwenhoek, 2012, 101, 73–104 CrossRef CAS PubMed.
- A. Mehling, U. F. Wehmeier and W. Piepersberg, Microbiology, 1995, 141, 2139–2147 CrossRef CAS PubMed.
- J. Berdy, J. Antibiot., 2005, 58, 1–26 CrossRef CAS PubMed.
- WHO, Laboratory, biosafety manual, World Health Organization, Geneva, 3rd edn, 2004 Search PubMed.
- P. J. Meechan, J. Gyuris, B. R. Petuch, M. M. Chartain and W. K. Herber, in Biological safety: Principles and practices, D. O. Fleming and D. L. Hunt, Wiley, Washington, 4th edn, 2006, DOI: DOI:10.1128/9781555815899.
- HSA, Health and Safety Authority, Dublin, 2020, pp. 1–58.
- L. C. Burnett, G. Lunn and R. Coico, Curr. Protoc. Microbiol., 2009, 1, 1A.1–1A.14 Search PubMed.
- Y. Ohnishi, J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori and S. Horinouchi, J. Bacteriol., 2008, 190, 4050–4060 CrossRef CAS PubMed.
- S. Omura, H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki and M. Hattori, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 12215–12220 CrossRef CAS PubMed.
- V. Barbe, M. Bouzon, S. Mangenot, B. Badet, J. Poulain, B. Segurens, D. Vallenet, P. Marliere and J. Weissenbach, J. Bacteriol., 2011, 193, 5055–5056 CrossRef CAS PubMed.
- N. Pantidos, J. Nanomed. Nanotechnol., 2014, 5, 1–10 Search PubMed.
- M. Harir, H. Bendif, M. Bellahcene, Z. Fortas, R. Pogni, In Basic biology and applications of Actinobacteria, IntechOpen, London, 2018, vol. 6, pp. 99–122 Search PubMed.
- N. I. Hulkoti and T. C. Taranath, Colloids Surf., B, 2014, 121, 474–483 CrossRef CAS PubMed.
- A. Javaid, S. F. Oloketuyi, M. M. Khan and F. Khan, BioNanoScience, 2017, 8, 43–59 CrossRef.
- K. S. Siddiqi, M. Rashid, A. Rahman, Tajuddin, A. Husen and S. Rehman, Biomater. Res., 2018b, 22, 1–9 Search PubMed.
- M. Sastry, A. Ahmad, M. I. Khan and R. Kumar, Curr. Sci., 2014, 85, 162–170 Search PubMed.
- E. A. Coronado, E. R. Encina and F. D. Stefani, Nanoscale, 2011, 3, 4042–4059 RSC.
- K. L. Kelly, E. Coronado, L. L. Zhao and G. C. Schatz, J. Phys. Chem. B, 2003, 103, 668–677 CrossRef.
- R. Brause, H. Möltgen and K. Kleinermanns, Appl. Phys. B, 2002, 75, 711–716 CrossRef CAS.
- D. D. Evanoff and G. Chumanov, J. Phys. Chem. B, 2004, 108, 13957–13962 CrossRef CAS.
- D. Paramelle, A. Sadovoy, S. Gorelik, P. Free, J. Hobley and D. G. Fernig, Analyst, 2014, 139, 4855–4861 RSC.
- K. Mukherjee, R. Gupta, G. Kumar, S. Kumari, S. Biswas and P. Padmanabhan, J. Genet. Eng. Biotechnol., 2018, 16, 527–536 CrossRef PubMed.
- S. A. Kumar, M. K. Abyaneh, S. W. Gosavi, S. K. Kulkarni, R. Pasricha, A. Ahmad and M. I. Khan, Biotechnol. Lett., 2007, 29, 439–445 CrossRef CAS PubMed.
- S. Talekar, A. Joshi, R. Chougle, A. Nakhe and R. Bhojwani, Nano-Struct. Nano-Objects, 2016, 6, 23–33 CrossRef CAS.
- R. Veerasamy, T. Z. Xin, S. Gunasagaran, T. F. W. Xiang, E. F. C. Yang, N. Jeyakumar and S. A. Dhanaraj, J. Saudi Chem. Soc., 2011, 15, 113–120 CrossRef CAS.
- J. T. Tai, C. S. Lai, H. C. Ho, Y. S. Yeh, H. F. Wang, R. M. Ho and D. H. Tsai, Langmuir, 2014, 30, 12755–12764 CrossRef CAS PubMed.
- S. Honary and F. Zahir, Trop. J. Pharm. Res., 2013, 12, 265–273 Search PubMed.
- A. T. Saeb, A. S. Alshammari, H. Al-Brahim and K. A. Al-Rubeaan, Sci. World J., 2014, 2014, 1–9 CrossRef.
- Y. Zhang, M. Yang, N. G. Portney, D. Cui, G. Budak, E. Ozbay, M. Ozkan and C. S. Ozkan, Biomed. Microdevices, 2008, 10, 321–328 CrossRef CAS PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS.
- S. Ninganagouda, V. Rathod, D. Singh, J. Hiremath, A. K. Singh, J. Mathew and M. ul-Haq, BioMed Res. Int., 2014, 2014, 1–9 CrossRef PubMed.
- C. H. N. Barros, S. Fulaz, D. Stanisic and L. Tasic, Antibiotics, 2018, 7, 1–24 CrossRef PubMed.
- R. Chauhan, A. Reddy and J. Abraham, Appl. Nanosci., 2014, 5, 63–71 CrossRef.
- M. S. Nélly, D. C. Eduardo, A. S. Taciana, V. Q. Patrick, R. A. Alyne, C. M. M. Ana, L. d. S. C. Ana, P. F. L. João, B. A. Ricardo, A. d. S. Durcilene, R. d. S. d. A. L. José and F. S. T. Maria, Afr. J. Biotechnol., 2017, 16, 2072–2082 CrossRef.
- S. Baker, P. M. N. Nagendra, B. L. Dhananjaya, K. K. Mohan, S. Yallappa and S. Satish, Enzyme Microb. Technol., 2016, 95, 128–136 CrossRef.
- S. Z. Naqvi, U. Kiran, M. I. Ali, A. Jamal, A. Hameed, S. Ahmed and N. Ali, Int. J. Nanomed., 2013, 8, 3187–3195 CrossRef.
- S. Calvin, XAFS for everyone, CRC Press, Boca Raton, 2013 Search PubMed.
- J. P. Berumen, E. Gallegos-Loya, H. Esparza-Ponce, R. Gonzalez-Valenzuela, C. Gonzalez-Valenzuela and A. Duarte-Moller, Phys. Sci. Int. J., 2009, 1, 138–147 Search PubMed.
- D. Koningsberger and R. Prins, X-ray absorption: principles, applications, techniques of EXAFS, SEXAFS, and XANES, Wiley, 1988 Search PubMed.
- C. A. Moubareck and D. H. Halat, Antibiotics, 2020, 9, 1–29 Search PubMed.
- S. Shivaji, S. Madhu and S. Singh, Process Biochem., 2011, 46, 1800–1807 CrossRef CAS.
- C. H. N. de Barros, G. C. F. Cruz, W. Mayrink and L. Tasic, Nanotechnol., Sci. Appl., 2018, 11, 1–14 CrossRef CAS PubMed.
- T. G. F. Souza, V. S. T. Ciminelli and N. D. S. Mohallem, J. Phys.: Conf. Ser., 2016, 733, 1–5 Search PubMed.
- R. I. MacCuspie, K. Rogers, M. Patra, Z. Suo, A. J. Allen, M. N. Martin and V. A. Hackley, J. Environ. Monit., 2011, 13, 1212–1226 RSC.
- H. S. Kim, Y. S. Seo, K. Kim, J. W. Han, Y. Park and S. Cho, Nanoscale Res. Lett., 2016, 11, 1–9 CrossRef CAS PubMed.
- Z. S. Pillai and P. V. Kamat, J. Phys. Chem. B, 2004, 103, 945–951 CrossRef.
- N. Thongkrachang, S. Chanama and M. Chanama, J. Public Health Dev, 2016, 14, 59–68 Search PubMed.
- E. Banin, D. Hughes and O. P. Kuipers, FEMS Microbiol. Rev., 2017, 41, 450–452 CrossRef CAS PubMed.
- C. Batchelor-McAuley, K. Tschulik, C. C. M. Neumann, E. Laborda and R. G. Compton, Int. J. Electrochem. Sci., 2014, 9, 1132–1138 Search PubMed.
- N. V. Ayala-Núñez, H. H. Lara Villegas, L. del Carmen Ixtepan Turrent and C. Rodríguez Padilla, NanoBiotechnology, 2009, 5, 2–9 CrossRef.
- L. C. Antunes, P. Visca and K. J. Towner, Pathog. Dis., 2014, 71, 292–301 CrossRef CAS PubMed.
- M. Chen, X. Yu, Q. Huo, Q. Yuan, X. Li, C. Xu and H. Bao, J. Nanomater., 2019, 2019, 1–7 Search PubMed.
- J. Shi, T. Sun, Y. Cui, C. Wang, F. Wang, Y. Zhou, H. Miao, Y. Shan and Y. Zhang, BMC Infect. Dis., 2020, 20, 1–9 CrossRef PubMed.
- T. Tacconelli and N. Magrini, Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics, WHO, 2017, pp. 1–7 Search PubMed.
- S. Neethu, S. J. Midhun, E. K. Radhakrishnan and M. Jyothis, Microb. Pathog., 2018, 116, 263–272 CrossRef CAS PubMed.
- S. Prabhu and E. K. Poulose, Int. Nano Lett., 2012, 2, 32–41 CrossRef.
- E. Izak-Nau, A. Huk, B. Reidy, H. Uggerud, M. Vadset, S. Eiden, M. Voetz, M. Himly, A. Duschl, M. Dusinska and I. Lynch, RSC Adv., 2015, 5, 84172–84185 RSC.
- O. Velgosova, E. Čižmárová, J. Málek and J. Kavuličova, Int. J. Miner. Metall. Mater., 2017, 24, 1177–1182 CrossRef CAS.
- T. M. Abdelghany, A. M. H. Al-Rajhi, M. A. Al Abboud, M. M. Alawlaqi, A. G. Magdah, E. A. M. Helmy and A. S. Mabrouk, BioNanoScience, 2017, 8(1), 5–16 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08238h |
| This journal is © The Royal Society of Chemistry 2022 |