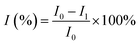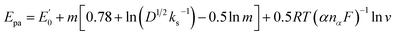Open Access Article
Open Access ArticleA novel electrochemical sensing platform based on the esterase extracted from kidney bean for high-sensitivity determination of organophosphorus pesticides†
Han Tao *ab,
Feng Liuab,
Chun Jic,
Yuangen Wu
*ab,
Feng Liuab,
Chun Jic,
Yuangen Wu ab,
Xiao Wangab and
Qili Shiab
ab,
Xiao Wangab and
Qili Shiab
aSchool of Liquor and Food Engineering, Guizhou University, Guiyang 550025, China. E-mail: hantaov@hotmail.com
bKey Laboratory of Fermentation Engineering and Biopharmacy of Guizhou Province, Guizhou University, Guiyang 550025, China
cSchool of Pharmaceutical Sciences, Guizhou University, Guiyang 550025, China
First published on 11th February 2022
Abstract
Similar to acetylcholinesterase, the activity of plant-derived esterase can also be inhibited by organophosphorus pesticides. Therefore, an electrochemical sensing platform using kidney bean esterase as a new detection enzyme was proposed for the highly sensitive determination of organophosphorus pesticides. Purified kidney bean esterase was obtained by an efficient and economical aqueous two-phase extraction method. Carboxylated graphene/carbon nanotube composites (cCNTs–cGR) and Au nanoparticles were used to provide a biocompatible environment to immobilize kidney bean esterase and also accelerate electron transport between the analyte and the electrode surface. Due to the good synergistic electrocatalytic effects of these nanomaterials, the biosensor exhibited an amplified electrocatalytic response to the oxidation of α-naphthalenol, which makes the sensor more sensitive. Based on the inhibitory effect of trichlorfon on kidney bean esterase activity, high sensitivity and low-cost detection of trichlorfon was achieved. Under optimum conditions, the inhibition of trichlorfon is proportional to its concentration in the range of 5 to 150 ng L−1 and 150 ng L−1 to 700 ng L−1 with an ultra-low detection limit of 3 ng L−1. Moreover, the validity of the prepared biosensor was verified by analyzing several actual agricultural products (cabbage and rice) with satisfactory recoveries ranging from 94.05% to 106.76%, indicating that kidney bean esterase is a promising enzyme source for the analysis of organophosphorus pesticides in food samples.
1 Introduction
Organophosphorus pesticides (OPPs), which represent 33% of available pesticides in developed countries and 50–60% in developing countries,1 are the most extensively used pesticides worldwide to protect crops. However, the excessive use of OPPs leads to residues in food2,3 and the natural environment.4 These OPP residues can enter the body through the food chain and irreversibly inhibit the activity of acetylcholinesterase (AChE), leading to the accumulation of acetylcholine in the nervous system and causing severe neurological dysfunction.5 Additionally, OPPs can induce cancer and type II diabetes6 and may affect the growth of children.7 Hence, OPPs are classified as a highly toxic class of chemical compounds by the World Health Organization (WHO). Many countries or organizations, especially developed countries and the European Union, have implemented monitoring programs to control the abuse of OPPs. Accordingly, the efficient detection of OPP residues in agricultural products has always been an essential topic in food safety field.8,9Currently, the standard methods for detecting OPPs are a series of chromatography-based methods, i.e., gas chromatography (GC),10 high-performance liquid chromatography (HPLC),11 or chromatography coupled with mass spectrometry.12,13 These methods are extremely sensitive and accurate, but they also have the disadvantages of complex pre-treatment steps, long analysis time, and require highly trained operators. More importantly, these instruments are expensive and inconvenient to carry. These drawbacks restrict their use for fast detection and on-site applications.14 However, the sale of agricultural products is often characterized by fragmentation and small scale, especially in underdeveloped and developing countries. In order to monitor OPPs residues in agricultural products in time and prevent foods with excessive levels of OPPs from entering the market, developing simple, rapid and sensitive analytical methods for OPPs residues detection in food samples is of great importance and urgency.
Electrochemical sensing is highly sensitive, easy to operate, and fast to detect.15–17 Moreover, electrochemical instruments are cheaper and smaller in size than chromatography-based devices, so they can be developed into portable devices to realize on-site detection.18 Electrochemical technology for OPPs detection has received extensive attention in recent years.19,20 Most electrochemical methods for detecting OPPs are based on enzyme inhibition, and the most commonly used enzyme is acetylcholinesterase (AChE).21 AChE is extracted from animal tissues and organs,22 such as the head of the housefly, the electric organ of the electric eel, and horse serum and duck plasma. The extraction process is complex and needs to be carried out at low temperature. So AChE is expensive and not suitable for widespread use.23 Meanwhile, recent studies have found that the activity of esterase derived from plants can also be inhibited by OPPs and its sensitivity to pesticides is similar to that of AChE.24–26 Even more appealing, plant-derived esterases come from more readily available plants and can be produced by a simpler extraction procedure and stored more conveniently.26 Thus, they are much easier to obtain than AChE and have a lower price. Therefore, using plant-derived esterase instead of AChE to determine OPPs is more economical and practical.
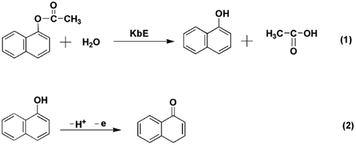
In this study, plant esterase extracted from white kidney bean (KbE) was selected as the detection enzyme to construct a new electrochemical OPPs biosensor. The KbE immobilized on the electrode catalyzed the hydrolysis of its substrate, α-naphthyl acetate, to produce α-naphthalenol. α-Naphthalenol molecules contain electroactive phenolic hydroxyl groups, which can be oxidized to produce an oxidation peak. In the presence of organophosphorus pesticides, the activity of KbE is inhibited, resulting in a decrease in the production of α-naphthalenol, thus the detection of pesticides can be achieved by monitoring the reduction of the oxidation peak current. To enhance the sensitivity of detection, negatively charged carboxyl-functionalized carbon nanomaterials and positively charged gold nanoparticles (AuNPs) were successively modified on the glass carbon electrode. AuNPs were also used for KbE immobilization. Based on the synergistic electrocatalytic effect of carbon nanomaterials and AuNPs, the electrochemical sensing signal was effectively improved. Hence, high sensitivity and low-cost detection of trichlorfon were realized. To the best of our knowledge, there have been no report on the application of KbE in OPPs electrochemical sensing. This work also provides a new idea for the application of plant-derived esterase in pesticide detection, and the proposed biosensor has the potential to be developed into a portable device for on-site pesticide monitoring.
2 Experimental
2.1 Chemical reagents and materials
Chitosan, PEG1000, (NH4)2SO4, HAuCl4·4H2O, PDDA (MW = 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–350
000–350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Shanghai Aladdin Reagent Co., Ltd. Trichlorfon, α-naphthyl acetate (α-NA), α-naphthalenol, fast blue B salt were purchased from Sigma-Aldrich (St. Louis, USA). Graphene oxide (GO) and carbon nanotubes (CNTs) were acquired from Xianfeng Nanomaterial Technology Co., Ltd. Wheat, white kidney bean, wheat bran, myotonin, corn were obtained from a local market. All other chemicals used were analytical grade. Doubly distilled water was used throughout all experiments.
000) were purchased from Shanghai Aladdin Reagent Co., Ltd. Trichlorfon, α-naphthyl acetate (α-NA), α-naphthalenol, fast blue B salt were purchased from Sigma-Aldrich (St. Louis, USA). Graphene oxide (GO) and carbon nanotubes (CNTs) were acquired from Xianfeng Nanomaterial Technology Co., Ltd. Wheat, white kidney bean, wheat bran, myotonin, corn were obtained from a local market. All other chemicals used were analytical grade. Doubly distilled water was used throughout all experiments.
2.2 Instrumentation
All electrochemical measurements were performed with a CHI660E electrochemical workstation (Chenhua, China). Scanning electron microscopy (SEM) images were obtained from SU8010 SEM (HITACHI, Japan). Transmission electron microscopy (TEM) image was taken with JEM-2100 TEM (Hitachi, Japan). Fourier transform infrared (FTIR) spectra were collected on Nicolet 6700 FT-IR spectrometer (Thermo Scientific, USA). UV-VIS spectra were collected by UV-2700 spectrophotometer (Shimadzu, Japan).2.3 Preparation of nanomaterials
AuNPs were prepared using positively charged PDDA as a protective and reducing agent according to literature.27 Typically, 500 μL PDDA (4 wt%) and 400 μL NaOH (0.5 M) in 80 mL water was brought to boiling. Then 200 μL HAuCl4 (10 mg mL−1) was quickly added, and the mixed solution was kept stirring and boiling until the colour of the solution changed to burgundy. The residual PDDA was removed by centrifugation for 20 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, followed by washed with water. The purified PDDA-capped gold nanoparticles (PDDA-Au) were redispersed in water to produce a colloidal suspension with positive charge.
000 rpm, followed by washed with water. The purified PDDA-capped gold nanoparticles (PDDA-Au) were redispersed in water to produce a colloidal suspension with positive charge.
For the preparation of carboxylated graphene (cGR), 12 g NaOH was added into 50 mL GO aqueous solution (2 mg mL−1) and reacted under ultrasonication for 2 h. Then 10 g monochloroacetic acid was added and ultrasonized again for 2 h. The resulting suspension was filtered and the residue was washed with water until the pH was nearly neutral, followed by drying under vacuum at 50 °C to obtain cGR.28 Carboxylated CNTs (cCNTs) were obtained by acid treatment with H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3 (3
HNO3 (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) at 80 °C for 3 h. The presence of carboxyl functional groups causes the cGR and cCNTs to be negatively charged.29
1 v/v) at 80 °C for 3 h. The presence of carboxyl functional groups causes the cGR and cCNTs to be negatively charged.29
2.4 Enzyme extraction and purification
Various plants, including wheat, white kidney bean, wheat bran, myotonin, corn and millet, were washed with water and dried naturally, then crushed into powder with a grinder. The crushed material was stirred in distilled water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 w/w) for 30 min and then centrifuged at 8000 rpm for 10 min at 4 °C. The supernatant was filtered through a 0.45 μm microporous membrane to obtain the crude enzyme solution.
5 w/w) for 30 min and then centrifuged at 8000 rpm for 10 min at 4 °C. The supernatant was filtered through a 0.45 μm microporous membrane to obtain the crude enzyme solution.
The crude enzyme was further purified using the double aqueous phase extraction method proposed by Yang et al.23 Briefly, in the first step, 27.0% PEG1000/13.0% NaH2PO4 (pH 5.0) was used as the ATPS conditions. Most of plant-esterase was partitioned to the top phase in the ATPS. After the bottom phase was discarded, (NH4)2SO4 was added to form the second ATPS (27.0% PEG1000/13.0% NaH2PO4/6.0% (NH4)2SO4 (pH 5.0)), and most of the plant-derived esterase shifted to the bottom phase. Subsequently, the bottom phase was collected and dialyzed in 0.05 M phosphate buffer (pH 6.5) for 48 h at 4 °C. Finally, the plant-esterase dialysate was lyophilized and then dissolved in PBS6.5 for later use. Since the isoelectric point of plant-derived esterase is 4.3–4.6,23 KbE is negatively charged at pH 6.5.
2.5 Electrodes modification
Prior to modification, the GCE was polished sequentially with 0.3 μm and 0.05 μm Al2O3 slurry and then sonicated in water, acetone and distilled water, respectively. The cleaned electrode was activated in 0.1 mol L−1 PBS 7.0 by 10 successive CV scans from −0.5 to +1.2 V at 0.05 V s−1. 6 μL of cCNTs–cGR composite solution was dropped onto the electrode surface and dried naturally. Then the modified electrode was immersed in PDDA-AuNPs solution for 60 min. Since PDDA-AuNPs are positively charged, they would be adsorbed on the negatively charged cCNTs–cGR and the resulting modified electrode was noted as Au/cCNTs–cGR/GCE. Afterward, 18 μL kidney beans esterase (KbE) solution was dropped on PDDA-AuNPs and dried at 4 °C. The modified electrode was washed carefully with water to remove the unstable KbE and the resulting electrode was denoted as KbE/Au/cCNTs–cGR/GCE. Finally, 6 μL CS (0.25 wt%) with good film-forming property and positive charge were dropped onto the modified electrode to prevent leakage of the enzyme, the as-fabricated electrode (labelled for CS/KbE/Au/cCNTs–cGR/GCE) was stored at 4 °C for later use. The preparation process of the electrodes is shown in Scheme 1(a). In addition, CS/KbE/cCNTs–cGR/GCE, CS/KbE/Au/GCE and CS/KbE/GCE were prepared for comparison.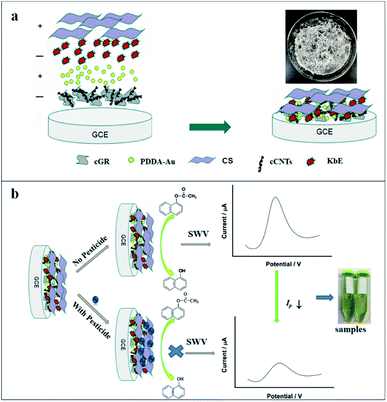 | ||
| Scheme 1 Schematic illustration of the biosensor fabrication process (a) and the principle for pesticide determination (b). Inset in (a) is photograph of the lyophilized KbE. | ||
2.6 Electrochemical measurements
A traditional three-electrode system was used for electrochemical experiments, with Ag/AgCl (3 M KCl) electrode as the reference electrode, Pt wire as the counter electrode and modified GCE as the working electrodes. Cyclic voltammogram (CV) measurements were performed in 0.1 mol L−1 PBS buffer solution using the potential range from 0.0 to +1.0 V at a rate of 100 mV s−1. The detection of trichlorfon (as a model compound of OPPs) was done through the square wave voltammetry (SWV) technique (from 0.2 to 0.8 V; frequency, 15 Hz; potential increment, 0.004 V; amplitude, 0.025 V) in 0.1 mol L−1 PBS.For the detection of trichlorfon, the prepared CS/KbE/Au/cCNTs–cGR/GCE biosensor was employed. The current responses in PBS containing 0.8 mM α-NA were recorded before and after incubating the electrode with different concentrations of trichlorfon, respectively. The inhibition ratio (I%), which was taken as the sensing signal towards trichlorfon, was calculated as follows:
2.7 Preparation of real samples
Cabbage samples were processed as reported.30 50 g cabbage pieces were homogenized in 50 mL PBS (0.1 M, pH 6.5). The cabbage supernatant was centrifuged at 8000 rpm for 15 min and the supernatant was collected and used as the actual sample. For rice samples.31 The rice was first ground into powder, then 10 mg rice powder was mixed with 40 mL PBS (0.1 M, pH 6.5) and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The supernatant was diluted 100 times with PBS and used as the actual rice sample. Different amounts of pesticide were added to the actual samples to assess the potential practical applicability of the developed biosensor through recovery tests.
000 rpm for 30 min. The supernatant was diluted 100 times with PBS and used as the actual rice sample. Different amounts of pesticide were added to the actual samples to assess the potential practical applicability of the developed biosensor through recovery tests.
3 Results and discussion
3.1 Material characterizations
The prepared nanomaterials were characterized by SEM and TEM, respectively. The results are presented in Fig. 1. As shown in Fig. 1b, the cCNTs were well dispersed in the thin flexible cGR sheets. Also, the TEM image in Fig. 1c shows that the cCNTs were uniformly distributed on the transparent and crimple cGR sheets without entanglement. Furthermore, by comparing Fig. 1a and b, it can be found that the cCNTs were inserted between the cGR sheet layers as spacers, thus effectively preventing the irreversible stacking between GR sheets. The one-dimensional cCNTs and two-dimensional cGR formed a homogeneous three-dimensional network nanohybrid, as shown in Fig. 1b. It is highly expected that the excellent electrocatalytic activity and conductivity of CNTs as well as the intrinsic high surface area of GR will improve the electrochemical performance of GCE. Fig. 1d shows the TEM image of PDDA-Au. The particle size of AuNPs was about 12 nm. The inset is digital photos of them dispersed in water, and the solution was typical wine red.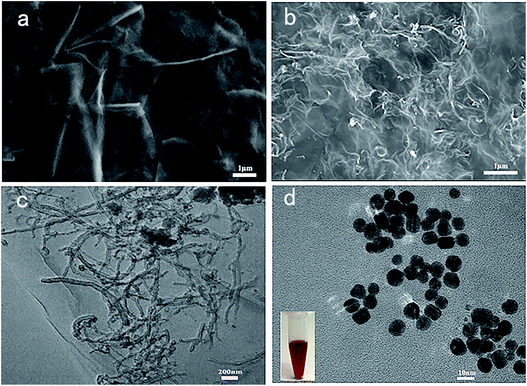 | ||
| Fig. 1 SEM images of cGR (a) and cCNTs–cGR (b); TEM images of cCNTs–cGR (c) and PDDA-Au (d) (inset: the photograph of PDDA-capped AuNPs). | ||
Fig. S1† shows the FT-IR spectra of the prepared cGR and cCNTs. For cGR (Fig. S2a†), the characteristic peaks at 1715 cm−1 and 3366 cm−1 are due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and O–H stretching vibrations of the carboxyl group (–COOH). 1382 cm−1 for O–H bending vibrations and 1056 cm−1 for C–O stretching of the alkoxy group are also observed. FTIR spectra of cCNTs (Fig. S2b†) also shows the characteristic absorption band at 1727 cm−1, corresponding to carbonyl stretching of carboxyl groups. The two weak bonds at 2800–2950 cm−1 are assigned to C–H stretching of cCNTs defects.32 The 1632 cm−1 band is due to the bending vibrations of water adsorbed between molecular layers.33 All above FTIR results confirm the presence of carboxyl groups in the prepared cGR and cCNTs.
O and O–H stretching vibrations of the carboxyl group (–COOH). 1382 cm−1 for O–H bending vibrations and 1056 cm−1 for C–O stretching of the alkoxy group are also observed. FTIR spectra of cCNTs (Fig. S2b†) also shows the characteristic absorption band at 1727 cm−1, corresponding to carbonyl stretching of carboxyl groups. The two weak bonds at 2800–2950 cm−1 are assigned to C–H stretching of cCNTs defects.32 The 1632 cm−1 band is due to the bending vibrations of water adsorbed between molecular layers.33 All above FTIR results confirm the presence of carboxyl groups in the prepared cGR and cCNTs.
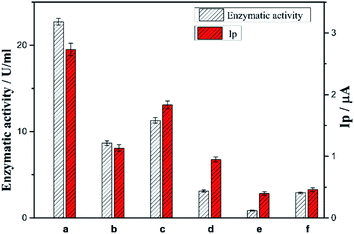
3.2 Selection of detection enzyme
As the critical biorecognition element, enzyme is an important factor affecting pesticide detection. Enzymes derived from different plants have different catalytic activities, which can specifically affect their sensitivity to the substrate, so the selection of enzyme sources was carried out first. Plant esterases from wheat, white kidney bean, wheat bran, myotonin, corn and millet were extracted using the method described above, and then their enzyme activities were determined separately. The results are shown in Fig. 2. It can be seen that the plant-derived esterases obtained from different plant sources have different enzyme activities. The activity of esterase is not only related to its content in the plant, but also to its amino acid composition, spatial conformation, and the essential groups of the enzymatic activity center and other factors. Among these plant-derived esterases, kidney bean esterase had the highest activity, while corn esterase had the lowest activity. Subsequently, these enzymes were modified on the electrode surface and their sensitivity towards the substrate α-NA were investigated. Fig. 2 also shows the electrochemical response of different plant-derived esterase to the same concentration of α-NA. It was found that enzyme with higher activity obtained more excellent electrochemical response signal to α-NA. Since the esterase extracted from kidney beans had the highest enzyme activity and electrochemical response, it was chosen as the enzyme for pesticide detection in the following study.The purity and Mw of the purified KbE were analysed by SDS-PAGE gel electrophoresis. The results are presented in Fig. 3a. It can be observed that many impurities were present in the crude enzyme (lane 2). After extraction by ATPS, most of the impurities were removed and only two clear bands were observed, indicating that the purification effect was good. Meanwhile, Mw of the purified KbE was between 25 and 35 kDa, which is consistent with the reported results.26 Besides, plant-derived esterase is protein and therefore has UV absorption properties. The UV absorption spectrum of purified enzyme solution is shown in Fig. 3b. An absorption peak was observed at 280 nm, which is the characteristic absorption peak of protein and is attributed to the absorption of light by tyrosine and tryptophan residues in protein molecules.
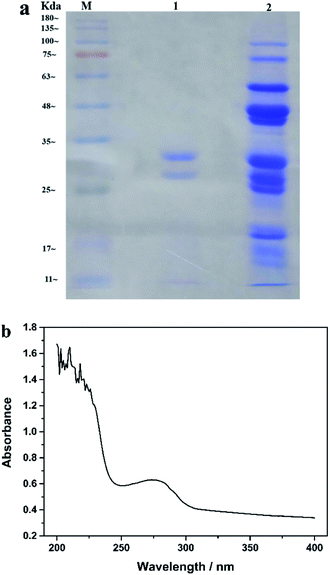 | ||
| Fig. 3 (a) SDS-PAGE images of different samples (M. marker; lane 1. Purified enzyme; lane 2. crude enzyme) (b) UV absorption spectroscopy of purified enzyme solution. | ||
3.3 Electrochemical responses of modified electrodes to α-NA
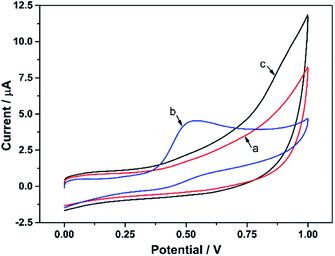
Fig. 4 shows the CV responses of the different modified electrodes in PBS with or without α-NA. For the electrode modified with KbE (CS/KbE/AuNPs/cCNTs–cGR/GCE), no redox peak appeared in blank PBS (curve a), but a significant oxidation peak appeared after the addition of α-NA (curve b). Meanwhile, the CS/AuNPs/cCNTs–cGR/GCE electrode without modified KbE showed no redox peak in PBS solution containing α-NA (curve c), indicating that the electrode modified material (cCNTs–cGR, AuNPs and CS) would not generate redox peaks in the scanning range. The above results demonstrate that KbE was successfully immobilized on the electrode and maintained its biocatalytic activity. The oxidation peak around 0.5 V was attributed to the oxidation of α-naphthalenol, an electroactive substance generated by the KbE-catalyzed α-NA hydrolysis reaction.83.4 Component effect of the KbE biosensor
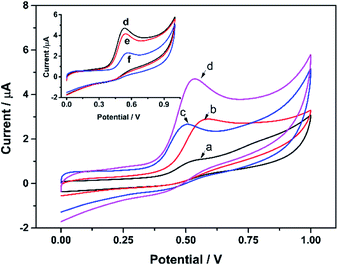
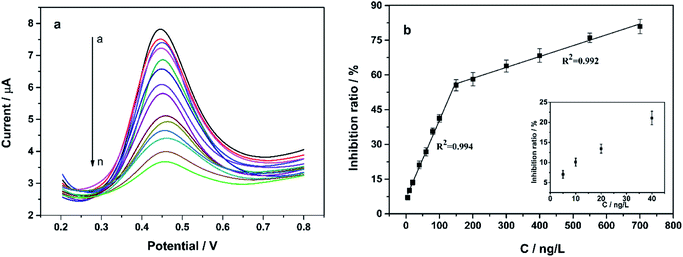
Fig. 5 shows the CV curves of different KbE modified electrodes in PBS containing α-NA. For CS/KbE/GCE, an irreversible peak at 0.52 V was observed owing to the oxidation of α-naphthalenol as described above (curve a). When KbE was immobilized on the cCNTs–cGR/GCE, the oxidation peak current (Ipa) increased significantly, the oxidation peak potential (Epa) shifted negatively and the peak shape was also improved (curve c). The improvement of the signal is due to the excellent conductivity and electrocatalytic activity of CNTs and GR.34 cCNTs–cGR nanocomposite with network structure formed a highly conductive electron pathway that can accelerate the electron transfer rate and thus increase the voltametric response of α-naphthalenol. Nano-Au is often used to immobilize enzymes in biosensors because of its excellent adsorption ability to protein. It can be found that PDDA-Au can also effectively promote response performance (curve c). This is because gold nanoparticles are also highly conductive and thus facilitate electron transfer. Besides, PDDA-Au has good biocompatibility,35,36 which allows KbE to maintain its biological activity well. The biggest Ipa was obtained on the CS/KbE/PDDA-Au/cCNTs–cGR/GCE (curve d), which suggests that cCNTs–cGR and PDDA-Au have a synergistic electrocatalytic effect and can significantly amplify the electrochemical oxidation signal of α-naphthalenol. The above experimental results suggest that the selected nanomaterials can not only maintain the catalytic activity of KbE, but also significantly enhance the response performance of KbE biosensors.Subsequently, the effect of OPPs on the biocatalytic activity of immobilized KbE was further investigated using trichlorfon as a model. As shown in the inset of Fig. 5, the Ipa of α-naphthalenol decreased after interaction with trichlorfon and higher concentrations of trichlorfon resulted in lower Ipa, which indicates a dose-dependent inhibition of KbE activity by trichlorfon. Accordingly, a new trichlorfon electrochemical sensing platform based on KbE can be established.
3.5 Optimization of experimental parameters
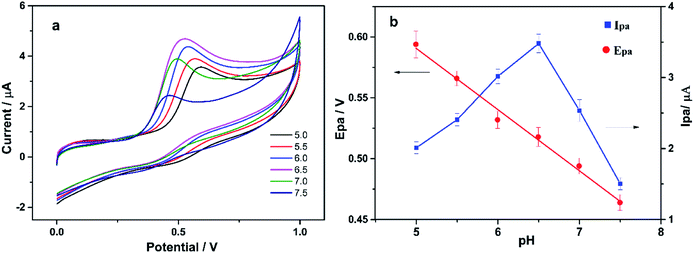 | ||
| Fig. 6 (a) CV curves of CS/KbE/PDDA-Au/cCNTs–cGR/GCE in PBS buffer containing 0.8 mM α-NA with different pH values; (b) the effect of pH value on Epa and Ipa, respectively. | ||
To further understand the electro-oxidation mechanism of α-naphthalenol on the KbE modified electrode, the effect of scan rate (ν) on Ipa and Epa was investigated. The results are displayed in Fig. S2.† As the scan rate (ν) increased, Ipa also increased and Epa shifted to the positive direction. In the range of 0.05 to 0.35 V s−1, Ipa was linearly related to the square root of the scan rate (ν1/2), and the linear regression equation was Ipa (μA) = 1.562 + 4.698v1/2 (R2 = 0.996), indicating a diffusion-controlled process. Also, the linear relationship between Epa and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v was Epa (V) = 0.610 + 0.035ln
v was Epa (V) = 0.610 + 0.035ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v (R2 = 0.990). The electron number transferred (nα) can be calculated using the following equation:
v (R2 = 0.990). The electron number transferred (nα) can be calculated using the following equation:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
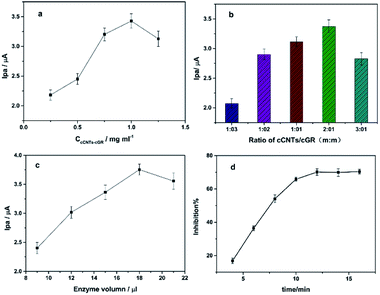
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Therefore, 6 μL of 1 mg mL−1 cCNT–cGR solution with a mass ratio of 2
1. Therefore, 6 μL of 1 mg mL−1 cCNT–cGR solution with a mass ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (cCNTs
1 (cCNTs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cGR) was chosen for the subsequent experiments.
cGR) was chosen for the subsequent experiments.
Incubation time with pesticide is a key experimental parameter for pesticide detection. The change of peak current before and after incubation was investigated by SWV. It was found that the peak current decreased after incubation with trichlorfon, indicating inhibition of enzyme activity. Fig. 7d shows that the inhibition rate increased with the increase of incubation time and reached basic stability at 12 min, indicating the binding of pesticide with active target groups in KbE reached a basic equilibrium. Therefore, 12 minutes was chosen as the optimal incubation time.
3.6 Detection of trichlorfon
Inhibition measurements were performed under the optimized variables, the results showed that the biosensor's response to α-NA decreased after interaction with trichlorfon (Fig. 8). This is because trichlorfon inhibited the activity of KbE, thus reduced the amount of α-naphthalenol produced by catalytic hydrolysis of α-NA by KbE. Trichlorfon showed concentration-dependent inhibition towards KbE activity. The inhibition ratio (I%) was proportional to the concentrations of trichlorfon from 5 to 150 ng L−1 and from 150 to 700 ng L−1. The linear equations were I% = 0.336C (ng L−1) + 6.855 (R2 = 0.994) from 5 to 150 ng L−1 and I% = 0.047C (ng L−1) + 49.054 (R2 = 0.992) from 150 to 700 ng L−1, respectively. In the high concentration interval, the activity of KbE was severely inhibited, and the structure and active center of the enzyme changed greatly, with fewer active sites that could work effectively, leading to a decrease in the response sensitivity and thus showing a lower slope. The same phenomenon of decreased sensitivity in high concentration interval has also been observed in many AChE-based electrosensing studies for pesticides.30,37 The limit of detection (LOD) is an important index to evaluate the sensitivity, which can be calculated by the following equation.38–40| LOD = kSb/S |
A comparison of analytical characteristics of the herein biosensor with previous reported methods for the detection of trichlorfon was listed in Tables S1 and S2.† Notably, the present CS/KbE/AuNPs/cCNTs–cGR/GCE exhibited comparable or lower detection limit. Such a high sensitivity can be attributed to several reasons. (1) The good synergistic electrocatalysis of AuNPs@cCNTs–cGR, which can effectively accelerate the electron transfer rate and thus amplify the voltammetric response signal. (2) The good biocompatibility of AuNPs, cCNTs–cGR and CS, which allows the catalytic activity of KbE to be well maintained. (3) High sensitivity of KbE to pesticides. Previous studies have reported that plant-derived esterases are highly sensitive to pesticides.26,41 The above experimental results demonstrate that highly sensitive detection of OPPs with plant-derived esterase is feasible.
3.7 Reproducibility and stability
The intra-batch reproducibility was evaluated by the RSD of the sensor response for five consecutive measurements in 0.8 mM α-NA with the same electrode. Similarly, the inter-batch reproducibility was assessed on five independently prepared electrodes. The RSDs were found to be 3.6% and 5.2%, respectively, indicating an acceptable operational reproducibility and fabrication reproducibility. When CS/KbE/AuNPs/cCNTs–cGR/GCE was not in use, it was stored at 4 °C in dry conditions. No apparent decrease in the current response for α-NA was observed in the first 5 day storage. After 15 days of storage, the sensor response still maintained 84.5% of the original current response, suggesting the storage stability of CS/KbE/AuNPs/cCNTs–cGR/GCE is acceptable.3.8 Interferences study
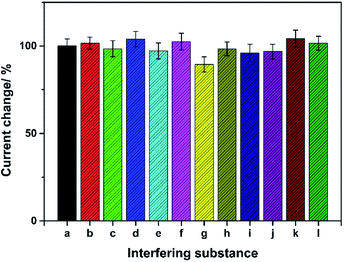
Interference studies were conducted to investigate the feasibility of the sensor for OPPs detection. Fig. 9 compares the signal change after incubation with 400 ng L−1 trichlorfon in the presence or absence of different interfering species. The results showed that when a 200-fold contents of K+, Na+, Fe3+, Cu2+, Zn2+, Cl−, SO42−, NO3−, citric acid, glucose coexisted with 400 ng L−1 of trichlorfon, the current signals did not change obviously, suggesting the good anti-interference ability. However, when 200-fold concentration of Pb2+ coexisted with 400 ng L−1 trichlorfon, the response signal decreased to 89.4% of the original.3.9 Practical application
To study the applicability of the proposed method, CS/KbE/AuNPs/cCNTs–cGR/GCE was finally applied for the detection of trichlorfon in rice and cabbage samples. The results show that trichlorfon was not detected in the original samples. Thus, these samples could be employed as blank samples and then these samples were treated by spiking with trichlorfon standards. Recovery results and the RSDs were listed in Table 1, demonstrated satisfactory data with recoveries ranging from 94.05% to 106.76% and RSD less than 5.16% (n = 3). The results clearly confirmed that the proposed CS/KbE/Au/cCNTs–cGR/GCE biosensor was highly accurate. Therefore, the plant-derived KbE is available for highly sensitive and accurate determination of OPPs.| Samples | Added (μg L−1) | Founded (μg L−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Cabbage | 40 | 37.62 | 94.05 | 4.51 |
| 80 | 85.09 | 106.37 | 4.82 | |
| 300 | 316.65 | 105.55 | 4.38 | |
| 600 | 628.26 | 104.71 | 3.83 | |
| Rice | 40 | 42.70 | 106.76 | 5.16 |
| 80 | 77.67 | 97.09 | 3.46 | |
| 300 | 284.19 | 94.73 | 4.33 | |
| 600 | 613.38 | 102.23 | 4.08 |
4 Conclusions
The enzyme is the core component for rapid determination of pesticide by enzymatic inhibition method. Enzymes used for pesticide detection should be sensitive, readily available and stable. Therefore, a novel biosensing electrode (CS/KbE/AuNPs/cCNTs–cGR/GCE) was constructed by using plant-derived KbE as a new detection enzyme. On the basis of the inhibition of KbE catalytic activity, complemented by the synergistic electrocatalytic signal amplification effect of carbon nanomaterials and nanogold, the determination of trace trichlorfon can be easily achieved with a detection limit as low as 3 ng L−1 (S/N = 3). It was the first time for KbE to be used as an enzyme source to develop a highly sensitive and low-cost electrochemical method for pesticide's detection. Compared with the large-size instrumental analysis method, the proposed electrochemical detection method has the advantages of simplicity, sensitivity and simple sample pre-treatment. This study also provides valuable theoretical and technical information for the practical application of plant-derived esterase in the electrochemical rapid detection technology for large-scale screening of pesticides in the field.Conflicts of interest
The authors declare they have no conflicts of interest.Acknowledgements
This work was financially supported by the Science and Technology Support Program of Guizhou Province (LH[2017]7294, [2019]2382), the National Natural Science Foundation of China (32160603, 31760486).References
- J. Yao, Z. Wang, L. Guo, X. Xu, L. Liu, L. Xu, S. Song, C. Xu and H. Kuang, TrAC, Trends Anal. Chem., 2020, 131, 116022 CrossRef CAS.
- E. F. S. European Food Safety Authority, EFSA J., 2019, 17, 5743 Search PubMed.
- P. Medina-Pastor, G. Triacchini and A. European Food Safety, EFSA J., 2020, 18, 6057 Search PubMed.
- Z. Li, J. Sun and L. Zhu, Sci. Total Environ., 2021, 765, 142757 CrossRef CAS PubMed.
- A. S. Tsagkaris, J. Pulkrabova and J. Hajslova, Foods, 2021, 10, 88 CrossRef CAS PubMed.
- M. M. Lasram, I. B. Dhouib, A. Annabi, S. El Fazaa and N. Gharbi, Toxicology, 2014, 322, 1–13 CrossRef CAS PubMed.
- J. Butler-Dawson, K. Galvin, P. S. Thorne and D. S. Rohlman, Neurotoxicology, 2016, 53, 165–172 CrossRef CAS PubMed.
- J. Cao, M. Wang, H. Yu, Y. She, Z. Cao, J. Ye, A. M. Abd El-Aty, A. Hacimuftuoglu, J. Wang and S. Lao, J. Agric. Food Chem., 2020, 68, 7298–7315 CrossRef CAS PubMed.
- D. Ibanez, M. Begona Gonzalez-Garcia, D. Hernandez-Santos and P. Fanjul-Bolado, Spectrochim. Acta, Part A, 2021, 248, 119174 CrossRef CAS PubMed.
- W. A. Collimore and G.-A. Bent, Environ. Monit. Assess., 2020, 192, 128 CrossRef CAS PubMed.
- T. Gao, J. Wang, Q. Wu, C. Wang and Z. Wang, Food Anal. Methods, 2020, 13, 690–698 CrossRef.
- S. Moinfar, L. A. Jamil, H. Z. Sami and S. Ataei, J. Food Compos. Anal., 2021, 95, 103695 CrossRef CAS.
- J. Zhan, R.-r. Zhang, X.-z. Shi, Z. Huang, G.-z. Cao, X.-f. Chen and L. Hu, J. Chromatogr., 2021, 1636, 461794 CrossRef CAS PubMed.
- J. Regueiro, O. Lopez-Fernandez, R. Rial-Otero, B. Cancho-Grande and J. Simal-Gandara, Crit. Rev. Food Sci. Nutr., 2015, 55, 839–863 CrossRef CAS PubMed.
- L. F. de Lima, A. L. Ferreira, C. C. Maciel, M. Ferreira and W. R. de Araujo, Talanta, 2021, 227, 122200 CrossRef CAS PubMed.
- K. S. Ranjith, A. T. E. Vilian, S. M. Ghoreishian, R. Umapathi, Y. S. Huh and Y. K. Han, Sens. Actuators, B, 2021, 344, 130202 CrossRef CAS.
- C. E. Guo, C. X. Wang, H. Y. Sun, D. M. Dai and H. T. Gao, RSC Adv., 2021, 11, 29590–29597 RSC.
- Q. W. Bao, G. Li, Z. C. Yang, P. Pan, J. Liu, R. R. Li, J. Wei, W. Hu, W. B. Cheng and L. Lin, Analyst, 2021, 146, 5610–5618 RSC.
- Y. Liu, X. Cao, Z. Liu, L. Sun, G. Fang, J. Liu and S. Wang, Analyst, 2020, 145, 8068–8076 RSC.
- X. Xie, B. Zhou, Y. Zhang, G. Zhao and B. Zhao, Chem. Phys. Lett., 2021, 767, 138355 CrossRef CAS.
- J. S. Van Dyk and B. Pletschke, Chemosphere, 2011, 82, 291–307 CrossRef CAS PubMed.
- K. A. Askar, A. C. Kudi and A. J. Moody, Appl. Biochem. Biotechnol., 2011, 165, 336–346 CrossRef CAS PubMed.
- L. Yang, D. Huo, C. Hou, K. He, F. Lv, H. Fa and X. Luo, Process Biochem., 2010, 45, 1664–1671 CrossRef CAS.
- C.-j. Hou, K. He, L.-m. Yang, D.-q. Huo, M. Yang, S. Huang, L. Zhang and C.-h. Shen, World J. Microbiol. Biotechnol., 2012, 28, 541–548 CrossRef CAS PubMed.
- J.-l. Wang, Q. Xia, A.-p. Zhang, X.-y. Hu and C.-m. Lin, J. Zhejiang Univ., Sci., B, 2012, 13, 267–273 CrossRef CAS PubMed.
- X. Yang, J. Dai, S. Zhao, R. Li, T. Goulette, X. Chen and H. Xiao, J. Sci. Food Agric., 2018, 98, 5095–5104 CrossRef CAS PubMed.
- K. Das, A. Uppal and R. K. Saini, Spectrochim. Acta, Part A, 2016, 152, 378–383 CrossRef CAS PubMed.
- Q. Huang, S. Liu, K. Li, I. Hussain, F. Yao and G. Fu, J. Mater. Sci. Technol., 2017, 33, 821–826 CrossRef CAS.
- D. Song, J. Xia, F. Zhang, S. Bi, W. Xiang, Z. Wang, L. Xia, Y. Xia, Y. Li and L. Xia, Sens. Actuators, B, 2015, 206, 111–118 CrossRef CAS.
- H.-F. Cui, W.-W. Wu, M.-M. Li, X. Song, Y. Lv and T.-T. Zhang, Biosens. Bioelectron., 2018, 99, 223–229 CrossRef CAS PubMed.
- N. Nesakumar, S. Sethuraman, U. M. Krishnan and J. B. B. Rayappan, Biosens. Bioelectron., 2016, 77, 1070–1077 CrossRef CAS PubMed.
- H. Tahermansouri and E. Biazar, New Carbon Mater., 2013, 28, 199–207 CrossRef CAS.
- X. Wang, M. Yang, Q. Liu, S. Yang, X. Geng, Y. Yang, H. Fa, Y. Wang and C. Hou, Anal. Sci., 2019, 35, 441–448 CrossRef CAS PubMed.
- G. Xing, B. Luo, J. Qin, X. Wang, P. Hou, H. Zhang, C. Wang, J. Wang and A. Li, Talanta, 2021, 232, 122477 CrossRef CAS PubMed.
- R. R. Liu, L. T. Song, Y. J. Meng, M. Zhu and H. L. Zhai, J. Phys. Chem. B, 2019, 123, 7570–7577 CrossRef CAS PubMed.
- C. Moonla, C. Nontapha, T. Ouiram, A. Preechaworapun and T. Tangkuaram, Electroanalysis, 2019, 31, 1605–1614 CrossRef CAS.
- H. D. Cancar, S. Soylemez, Y. Akpinar, M. Kesik, S. Goker, G. Gunbas, M. Volkan and L. Toppare, ACS Appl. Mater. Interfaces, 2016, 8, 8058–8067 CrossRef PubMed.
- Y. Liu, X. Hu, Y. Xia, F. Zhao and B. Zeng, Anal. Chim. Acta, 1800, 1190, 339245 CrossRef PubMed.
- T. S. K. Sharma and K.-Y. Hwa, J. Hazard. Mater., 2021, 410, 124659 CrossRef CAS PubMed.
- X.-P. Wei, R.-Q. Zhang, L.-B. Wang, Y.-L. Luo, F. Xu and Y.-S. Chen, J. Mater. Chem. C, 2019, 7, 119–132 RSC.
- L. Dong, Y. Ren, J. J. Li, H. X. Wu, C. J. Hou, H. B. Fa, M. Yang, S. Y. Zhang and D. Q. Huo, J. Appl. Spectrosc., 2018, 85, 535–542 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: FTIR spectra, effect of scan rate, comparison of methods. See DOI: 10.1039/d1ra08129b |
| This journal is © The Royal Society of Chemistry 2022 |