Open Access Article
Open Access ArticleAntibacterial effect of copper nanoparticles produced in a Shewanella-supported non-external circuit bioelectrical system on bacterial plant pathogens†
Huong Thu Luong a,
Canh Xuan Nguyen*b,
Thuong Thuong Lama,
Thi-Hanh Nguyenc,
Quang-Le Dangd,
Ji-Hoon Lee
a,
Canh Xuan Nguyen*b,
Thuong Thuong Lama,
Thi-Hanh Nguyenc,
Quang-Le Dangd,
Ji-Hoon Lee e,
Hor-Gil Hurf,
Hoa Thi Nguyen
e,
Hor-Gil Hurf,
Hoa Thi Nguyen g and
Cuong Tu Ho
g and
Cuong Tu Ho *ag
*ag
aInstitute of Environmental Technology, Vietnam Academy of Science and Technology (VAST), 18 Hoang Quoc Viet Str., Cau Giay Dist., Hanoi, 10072, Vietnam. E-mail: hotucuong@gmail.com
bVietnam National University of Agriculture, Trau Quy, Gia Lam, Hanoi, Vietnam. E-mail: Xuancanh79@yahoo.com
cInstitute of Chemistry, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Str., Cau Giay Dist., 10072, Vietnam
dR&D Center of Bioactive Compounds, Vietnam Institute of Industrial Chemistry (VIIC), No. 2 Pham Ngu Lao, HoanKiem, Hanoi, Vietnam
eDepartment of Bioenvironmental Chemistry, Jeonbuk National University, 567 Baekje-daero, Deokjin-gu, Jeonju-si, 54896, Republic of Korea
fSchool of Environmental and Earth Science, Gwangju Institute of Science and Technology, 123 Cheomdangwagi-ro, Buk-gu, Gwangju, 61005, Republic of Korea
gGraduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Str., Cau Giay Dist., Hanoi, 10072, Vietnam
First published on 3rd February 2022
Abstract
The use of copper nanoparticles for the inhibition of plant pathogens Ralstonia solanacearum, which causes wilt disease, and Xanthomonas axonopodis, which causes citrus canker, was investigated in this study. To avoid the inhibiting effect of Cu2+ ions on the bacterial cells, the copper nanoparticles were synthesized in the cathode chamber of a non-external circuit bioelectrochemical system (nec_BES) inoculated with Shewanella sp. HN-41 at the anode. The electrons produced by the oxidation of lactate by Shewanella sp. HN-41 were directly transferred to the anolyte in the cathode via a graphite electrode connecting the anode and cathode chambers. SEM images of the produced particles revealed that the copper nanoparticles were aggregated into spherical shapes with an average size of 2.9 μm from smaller particles with a size range from 30 nm to approximately 190 nm. X-ray diffraction demonstrated that the copper nanoparticles were mainly in the form of a single-phase crystal mixture of atacamite (Cu2Cl(OH)3) and paracatamite (Cu2Cl(OH)3). Finally, for the application of synthesized nanoparticles, an agar diffusion test was applied to assess the antibacterial activity of the formed copper nanoparticles in propylene glycol solvent against R. solanacearum and X. axonopodis. The results showed that the nanoparticles damaged the cells of R. solanacearum, with a half maximum inhibition (IC50) value of 42 ppm, but did not damage X. axonopodis cells.
Introduction
Xanthomonas axonopodis is a Gram-negative, aerobic, slender, rod-shape bacterium that has a single polar flagellum. It causes citrus canker disease in citrus trees such as lime, orange, grapefruit, mandarin and kumquat. The major symptom of citrus canker is the formation of lesions on leaves, stems, thorns, and fruits. This pathogen invades host plant tissues through stomates or wounds on leaves or other green parts.1 Ralstonia solanacearum or Pseudomonas solanacearum is a Gram-negative, non-spore-forming bacterium that causes bacterial wilt in many host plants including banana, tomato, pepper, eggplant, potato and geranium.2 It enters roots via wounds or lateral root emergence points.3 These plant pathogens affect food crops, causing significant losses for farmers and threatening food security.Nanotechnology holds many potential solutions for sustainable agriculture. Compared to bulk metal materials, nanomaterials show many advantages in terms of their ultraviolet-visible sensitivity, high surface-to-volume ratios, and their electrical, catalytic, thermal and quantum properties. Because of their quantum effects and high surface area-to-volume ratios, nanomaterials can affect bacteria.4 Nanomaterials have been applied in agriculture as agents for plant growth, fertilizers, pesticides to control insects, fungi and weeds, and as antimicrobial agents. Recently, nanomaterials have been studied for their effects on plant growth and antimicrobial activities. According to a study by Alaa Y. Ghidan in 2019, zinc showed the ability to increase the synthesis of the auxin plant hormone, indoleacetic acid (IAA).5 Silver is widely used for its antibacterial activity, including as a disinfectant and sterilizing agent for water and active food-packing. Gold can also inhibit many fungal and bacterial species.5 Copper-containing compounds such as CuSO4, CuCO3, and Cu(OH)2 are the traditional inorganic antibacterial materials.5 CuO nanoparticles have been shown to inhibit the growth of microorganisms and exert antiviral and antimicrobial properties; as such, they are used in clothing, bedding, face masks, wound dressing, socks, and other items.6,7 In recent years, there have been many studies on the use of copper nanoparticles in the field of agriculture, and their inhibition of bacterial pathogens in plants is gaining more interest.
Copper nanoparticles can be synthesized using physical, chemical, and biological methods. Microemulsion is the most popular method. However, this method uses a large number of surfactants, which increases the cost. Aerosol techniques, laser ablation, and radiolysis have high efficiency, though they consume a lot of energy.8 Therefore, these methods are less widely applied in industrial production. Another physical method is electron irradiation in the microwave range (wavelength: 0.3–300 GHz).9 Chemical methods, such as ascorbic acid addition by the formation of copper oxide to form nanoparticles or adding strong reducing agents to copper ion micelles, can lead to the disintegration of these ions into copper nanoparticles.10 The above methods are not only intensive and costly, but also use a variety of chemicals that cause serious environmental issues. Therefore, biological methods have emerged as an alternative method for the safe and sound synthesis of nanoparticles. Among them, bacterial strains that have the electrochemical11,12 ability to reduce metals, for instance Shewanella and Geobacter, are potential tools for the synthesis of nanoparticles. Shewanella sp. HN-41, a new strain of bacteria closely related to Shewanella oneidensis MR-1, can synthesize AsS nanotubes,13 and Se nanoparticles,14 and can mediate the formation of nanowires and nano-ribbon particles of Se.15 However, the purification of nanoparticles from cells and the toxicity of metals are factors that limit the further application and production of nanomaterials.
Previous studies with bioelectrochemical systems (BESs) showed that Cu ions were removed from a cathodic solution to produce pure and aggregated Cu or CuO particles.16,17 Therefore, in this study, we applied a new design of BES,18,19 which is composed of two chambers (anode and cathode) connected by a graphite rod to evaluate the possibility and chemical structure of synthesized Cu nanoparticles and also the antibacterial activities of the produced nanoparticles. The produced particles were characterized by SEM, TEM, and XRD, and tested for their antimicrobial activities against plant pathogens (X. axonopodis and R. solanacearum).
Materials and methods
nec_BES system configuration
The system was designed with the anodic and cathodic chambers separated by silicon rubber. These two chambers were directly connected with each other by a 13 cm-long graphite charcoal electrode, which can transport electrons from the anode to the cathode. The anodic chamber was bubbled with nitrogen to remove oxygen and then closed air tight by a screw cap for retaining anaerobic conditions. The cathodic chamber was placed under aerobic conditions (Fig. 1).Preparation of cathodic and anodic chambers
Shewanella sp. HN-41 used in this experiment was kindly donated by the Applied and Environmental Microbiology Laboratory, School of Earth Science and Environmental Engineering, Gwangju Institute of Science and Technology, South Korea. Before adding the bacterial cells into the BES system, it was cultured in Luria–Bertani agar medium, at 30–32 °C for 24 hours to obtain biomass. The biomass was collected on the agar surface and resuspended in mineral media before it was injected into the anode chamber. The anodic cell contained 80 mL of sterilized mineral media, which was prepared with distilled water and the following components: (1) NaHCO3 – 2.5 g L−1; (2) CaCl2 – 0.06 g L−1; (3) NH4Cl – 1 g L−1; (4) MgCl2·H2O – 0.2 g L−1; (5) KCl – 0.1 g L−1; (6) beta-glycerolphosphoric acid disodium salt 0.06 g L−1; (7) NaCl – 10 g L−1; (8) HEPES – 7.2 g L−1; (9) yeast extract – 0.1 g L−1; (10) trace elements solution – 10 mL L−1 (including (mg L−1): nitriloacetic acid – 1500, FeCl2·4H2O – 200, MgCl2·6H2O – 100, Na2WO4·2H2O – 20, MnCl2·4H2O – 100, CoCl2·6H2O – 100, CaCl2·2H2O – 1000, ZnCl2 – 50, CuCl2·2H2O – 2, boric acid – 5, Na2MoO4·2H2O – 10, NaCl – 1000, Na2SeO3 – 17, NiCl2·6H2O – 24); and (11) resazurin 0.05% – 0.1 mL L−1. The pH of the medium solution was adjusted to pH 7.6.14 After sterilizing, mineral media were bubbled with filtered nitrogen gas for 5 minutes, as reported previously, to maintain the dissolved oxygen at 0.24 mg L−1.20 Sodium lactate (10 mM) as a substrate, which was added to the anode chamber later, was filtered and dissolved in distilled and sterilized water.For the aerobic cathodic cell, 75 mL of 5 mM copper dichloride solution was prepared with distilled water without bubbling nitrogen and was added into a glass beaker for the cathodic chamber (Fig. 1).
Operation of nec_BES for nanoparticle production
The experiment was carried out in batch mode in triplicate for 7 days. The variation of pH value, substrate concentration, and Cu2+ ion concentration was monitored at 0, 3, 5, and 7 days. The sodium lactate concentration was determined by high-performance liquid chromatography (HPLC) using a UV detector at a wavelength of 210 nm.At the end of the experiment, the cathode solution was collected for producing nanoparticles. The cathodic solution was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to collect a precipitate, which was rinsed with distilled water by the decantation centrifugation method. Then, the precipitated copper nanoparticles were characterized using SEM and XRD. The copper particles were dispersed in propylene glycol and stored in the refrigerator for biological tests.
000 rpm to collect a precipitate, which was rinsed with distilled water by the decantation centrifugation method. Then, the precipitated copper nanoparticles were characterized using SEM and XRD. The copper particles were dispersed in propylene glycol and stored in the refrigerator for biological tests.
Antibacterial effect of copper nanoparticles
R. solanacearum and X. axonopodis were used from the culture collection in the laboratory of Dept. of Environmental Microbiology – Institute of Environmental Technology. The two bacterial species were cultured on MPA (meat peptone agar) medium at 30 °C for 24 hours to obtain a biomass. Two methods used were the agar diffusion test and the bacterial growth inhibition test. For the agar diffusion test, agar plates were spread with bacteria, and then holes of 10 mm in diameter were perforated and filled with different concentrations of nanoparticles. The plates were incubated overnight at 37 °C. The antibacterial resistance was calculated using the formula (D − d); where D is the diameter of the zone without bacteria, and d is the diameter of the agar hole.For the bacterial lethal dose test, copper nanoparticles were added into different 200 mL glass flasks containing 100 mL meat peptone broth (MPB) medium with different concentrations of 10, 25, 50, 100, 200 ppm from a stock solution of 1000 ppm. The stock solution was prepared as follows: first, the nanoparticles were concentrated by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, then dissolved in polyethylene glycol. The concentration of the stock solution was calculated by drying 10 μL of suspended nanoparticles in the oven at 60 °C overnight to obtain the weight balance (mg L−1 or ppm), and then the concentration of the stock solution was adjusted with polyethylene glycol to 1000 ppm. After that, bacteria were inoculated into the different media and bacterial growth was recorded at 0, 8, 24, and 48 hours by measuring the optical density of the medium at a wavelength of 600 nm on a UV-Vis spectrophotometer (Shimadzu 2450, Tokyo, Japan). The biomass was estimated by integrating the curved growth of cells (optical density of cells at 600 nm) and the value of inhibitory efficiency (%) was calculated based on the integral areas of the growth curves obtained for each of the experimental and control conditions by using the equation Ii (%) = 100 × (Ac − Ai)/Ac, where Ii is the percentage of growth inhibition obtained at concentration i, Ai is the integral area of the growth curve obtained at concentration i, and Ac is the integral area of the growth curve obtained for the control. The IC50 values were obtained from the sigmoidal curve using the equation: y = y0 + a/(1 + exp(−(x − x0)/b)), where y is the inhibitory efficiency, and x is the Cu nanoparticle concentration fitted by regression analysis by open-source SciDAVis software. Then, the half-maximum inhibitory concentration value (IC50) was calculated with response–dose performance based on the fitted sigmoidal curve.
000 rpm, then dissolved in polyethylene glycol. The concentration of the stock solution was calculated by drying 10 μL of suspended nanoparticles in the oven at 60 °C overnight to obtain the weight balance (mg L−1 or ppm), and then the concentration of the stock solution was adjusted with polyethylene glycol to 1000 ppm. After that, bacteria were inoculated into the different media and bacterial growth was recorded at 0, 8, 24, and 48 hours by measuring the optical density of the medium at a wavelength of 600 nm on a UV-Vis spectrophotometer (Shimadzu 2450, Tokyo, Japan). The biomass was estimated by integrating the curved growth of cells (optical density of cells at 600 nm) and the value of inhibitory efficiency (%) was calculated based on the integral areas of the growth curves obtained for each of the experimental and control conditions by using the equation Ii (%) = 100 × (Ac − Ai)/Ac, where Ii is the percentage of growth inhibition obtained at concentration i, Ai is the integral area of the growth curve obtained at concentration i, and Ac is the integral area of the growth curve obtained for the control. The IC50 values were obtained from the sigmoidal curve using the equation: y = y0 + a/(1 + exp(−(x − x0)/b)), where y is the inhibitory efficiency, and x is the Cu nanoparticle concentration fitted by regression analysis by open-source SciDAVis software. Then, the half-maximum inhibitory concentration value (IC50) was calculated with response–dose performance based on the fitted sigmoidal curve.
To observe the effect of nanoparticles on the bacterial cells, pathogen cells were incubated with MPA medium with 100 ppm of the produced nanoparticles for 48 h. The cell structure of dead and live (control without Cu nanoparticle addition) cells were observed using SEM and cross-section TEM images after the incubation period.
Analytical method
For lactate concentration analysis, the filtrate sample of 10 mL was injected into a high-performance liquid chromatography (HPLC) instrument (Agilent Infinity 1260, USA) equipped with an SPD-10A UV detector (Shizuma, Tokyo, Japan) and a ShodexRSpack KC-811 (8.0 mm ID × 300 nm) column (Shodex, Tokyo, Japan). HPLC was run at the isocratic flow regime of 5 mM H2SO4 mobile phase at the rate of 0.5 mL min−1. Lactate was detected by a UV-Vis detector at the wavelength of 210 nm. Sodium lactate concentration was calculated using a calibration curve with different concentrations of sodium lactate standard solution using HPLC.Copper ion concentration in the cathode chamber was analyzed using an atomic absorption spectrometer (AAS; PerkinElmer 3300, PerkinElmer, Shelton, CT). One mL of cathodic sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for supernatant for AAS analysis. The concentration of copper ion was calculated based on the prepared standard curve of copper ion in the AAS.
000 rpm for supernatant for AAS analysis. The concentration of copper ion was calculated based on the prepared standard curve of copper ion in the AAS.
For the morphological structure of nanoparticles and cells, 10 μL of either the washed precipitate of cathodic products or bacterial cells was deposited on a silicon wafer, dried at room temperature overnight, and analyzed by scanning electron microscopy (Hitachi S4800, Japan) at 15 kV. The crystal structure of dried nanoparticles was analyzed using an X-ray diffractometer (D8 ADVANCE, Bruker inc., Germany), equipped with Cu Kα radiation (λ = 1.54056 Å). ImageJ software was used to determine the nanoparticle size based on the SEM images.
For TEM analysis, the control and experiment samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to collect cells. The collected samples were treated with 2.5% glutaraldehyde/0.2 M cacodylate overnight, then washed with 0.1 M cacodylate and fixed by 1% OsO4 in 1 M cacodylate for 1 h at room temperature. The fixed samples were dehydrated with ethanol at 50 °C, 70 °C, 80 °C, 90 °C, and 100 °C, and then the residual ethanol was removed from the samples with propylene oxide. The samples were embedded with the addition of epon (at a 1
000 rpm to collect cells. The collected samples were treated with 2.5% glutaraldehyde/0.2 M cacodylate overnight, then washed with 0.1 M cacodylate and fixed by 1% OsO4 in 1 M cacodylate for 1 h at room temperature. The fixed samples were dehydrated with ethanol at 50 °C, 70 °C, 80 °C, 90 °C, and 100 °C, and then the residual ethanol was removed from the samples with propylene oxide. The samples were embedded with the addition of epon (at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with propylene oxide). Embedded bacterial cells were cross-sectioned onto a thin slide (300–400 Å) by microtome, and then placed on a copper grid with collodion film. Finally, the cross-sectioned cells were dyed with uranyl acetate and citrate lead for 10 and 5 minutes, respectively. Images of the cross-sectioned cells were obtained by transmission electron microscopy (JEOL 1010, Japan) at an acceleration voltage of 200 kV.
1 ratio with propylene oxide). Embedded bacterial cells were cross-sectioned onto a thin slide (300–400 Å) by microtome, and then placed on a copper grid with collodion film. Finally, the cross-sectioned cells were dyed with uranyl acetate and citrate lead for 10 and 5 minutes, respectively. Images of the cross-sectioned cells were obtained by transmission electron microscopy (JEOL 1010, Japan) at an acceleration voltage of 200 kV.
Results and discussion
Lactate and pH changes in the anode chamber
The sodium lactate concentration at the anode showed a significant decrease over time. The initial substrate concentration was reduced from 10 mM to 1 mM after 5 days in all 3 batches of the nec_BES system, while in the control, the initial lactate concentration remained (Fig. 2A). In addition, the pH value of the anodic solution declined from 7.7 to 7.4 after 5 days in the nec_BES inoculated with Shewanella sp. HN-41, implying microbial viability and activity (Fig. 2B). This means that Shewanella consumed lactate and the substrate was fermented to generate acetic acid according to eqn (1). During bacterial metabolism, the pH value typically decreases due to the production of protons. However, HEPES buffer in the medium of the anode kept the pH value of the anode medium solution constant. No electron acceptor was added to the anode chamber except for the graphite carbon electrode, and it was assumed that electrons produced from the metabolism of the substrate were transferred to the electron acceptor in the cathodic solution via the shared electrode.21| CH3CH2OCOO− + H2O → CH3COO− + CO2 + 4e− + 4H+ | (1) |
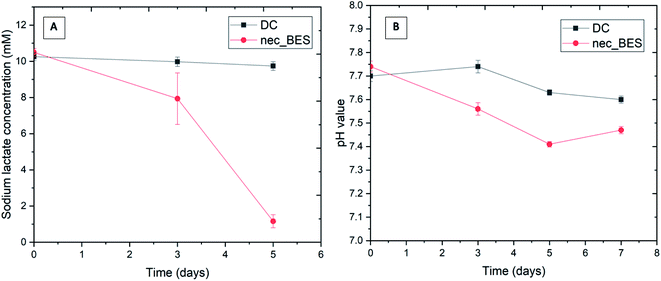 | ||
| Fig. 2 Changes in sodium lactate concentration (A) and pH value (B) in anolyte (DC: control sample (without bacteria added), nec_BES: experimental sample). | ||
Formation of copper nanoparticles in the cathodic solution
Blue lustrous precipitation appeared in cathodic solution and electrode surface after three days of the experiment. In the cathode chamber of the control system, precipitation was not observed. In the catholyte of the experimental systems, Cu2+ ion concentration declined by about 18%, from 4.9 mM to 4 mM (Fig. 3). In a previous experiment, a conventional dual-chamber MFC supported by Shewanella putrefaciens was capable of removing Cu2+ ions by up to 79.9% (at a concentration of 30 ppm Cu2+ solution) and 84.8% (at a concentration of 50 ppm Cu2+ solution).16 Another study using dual-chamber MFCs with a proton exchange membrane for the removal of Cu2+ from CuSO4 solution showed that copper nanoparticles were created in the form of Cu2O and Cu with an efficiency up to 96% after 15 days.17 In our nec_BES, the lower efficiency was probably due to the high internal resistance of the system.16,17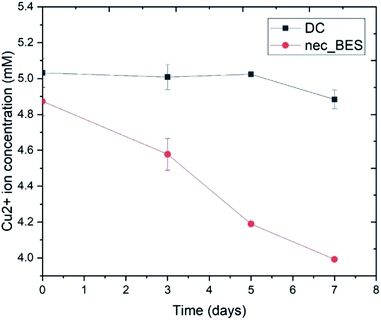 | ||
| Fig. 3 Changes in Cu2+ ion concentration in catholyte (DC: control sample (without bacteria added), nec_BES: experimental sample). | ||
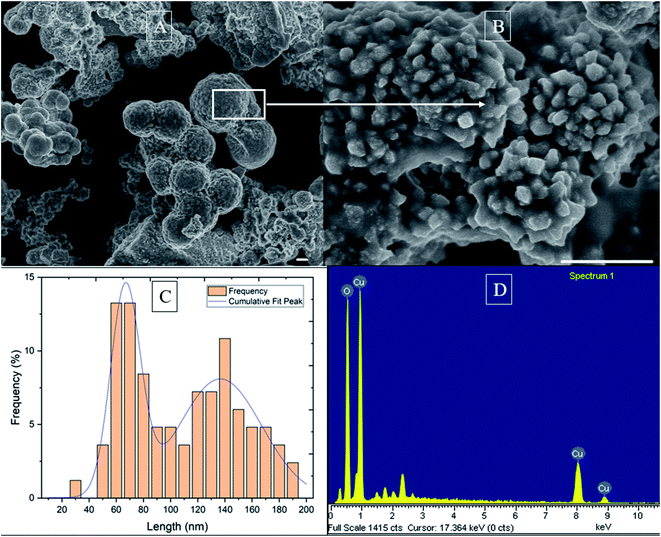
The precipitates were characterized for their morphological grain structures by SEM and XRD analysis. The SEM results showed that nanoparticles were observed as rough and spherical shapes with an average size of 2.9 μm (Fig. 4A). However, high-resolution SEM analysis revealed smaller and irregularly-shaped nanoparticles with an average size of 104 nm on the surface of the aggregated, bigger nanoparticles. These irregularly shaped nanoparticles had many sharp edges with widths ranging from 30 to 190 nm. Further size analysis of these irregular nanoparticles, in which nanoparticle size distribution was fitted with a Gaussian model (R2 = 0.92), showed two peaks at 66 nm and 135 nm (Fig. 4C).
The results of X-ray energy dispersive spectroscopy analysis showed the existence of the elements Cu and O in the nanoparticles (Fig. 4D). The pH value observed after one week showed an increase from 4.47 to 7.83, compared with the remaining of pH value in the control (changing from 4.5–4.45; data not shown). The increase of pH followed eqn (2)–(4).22 It was assumed that oxygen in the cathode chamber had received electrons from the graphite electrode to form OH−. Thereby, in the cathode chamber, the reaction between Cu2+, OH− and Cl− ions occurred to form copper nanoparticles.
| O2 + 2H+ + 2e− ⇌ H2O2 | (2) |
| H2O2 + 2H+ + 2e− ⇌ H2O | (3) |
| H2O ⇌ H+ + OH− | (4) |
The XRD image of nanoparticles in Fig. 5 shows the crystal mixture of particles. The peak was compared and indexed with peaks in the PDF cards of ICDD (PDF no. 00-045-0029, PDF no. 01-070-0821, and PDF no. 01-074-9208). Strong peaks at 15.7°, 16.4°, and 17.9° correspond to the typical planes of (111) from PDF no. 00-045-0029, (021) from PDF no. 01-070-0821, and (101) from PDF no. 01-074-9208, respectively (Fig. 5). It is interesting that the other peaks with large peak widths at the base were typical for paratacamite and atacamite, possibly demonstrating that the mixture of crystals was mainly paracatamite and atacamite (Fig. 5). A previous study in the synthesis of paratacamite and atacamite confirmed that an excess of OH− will result in a stable single phase of paratacamite and atacamite in the solution at 25 °C from the reaction between NaOH and CuCl2.23
 | ||
| Fig. 5 XRD image of Cu nanoparticles produced from nec_BES system supported by Shewanella sp. HN-41. | ||
Antibacterial activities of synthesized copper nanoparticles
An agar diffusion test for assessing the antibacterial activity of copper nanoparticles with X. axonopodis and R. solanacearum showed a clear zone of antimicrobial activity against R. solanacearum at concentrations above 25 ppm but no clear zone for X. axonopodis (Fig. 6). No clear zone was observed for the control of polyethylene glycol. | ||
| Fig. 6 Effect of different Cu nanoparticle concentrations (10, 25, 50, 100, 200 and 500 ppm) on bacteria (2012.001: Xanthomonas axonopodis; 2012.002: Ralstonia solanacearum; PG: polyethylene glycol). | ||
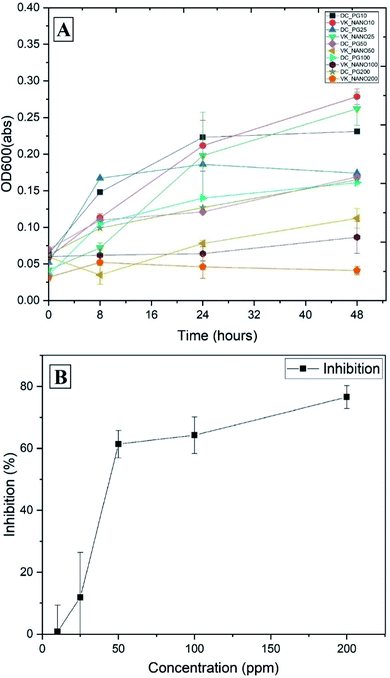
A further experiment with R. solanacearum was set up to observe the effects of copper nanoparticles at various concentrations. At concentrations of 50, 100, 200, and 500 ppm against R. solanacearum, the diameters of inhibition zones were 4, 6, 10, and 14 mm, respectively. The growth curves of R. solanacearum at different concentrations of copper nanoparticles are shown in Fig. 7A. At concentrations of 10 and 25 ppm nanoparticles, the densities of bacterial cells were similar to that of the control sample. At increasing concentrations (50, 100, and 200 ppm) of copper nanoparticles, the bacterial biomass was reduced by approximately 61%, 64%, and 77% in comparison with the control biomass, respectively (Fig. 7B). Using SciDAVis software, the inhibition curve was fitted to a sigmoidal curve and the fitted equation was y = −1.24 + 71.67/(1 + exp (−(x − 35.9)/7.31)), R2 = 0.98. The IC50 value was estimated to be 42 ppm based on this equation.
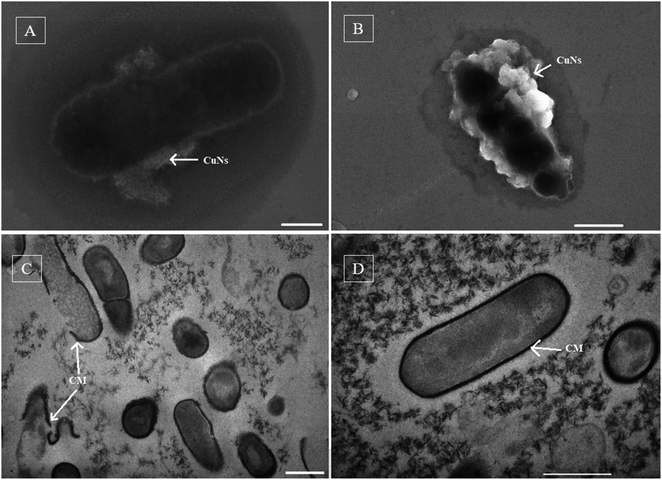
The antimicrobial mechanism of copper nanoparticles was investigated by the treatment of pathogen cells (both R. solanacearum and X. axonopodis) with 100 ppm of copper nanoparticles in the MPB (liquid medium) for 48 h. The cells collected after the experiment were observed by SEM, copper nanoparticles were found to be attached to the cell surfaces of both cells (Fig. 8A and B). The SEM image of R. solanacearum cells shows the distortion of the cell wall (Fig. 8B) in comparison with the control (Fig. 8D), but this distortion was not observed in the case of X. axonopodis (Fig. 8A). Further studies investigating the TEM images of cross-sections of R. solanacearum cells demonstrated that the particles were not deposited inside of the cells but peeled off the cell membranes (Fig. 8C).
In our experiment, R. solancearum cells responded differently to the lower (10 or 25 ppm) and higher (50, 100, or 200 ppm) concentrations of Cu nanoparticles, due to the essential nature of copper. Cu is involved in essential enzymatic activities of bacteria, particularly in cellular respiration (cytochrome C oxidase and NADH dehydrogenase), but it is supposed that Cu homeostasis is focused on protection from Cu rather than Cu utilization.24 Slavin et al. (2017) also argued that the lower toxicity of Cu nanoparticles compared to Ag nanoparticles was due to the essentiality of Cu in physiological systems. The 4 and 5 mM CuSO4 solutions did not kill E. coli, and these concentrations appeared to be bacteriostatic but not bactericidal.25 Thus, at low concentration, Cu nanoparticles in this experiment were able to act as an essential source of Cu for cell growth rather than killing the cells.
Previous studies using various microscopy approaches, including atomic force microscopy, transmission electron microscopy and laser confocal microscopy, showed that nanoparticles were absorbed on the cell membrane, leading to depolarization. The cell wall, whose typical negative charge was changed by copper nanoparticles, became easily broken.26,27 Other researchers reported copper cations released from nanoparticles were able to interact with lipids, causing their peroxidation and pore opening in cell membranes, compromising the integrity of bacterial cells.25 Using E. coli as a model bacteria for investigating the interaction between cells and nano-Mg(OH)2 slurries, Pan et al. (2013)28 found that the aggregates of these nanoparticles play an important role in biocidal activities. The toxicity of Mg(OH)2 aggregates depends on their size, and particularly, the bigger size has a stronger effect on the cells. Interestingly, the particles caused the damage of E. coli cell membranes and were not found inside of the cells.28,29 In our study, the produced nanoparticles, in the form of Cu2Cl(OH)3 aggregated into large particles (>1 μm), were supposed to be attracted first to the cell wall, then interact abrasively26 with the cell wall, and finally peel off the cell wall. The crystal structure of small nanoparticles with sharp edges possibly made it easier for peeling off the cell membrane.
It is interesting that both X. axonopodis and R. solanacearum are Gram-negative bacteria with a layer of lipopolysaccharide in their outer membranes, but their responses to the Cu nanoparticles were different in agar diffusion. However, when incubating X. axonopodis, R. solanacearum, and Bacillus subtilis (Gram positive cells) with 200 ppm of Cu nanoparticles or Cu ions in the liquid medium for 48 h, it was observed that all bacteria were inhibited by Cu ions and Cu nanoparticles (ESI†). Cu ions also appeared to have a lower influence on the cell growth than Cu nanoparticles. In addition, the small sizes of the Cu nanoparticles surrounding the X. axonopodis cell in the SEM image suggest that the aggregates of the Cu nanoparticles were hindered by the cell wall, resulting in the decrease of the killing effect by the aggregates of Cu nanoparticles (Fig. 8A). Other factors, such as ligands in the lipopolysaccharide and exopolysaccharide that interacted with the nanoparticles, also trapped nanoparticles and decreased their toxicity.27 Therefore, the nature of the outer membrane and exopolysaccharide in different bacteria possibly plays an important role in the detoxification of Cu nanoparticles.
Conclusion
In this study, a simple non-external circuit bioelectrochemical system containing Shewanella sp. HN-41 successfully produced Cu nanoparticles in the form of a crystal mixture (acatamite and paratacamite). The small particles under 100 nm were aggregated into larger particles with sizes of over 1 μm, which killed R. solanacearum cells more effectively than Cu ions. The produced Cu nanoparticles exhibited an IC50 value of 42 ppm to R. solanacearum, and also inhibited the growth of both Gram-negative and positive cells (R. solanacearum, X. axonopodis, and B. subtilis, respectively) in the liquid medium. Therefore, the nec_BES might be suggested as a potential tool for utilizing organic waste to produce effective nanoparticles for controlling plant pathogens.Conflicts of interest
The manuscript contains data and writing with no conflicts of interest.Data availability
All data used in the manuscript are original. If you have any interest in the data, please contact the corresponding author via email: E-mail: hotucuong@gmail.com.Acknowledgements
This research is funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 104.03-2016.45.References
- J. H. Graham, T. R. Gottwald, J. Cubero and D. S. Achor, Mol. Plant Pathol., 2004, 5, 1–15 CrossRef PubMed.
- M. Fegan, A. C. Haward, A. C. H. Mark Fegan, D. Liu, M. Fegan, A. Hayward and A. C. H. Mark Fegan, in Manual of Security Sensitive Microbes and Toxins, CRC press, 2014, vol. 34, pp. 807–818 Search PubMed.
- J. Yao and C. Allen, J. Bacteriol., 2007, 189, 6415–6424 CrossRef CAS PubMed.
- M. I. Din and R. Rehan, Anal. Lett., 2017, 50, 50–62 CrossRef CAS.
- A. Y. Ghidan and T. M. A. Antary, in Applications of Nanobiotechnology, ed. M. Stoytcheva and R. Zlatev, IntechOpen, London, 2019, pp. 115–142 Search PubMed.
- G. Borkow, S. S. Zhou, T. Page and J. Gabbay, PLoS One, 2010, 5(6) DOI:10.1371/journal.pone.0011295.
- G. Borkow and J. Gabbay, FASEB J., 2004, 18, 1728–1730 CrossRef CAS PubMed.
- R. M. Tilaki, A. I. Zad and S. M. Mahdavi, Appl. Phys. A: Mater. Sci. Process., 2007, 88, 415–419 CrossRef CAS.
- H. Wang, J. Z. Xu, J. J. Zhu and H. Y. Chen, J. Cryst. Growth, 2002, 244, 88–94 CrossRef CAS.
- E. M. Egorova and A. A. Revina, Colloids Surf., A, 2000, 168, 87–96 CrossRef CAS.
- S. P. S. Badwal, S. S. Giddey, C. Munnings, A. I. Bhatt and A. F. Hollenkamp, Front. Chem., 2014, 2, 1–28 CAS.
- B. Min, S. Cheng and B. E. Logan, Water Res., 2005, 39, 1675–1686 CrossRef CAS PubMed.
- J. H. Lee, M. G. Kim, B. Yoo, N. V. Myung, J. Maeng, T. Lee, A. C. Dohnalkova, J. K. Fredrickson, M. J. Sadowsky and H. G. Hur, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20410–20415 CrossRef CAS PubMed.
- K. Tam, C. T. Ho, J. H. Lee, M. Lai, C. H. Chang, Y. Rheem, W. Chen, H. G. Hur and N. V. Myung, Biosci., Biotechnol., Biochem., 2010, 74, 696–700 CrossRef CAS PubMed.
- C. T. Ho, J. W. Kim, W. B. Kim, K. Song, R. a. Kanaly, M. J. Sadowsky and H.-G. Hur, J. Mater. Chem., 2010, 20, 5899 RSC.
- A. Sumisha, J. Ashar, A. Asok, S. Karthick and K. Haribabu, Sep. Sci. Technol., 2019, 1–9 Search PubMed.
- H. Tao, M. Liang, W. Li, L. Zhang, J. Ni and W. Wu, J. Hazard. Mater., 2011, 189, 186–192 CrossRef CAS PubMed.
- C. T. Ho, T. T. Lam, H. T. Nguyen, C. X. Nguyen, Q. Le Dang, J. Lee, Y. Yang and H.-G. Hur, Nanomater. Nanotechnol., 2021, 11, 184798042110561 CrossRef.
- C. T. Ho, T. H. Nguyen, T. T. Lam, D. Q. Le, C. X. Nguyen, J. hoon Lee and H. G. Hur, Electron. J. Biotechnol., 2021, 54, 1–7 CrossRef CAS.
- J. H. Lee, Y. Roh, K. W. Kim and H. G. Hur, Geomicrobiol. J., 2007, 24, 31–41 CrossRef CAS.
- Y. V. Nancharaiah, S. Venkata Mohan and P. N. L. Lens, Bioresour. Technol., 2015, 195, 102–114 CrossRef CAS PubMed.
- I. Katsounaros, S. Cherevko, A. R. Zeradjanin and K. J. J. Mayrhofer, Angew. Chem., Int. Ed., 2014, 53, 102–121 CrossRef CAS PubMed.
- A. M. Pollard, R. G. Thomas and P. A. Williams, Mineral. Mag., 1989, 53, 557–563 CrossRef CAS.
- R. A. Festa and D. J. Thiele, Curr. Biol., 2011, 21, R877–R883 CrossRef CAS PubMed.
- R. Hong, T. Y. Kang, C. A. Michels and N. Gadura, Appl. Environ. Microbiol., 2012, 78, 1776–1784 CrossRef CAS PubMed.
- P. K. Stoimenov, R. L. Klinger, G. L. Marchin and K. J. Klabunde, Langmuir, 2002, 18, 6679–6686 CrossRef CAS.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, J. Nanobiotechnol., 2017, 15, 1–20 CrossRef PubMed.
- X. Pan, Y. Wang, Z. Chen, D. Pan, Y. Cheng, Z. Liu, Z. Lin and X. Guan, ACS Appl. Mater. Interfaces, 2013, 5, 1137–1142 CrossRef CAS PubMed.
- Y. H. Leung, A. M. C. Ng, X. Xu, Z. Shen, L. A. Gethings, M. T. Wong, C. M. N. Chan, M. Y. Guo, Y. H. Ng, A. B. Djurišić, P. K. H. Lee, W. K. Chan, L. H. Yu, D. L. Phillips, A. P. Y. Ma and F. C. C. Leung, Small, 2014, 10, 1171–1183 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08187j |
| This journal is © The Royal Society of Chemistry 2022 |