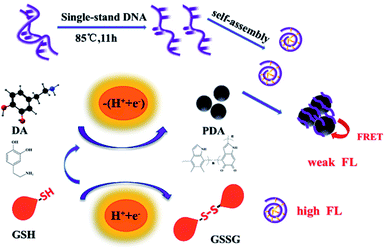
Open Access Article
Open Access ArticleTurn-on signal fluorescence sensor based on DNA derived bio-dots/polydopamine nanoparticles for the detection of glutathione†
Xiaoxiao Chen ab,
Pu Liab,
Gaojun Wuab,
Zhe Wangab and
Chaobiao Huang
ab,
Pu Liab,
Gaojun Wuab,
Zhe Wangab and
Chaobiao Huang *ab
*ab
aXingzhi College, Zhejiang Normal University, Lanxi 321100, China. E-mail: hcb@zjnu.cn
bCollege of Chemistry and Life Science, Zhejiang Normal University, Jinhua 321004, China
First published on 12th January 2022
Abstract
A convenient, fast, sensitive and highly selective fluorescence sensor for the detection of glutathione (GSH) based on DNA derived bio-dots (DNA bio-dots)/polydopamine (PDA) nanoparticles was constructed. The fluorescent switch of DNA bio-dots was induced to turn off because of fluorescence resonance energy transfer (FRET) reactions between DNA bio-dots and PDA. The presence of GSH blocked the spontaneous oxidative polymerization of dopamine (DA) to PDA, leading the fluorescent switch of DNA bio-dots to be “turned on”. The degree of fluorescence recovery of DNA bio-dots is linearly correlated with the concentration of GSH within the range of 1.00–100 μmol L−1, and the limit of detection (LOD) is 0.31 μmol L−1 (S/N = 3, n = 9). Furthermore, the fluorescence sensor was successfully used to quantify GSH in human urine and glutathione whitening power, indicating the fluorescence sensor has potential in the detection of human body fluids and pharmaceutical preparations.
1. Instruction
Glutathione (GSH) is the most abundant sulfhydryl compound in biological cells, and is a small molecular peptide composed of three amino acids (L-glutamate, L-cysteine and glycine). At the same time, as the most important antioxidant in the body, it can effectively protect proteins, DNA and other biomolecules in cells from loss by oxidation.1 GSH in cells is converted from reduced glutathione to oxidized glutathione (GSSG) when biological tissues are stimulated by oxidation. This results in altered levels of oxidized and reduced glutathione in the body, which affects cell growth and function.2 Abnormal levels of GSH and GSSG are associated with many clinical conditions, such as Alzheimer's disease, liver damage, and heart disease.3,4 Therefore, GSH is widely used in the pharmaceutical, cosmetics and other industries. Based on its important biological functions, the measurement of its internal parameters is greatly important for health diagnosis.The methods currently used for the detection of GSH are colorimetry,5–7 electrochemical analysis,8–10 high performance liquid chromatography11,12 and fluorescence analysis.13–15 Among these analytical methods, the fluorescence analysis method overcomes the defects of other methods such as complex operation, high equipment requirements, poor sensitivity and selectivity, and being time-consuming, which makes it more promising for detecting GSH. Therefore, it is of great significance to design a simple and efficient fluorescence analysis strategy for the detection of GSH.
DNA bio-dots, as a new luminescent material, has attracted much attention due to its low cytotoxicity, excellent dispersion, stable fluorescence and good biocompatibility.16,17 It has been proved that DNA bio-dots made from cytosine-rich DNA forms sp2 carbon sample centers through the accumulation of aromatic cytosine base pairs.3,18,19 Not only does it have a luminescent behavior similar to carbon quantum dots (CQDs), but it also owns a DNA-like structure.18,20 These characteristics make it easier to extend to the construction of fluorescent biosensors.
Polydopamine (PDA), as a new type of nanomaterial which is polymerized by several dopamine molecules, exhibits good biocompatibility and dispersion.19,21 In addition, PDA has a wide band absorbance in the UV-vis spectrum, which makes it a widely used fluorescent quenching agent.22 Studies have pointed out that the spontaneous oxidative polymerization from DA to PDA can be effectively inhibited in the presence of reductive substances.23
Hence, we developed a fluorescent sensor based on the FRET between DNA bio-dots and PDA for detecting GSH. As shown in Scheme 1, single-stranded DNA was produced by simply hydrothermal treatment of deoxyribonucleic acid (from salmon sperm). Then the aromatic cytosine bases in the single strand of DNA can spontaneously accumulate to form the sp2 carbon-like centers. The highly hydrophilic PO41− group on the skeleton was located outside the center of the sp2 type carbon and wrapped them to form shaped DNA bio-dots. DA can spontaneously oxidize and polymerize to form PDA. Then DNA bio-dots deposited on the surface of PDA, which may be caused by “π–π” stacking.19 Since there was the fluorescence resonance energy transfer between PDA and DNA bio-dots, the fluorescence state of DNA bio-dots (as donor) was turned off by PDA (as acceptor). And the fluorescence quenching mechanism between PDA and DNA bio-dots was proved to follow dynamic quenching. In the presence of GSH, the fluorescence state of DNA bio-dots would be “turned on”. It was because that the mercaptan group of GSH would donate the reduction equivalent (H+ + e−) to unstable radical intermediates (such as dopamine) and reversible intermediates, in other words, the presence of GSH can effectively inhibit the formation of PDA.
2. Experiment
2.1. Material and reagents
Glutathione (GSH), Alanine (Ala), Histidine (His), Arginine (Arg), Valine (Val), Glycine (Gly), Tryptophan (Try), Ornithine (Orn), Methionine (Met), Dithiothreitol (DTT), 3-mercaptopropionic acid (MPA), glucose (C6H12O6), dopamine (DA) and salmon extract low molecular weight double stranded DNA were purchased from Aladdin Co., Ltd. (Shanghai, China). Glutathione whitening powder was purchased from Xinrui Biotechnology Co., Ltd. (Jiangsu, China). The water used in experiments is ultrapure water.2.2. Apparatus
The FT-IR spectra was recorded on Nexus 670 infrared spectrometer (Reonicolai Inc., New York, USA). The UV-vis absorption spectrum was obtained with Lambda 950 spectrophotometer (PerkinElmer Inc., Waltham, USA). The morphological evaluations was characterized by JEM-2100F transmission electron microscopy (JEOL, Tokyo, Japan). Fluorescence spectra was recorded using RF-6000 Fluorescence Spectrometer (Shimadzu, Japan).2.3. Synthesis of DNA bio-dots
The synthesis of DNA bio-dots was modified as previous report.24 Salmon sodium deoxyribonucleic acid was dissolved in ultrapure water which had been removed oxygen through N2 for 15 min to obtain the homogeneous DNA solution with a concentration of 1.0 mg mL−1. Then the DNA solution was refluxed at 85 °C for 11 h with the protection of N2. At the end, it was quickly added to the same volume of acetone being sonicated. After ultrasonic treatment for 1 h, the mixed solution was removed acetone by rotary evaporation. The resulting solution was filtered through a 0.22 μm membrane to obtain the DNA bio-dots aqueous solution and kept at 4 °C for further use.2.4. Fluorescence quenching investigation
To explore the fluorescence quenching efficiency of PDA to DNA bio-dots, different amounts of DA were added to 200 μL DNA bio-dots and diluted to 2 mL by Tris–HCl buffer (50 mmol L−1, pH 8.5), where the final concentration of DA was from 0.00 to 20.0 μmol L−1. These mixtures were detected by fluorescence spectroscopy after incubated at room temperature for 20 min. In addition, different concentrations of GSH were added into the above fluorescence quenching system and determined the fluorescence spectrum to explore the anti-quenching effect of GSH.2.5. Detection of GSH
DNA bio-dots (200 μL) was placed in a centrifuge tube and mixed with a series of GSH aqueous solution at different concentrations. Then DA solution (5.0 mmol L−1) was added to the mixture, diluting with Tris–HCl buffer (50 mmol L−1, pH 8.5) to 2 mL (where the final concentration of GSH were 0.00, 0.10, 0.50, 1.00, 2.00, 4.00, 6.00, 10.0, 14.0, 16.0, 20.0, 30.0, 50.0, 70.0, 100, 130 and 150 μmol L−1). After 20 min, the final mixed solution was measured under the excitation wavelength of 338 nm.3. Results and discussion
3.1. Characterization
The morphology of DNA bio-dots, PDA and DNA bio-dots@PDA was evaluated by transmission electron microscope (TEM). As shown in Fig. 1a–c, DNA bio-dots, PDA and DNA bio-dots@PDA were all spherical in shape and displayed wonderful dispersion. According to the results of particle size calculation, the particle size of DNA bio-dots (Fig. 1d), PDA (Fig. 1e) and DNA bio-dots@PDA (Fig. 1f) was 2.5, 10.2 and14.4 nm, respectively. Compared with the particle size of DNA bio-dots and PDA, the particle size of DNA bio-dots@PDA had slightly increased which may indicated that DNA bio-dots had been assembled on the surface of PDA.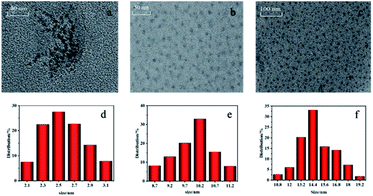 | ||
| Fig. 1 TEM and particle size distribution images of DNA bio-dots (a, d), PDA (b, e) and DNA bio-dots@PDA (c, f). | ||
The surface chemical state of DNA bio-dots was analyzed by XPS. According to the XPS spectrum in Fig. 2, there were four prominent peaks of C 1s, N 1s, O 1s and P 2p at 284.49, 405.46, 532.89 and 133.56 eV.16,17,24 Among them, the four peaks at 284.83, 286.17, 287.62 and 286.65 eV obtained by C 1s peak division fitting (Fig. 2b) corresponded to C–C, C–N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, respectively.25–28 The high resolution N 1s spectrum (Fig. 2c) performed two characteristic peaks at 399.93 and 398.57 eV, which was attributed to
O and C–O, respectively.25–28 The high resolution N 1s spectrum (Fig. 2c) performed two characteristic peaks at 399.93 and 398.57 eV, which was attributed to ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– and –NH2.24 The high-resolution XPS spectra of O 1s (Fig. 2d) exhibited four peaks at 530.93, 531.98, 532.56 and 533.08 eV, which was assigned to PO41−, C
N– and –NH2.24 The high-resolution XPS spectra of O 1s (Fig. 2d) exhibited four peaks at 530.93, 531.98, 532.56 and 533.08 eV, which was assigned to PO41−, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, O–N and C–O.17,29 These results indicated that DNA bio-dots was rich in nitrogenous and oxygen-containing groups and owned the original phosphate skeleton of DNA.
O, O–N and C–O.17,29 These results indicated that DNA bio-dots was rich in nitrogenous and oxygen-containing groups and owned the original phosphate skeleton of DNA.
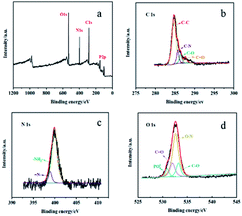 | ||
| Fig. 2 XPS spectrum of DNA bio-dots (a) and high-resolution XPS spectra of C 1s (b), N 1s (c), and O 1s (d). | ||
In addition, the FI-IR spectrum of DNA bio-dots further verified that its surface was rich in nitrogenous and oxygen-containing groups. As shown in Fig. S1b,† the broad peak from 2500 to 3750 cm−1 testified the presence of –COOH, –OH and –NH2 groups.20,30,31 The characteristic absorption peaks at 1720 cm−1 was caused by C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration.32,33 The peaks at 1430 and 1367cm−1 were attributed to C–O deformation vibration and C–OH tensile vibration.25,29,34 The peak at 1030 cm−1 was attributed to the C–N in-plane stretching vibration of amine groups.16,35 The obvious difference between the spectra of DNA bio-dots (Fig. S1b†) and the spectra of double-stranded DNA (Fig. S1a†) further revealed the successful preparation of DNA bio-dots.24 The fluorescence characteristics of DNA-bio-dots was also explored. Fig. S2A† showed that the optimal excitation wavelength (curve a) of DNA-bio-dots was 338 nm and the optimal emission wavelength (curve b) of that was 425 nm. At the same time, the ultraviolet absorption peak (curve d) of DNA-bio-dots was located near 321 nm as seen from Fig. S2B.† Then the fluorescence quantum yield of DNA-bio-dots was calculated to be about 10.6% using quinine sulfate (0.1 mol L−1 H2SO4) as the standard material. Besides, it is easy to find that the narrow emission spectra of DNA bio-dots showed a good spectral overlap with the broad absorption of PDA (curve c), indicating the FRET between PDA and DNA bio-dots. And the fact that the fluorescence lifetime of DNA bio-dots in the presence of PDA was significantly shorter than that in the absence of PDA (Fig. 3) confirmed that the existence of PDA can attenuate the fluorescence lifetime of DNA bio-dots, resulting the fluorescence quenching of DNA bio-dots. Therefore, the fluorescence quenching of DNA bio-dots by PDA followed the dynamic quenching mechanism. Finally, in order to further investigate whether DNA-bio-dots@PDA was successfully assembled, we studied the FI-IR spectroscopy of PDA (curve a), DNA bio-dots@PDA (curve b) and DA (curve c) as Fig. S3† shown. It can be found that the weak absorption peaks of C–H bond stretching vibration near 2966 and 2896 cm−1 could be observed in curve a and b, and the absorption intensity of these peaks in curve b was stronger than that of curve a. The absorption peaks at 1700 and 500 cm−1 vastly changed for curves a and b, which were significantly different from that of curve c. These results confirmed the disappearance of fine structure of DA and the production of PDA and DNA bio-dots@PDA.36
O stretching vibration.32,33 The peaks at 1430 and 1367cm−1 were attributed to C–O deformation vibration and C–OH tensile vibration.25,29,34 The peak at 1030 cm−1 was attributed to the C–N in-plane stretching vibration of amine groups.16,35 The obvious difference between the spectra of DNA bio-dots (Fig. S1b†) and the spectra of double-stranded DNA (Fig. S1a†) further revealed the successful preparation of DNA bio-dots.24 The fluorescence characteristics of DNA-bio-dots was also explored. Fig. S2A† showed that the optimal excitation wavelength (curve a) of DNA-bio-dots was 338 nm and the optimal emission wavelength (curve b) of that was 425 nm. At the same time, the ultraviolet absorption peak (curve d) of DNA-bio-dots was located near 321 nm as seen from Fig. S2B.† Then the fluorescence quantum yield of DNA-bio-dots was calculated to be about 10.6% using quinine sulfate (0.1 mol L−1 H2SO4) as the standard material. Besides, it is easy to find that the narrow emission spectra of DNA bio-dots showed a good spectral overlap with the broad absorption of PDA (curve c), indicating the FRET between PDA and DNA bio-dots. And the fact that the fluorescence lifetime of DNA bio-dots in the presence of PDA was significantly shorter than that in the absence of PDA (Fig. 3) confirmed that the existence of PDA can attenuate the fluorescence lifetime of DNA bio-dots, resulting the fluorescence quenching of DNA bio-dots. Therefore, the fluorescence quenching of DNA bio-dots by PDA followed the dynamic quenching mechanism. Finally, in order to further investigate whether DNA-bio-dots@PDA was successfully assembled, we studied the FI-IR spectroscopy of PDA (curve a), DNA bio-dots@PDA (curve b) and DA (curve c) as Fig. S3† shown. It can be found that the weak absorption peaks of C–H bond stretching vibration near 2966 and 2896 cm−1 could be observed in curve a and b, and the absorption intensity of these peaks in curve b was stronger than that of curve a. The absorption peaks at 1700 and 500 cm−1 vastly changed for curves a and b, which were significantly different from that of curve c. These results confirmed the disappearance of fine structure of DA and the production of PDA and DNA bio-dots@PDA.36
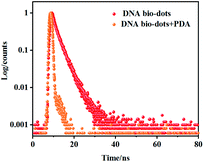 | ||
| Fig. 3 The luminescence time decay resolution curves of DNA bio-dots in the absence and presence of PDA. | ||
3.2. Optimum proposal
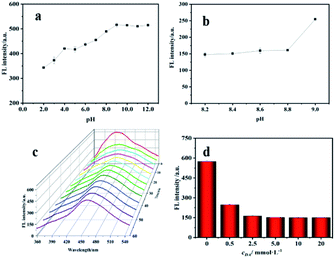
The pH, incubation time and DA concentration for fluorescence quenching reaction were optimized. Fig. 4a revealed that the fluorescence intensity of DNA bio-dots increased with the increase of pH, and the intensity remained basically stable when pH exceeded 9.0. This could be explained by that the negative charge on the surface of DNA bio-dots would combine with hydrogen ions, resulting the accumulation of DNA bio-dots and thus leading to fluorescence quenching when the solution was acidic,37 and the existence of amino groups on the surface of DNA bio-dots would make fluorescence intensity remain relatively stable under alkaline condition.35 Based on what studies have pointed that the spontaneous oxidation polymerization efficiency of DA performed best around the alkaline condition with pH 8.5,38 we selected the pH in the range of 8.0–9.0 to investigate the optimum pH for fluorescence quenching of DNA bio-dots by PDA. DNA bio-dots (200 μL) was mixed with DA (200 μL, 2.5 mmol L−1) and then diluted to 2 mL with different pH value of Tris–HCl buffer. The fluorescence intensity of these mixtures was detected after incubated for 30 min. As shown in Fig. 4b, the fluorescence intensity performed little difference as the pH ranged from 8.2 to 8.8. In addition, considering that the optimal fluorescence emission of DNA bio-dots was around pH 9.0, we finally selected pH 8.5 as the final optimal pH for subsequent experiments. As shown in Fig. 4c, the fluorescence intensity decreased gradually with the extension of reaction time and reached the quenching equilibrium after incubated 20 min. Thus, 20 min was chosen as the optimum reaction time for further experiments. As seen from Fig. 4d, when the concentration of DA increased from 5.0 mmol L−1 to 20 mmol L−1, the fluorescence intensity remained unchanged. Therefore, 5.0 mmol L−1 was considered to be the optimum concentration of DA for further study.3.3. Determination of GSH
Next, we investigated the performance of the fluorescence sensor for detecting GSH under the optimal conditions. As shown in Fig. 5a, the fluorescence intensity of sensing system gradually recovered with the increase of GSH concentration. Fig. 5b showed a positive linear relationship between (F − F0)/F0 and the concentration of GSH when the concentration of GSH was 1.00–100 μmol L−1, the linear equation is (F − F0)/F0 = 0.0353cGSH + 0.0568 (R2 = 0.9943) (where F and F0 are the fluorescence intensity in the presence and absence of GSH), and the LOD was 0.31 μmol L−1. The performance of the fluorescence analysis method was contrasted with many other analysis methods for GSH that were previously reported (Table 1). Compared with HPLC, the LOD of this method was comparable to or slightly worse. But this method overcomes the shortcomings such as laborious time, cumbersome technology and complex processing procedures. In contrast, from the perspective of simplicity and convenience, this method has more potential for rapid detection of GSH which can provide reference for the preliminary diagnosis of related diseases or the detection of glutathione content in pharmaceutical preparations. In addition, compared with others methods, the fluorescence sensor has a lower LOD, reveling its potential for detecting GSH.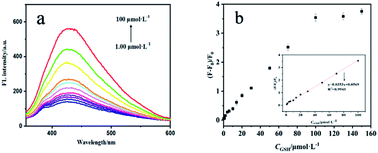 | ||
| Fig. 5 Fluorescence spectra of sensing system in the presence of different concentrations of GSH (a). Linear calibration curve for detection of GSH (b). | ||
| Methods | Linear range (μmol L−1) | LOD (μmol L−1) | Ref. |
|---|---|---|---|
| a EL: electrochemical analysis, HPLC: high performance liquid chromatography, FL: fluorescence analysis. | |||
| EL | 2.00–200.0 | 0.34 | 36 |
10.0–500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.73 | 39 | |
| HPLC | 0.10–4000 | 0.34 | 40 |
| 0.25–10.0 | 0.10 | 41 | |
| Colorimetry | 2.00–200 | 0.67 | 42 |
| 5.00–75.0 | 1.20 | 43 | |
| FL | 50.0–400 | 7.83 | 44 |
| 1.00–100 | 0.31 | This work | |
3.4. Selectivity for sensor
To test the sensitivity of the fluorescence sensor to GSH, some analogues that have similar structures or properties to GSH (C6H12O6, DTT, MPA, Ala, His, Arg, Val, Gly, Try, Orn and Met) were selected for the interference study. The concentration of analogues was 100 μmol L−1 which was 10 times than that of GSH. As shown in Fig. 6, the fluorescent sensor exhibits good selectivity against the selected possible interference, proving the feasibility of the proposed DNA bio-dots-based fluorescence sensor.3.5. Detection of GSH and recovery
The fluorescence sensor was used to detect GSH in human urine and glutathione whitening power. The urine sample was pretreated by simple centrifugation and the supernatant was filtered by 0.22 μm porous membrane for fluorescence detection. Furthermore, in order to exclude the possible interference of some oxidants in urine sample, we set up a controlled experiment to research the effect of urine sample on fluorescence quenching before the experiments. Taking two equal volumes of buffer solution. One of them contained DNA bio-dots (200 μL), DA (5.0 mmol L−1) and urine sample (200 μL) and the other contained equal amounts of DNA bio-dots and DA. After incubated for 20 min, the fluorescence intensity of the two groups was detected. And the above experiment was repeated for three times. As the results shown, the fluorescence intensity of the two groups was basically similar, suggesting that some oxidants which may exist in urine samples have negligible effects on the spontaneous oxidation of DA. Then the content of GSH in glutathione whitening powder was detected. Firstly, glutathione whitening powder was accurately weighed 0.0307 g and dissolved in ultrapure water. Then fixing the total volume to 100 mL. Finally, the prepared liquid to be tested was diluted step by step for fluorescence detection. As shown in Table 2, human urine was detected the absence of GSH, the content of GSH in glutathione whitening powder was 96.7 ± 0.5% which was basically consistent with the labeled quantity (98.7%), and the recovery rate was calculated as 97.9–104.6%, indicating that the fluorescence sensor has the promising potential for detecting GSH in human body fluids and pharmaceutical preparations.| Sample | Detected/μmol L−1 | Added/μmol L−1 | Found/μmol L−1 (![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± SD, n = 3) ± SD, n = 3) |
Recovery/% (![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± SD, n = 3) ± SD, n = 3) |
|---|---|---|---|---|
| Human urine | 0.00 | 5.00 | 4.89 ± 0.16 | 98.3 ± 3.3 |
| 30.00 | 30.73 ± 0.78 | 102.5 ± 2.7 | ||
| 60.00 | 59.54 ± 0.89 | 99.2 ± 1.5 | ||
| GSH whitening power | 0.97 ± 0.03 | 1.00 | 1.97 ± 0.06 | 97.9 ± 3.1 |
| 20.89 ± 0.49 | 20.00 | 40.92 ± 0.21 | 104.6 ± 2.4 | |
| 39.56 ± 0.67 | 40.00 | 79.68 ± 0.35 | 99.2 ± 1.7 |
4. Conclusions
In conclusion, we successfully proposed a convenient, fast and sensitive fluorescence sensor for the detection of GSH based on the FRET between DNA bio-dots and PDA. Throughout GSH hindering the DNA bio-dots fluorescence “switch” induced by PDA from turning off, it is simple and efficient to carry out the detection of GSH. At the same time, the fluorescence sensor was successfully applied to detect GSH in human urine and pharmaceutical preparations such as glutathione whitening powder. The novel technique has the potential to be extended to other samples for the detection of GSH.Ethical statement
This study was performed in compliance with the Ethical Committee of Zhejiang Normal University and was conducted in accordance with Helsinki declaration. All human subjects have been informed the situation and have consented to the use of urine samples in this study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The present research was partially supported by the National Natural Science Foundation of China (21575129 and 31970755).Notes and references
- A. Tsiasioti and P. D. Tzanavaras, Talanta, 2020, 222, 121559 CrossRef PubMed.
- A. Hamad, M. Elshahawy, A. Negm and F. R. Mansour, Rev. Anal. Chem., 2019, 38, 20190019 CAS.
- M. Alisik, T. Alisik, B. Nacir, S. Neselioglu, I. G. Isik, P. Koyuncu and O. Erel, Clin. Biochem., 2021, 94, 56–64 CrossRef CAS PubMed.
- Y. Shi, Y. Pan, H. Zhang, Z. Zhang, M. J. Li, C. Yi and M. Yang, Biosens. Bioelectron., 2014, 56, 39–45 CrossRef CAS PubMed.
- Z. Xian, L. Zhang, Y. Yu, B. Lin and Y. Cao, Microchim. Acta, 2021, 188, 65 CrossRef CAS.
- Y. Li, X. Liu and A. M. Zhang, Spectrochim. Acta, Part A, 2017, 173, 880–885 CrossRef CAS PubMed.
- Z. Mohammadpour, F. M. Jebeli and S. Ghasemzadeh, Microchim. Acta, 2021, 188, 1–11 CrossRef PubMed.
- J. Xie, D. Cheng, P. Li, Z. Xu and S. Yao, ACS Appl. Nano Mater., 2021, 4, 4853–4862 CrossRef CAS.
- S. Selvarajan, N. R. Alluri, S. J. Chandrasekhar and J. B. Kim, Biosens. Bioelectron., 2017, 91, 203–210 CrossRef CAS PubMed.
- V. B. Boongaling, N. Serrano, J. G. Guzman, J. M. Santander and J. D. Cruz, J. Electroanal. Chem., 2019, 847, 113184 CrossRef.
- K. Adrianna, O. Patrycja, B. Kamila, G. Rafa and C. G. Yna, J. Sep. Sci., 2018, 41, 3241–3249 CrossRef PubMed.
- F. Nuhu, A. Gordon, R. Sturmey, A. M. Seymour and S. J. Bhandari, Molecules, 2020, 25, 4196 CrossRef CAS PubMed.
- K. F. Wiktor, K. Agata, A. Flis, S. Dominika, P. Elżbieta, B. Dariusz, W. Sławomir, O. Joanna and S. Tomasz, Analyst, 2021, 146, 1897–1960 RSC.
- T. Wang and D. Xiao, Microchim. Acta, 2021, 188, 193 CrossRef CAS.
- Y. Cao, J. Liu, L. Zou, B. Ye and G. Li, Anal. Chim. Acta, 2020, 1145, 46–51 CrossRef PubMed.
- T. Song, X. Zhu, S. Zhou, G. Yang, W. Gan and Q. Yuan, Appl. Surf. Sci., 2015, 347, 505–513 CrossRef CAS.
- G. Cheng, W. Zhang, Z. Ying, Q. Ge and C. Huang, Anal. Methods, 2015, 7, 6274–6279 RSC.
- Z. M. Li, L. Kuang, Q. Zhang, J. D. Qiu, A. Z. Xu and A. Liang, Sens. Actuators, B, 2017, 244, 1031–1036 CrossRef.
- Y. J. Yan, L. Lin, M. Fang, J. Ma, X. H. Lu and P. S. Lee, Small, 2013, 9, 596–603 CrossRef PubMed.
- C. Hu, Y. Liu, Y. Yang, J. Cui and J. Rong, J. Mater. Chem. B, 2012, 1, 39–42 RSC.
- K. Y. Ju, Y. Lee, S. Lee, S. B. Park and J. K. Lee, Biomacromolecules, 2011, 12, 625–632 CrossRef CAS PubMed.
- X. Ji, B. Yi, Y. Xu, Y. Zhao, H. Zhong and C. Ding, Talanta, 2017, 169, 8–12 CrossRef CAS PubMed.
- S. Ma, Y. X. Qi, X. Q. Jiang, J. Q. Chen, Q. Y. Zhou, G. Shi and M. Zhang, Anal. Chem., 2016, 88, 11647–11653 CrossRef CAS PubMed.
- Q. Li, L. Zhang, J. Bai, Z. Liu, R. Liang and J. Qiu, Biosens. Bioelectron., 2015, 74, 886–894 CrossRef CAS PubMed.
- Z. Ma, H. Ming, H. Huang, Y. Liu and Z. Kang, New J. Chem., 2011, 36, 861–864 RSC.
- M. Yuan, R. Zhong, H. Gao, W. Li, X. Yun, J. Liu, X. Zhao, G. Zhao and F. Zhang, Appl. Surf. Sci., 2015, 355, 1136–1144 CrossRef CAS.
- M. C. O. Liebana, N. X. Chung, R. Limpens, L. Gomez, J. L. Hueso, J. Santamaria and T. Gregorkiewicz, Carbon, 2017, 117, 437–446 CrossRef.
- P. L. Gao, W. Wang, M. Zheng and Z. G. Xie, Chem. Eng. J., 2020, 381, 122665 CrossRef CAS.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 125, 4045–4049 CrossRef.
- M. Arvand and A. A. Mirroshandel, Food Chem., 2019, 280, 115–122 CrossRef CAS.
- S. T. Kalajahi, B. Rasekh, F. Yazdian, J. Neshati and L. Taghavi, Environ. Sci. Pollut. Res., 2020, 27, 40537–40551 CrossRef CAS.
- M. Han, L. P. Wang, S. H. Li, L. Bai, Y. J. Zhou, Y. Sun, H. Huang, H. Li, Y. Liu and Z. H. Kang, Talanta, 2017, 174, 265–273 CrossRef CAS.
- P. Thitarat, J. Panichakorn, I. Insik and P. Peerasak, Sens. Actuators, B, 2019, 299, 126936 CrossRef.
- L. Lai, L. Chen, Z. Da, S. Li, J. Liu, S. H. Lim, C. K. Poh, Z. Shen and J. Lin, Carbon, 2011, 49, 3250–3257 CrossRef CAS.
- F. Du, F. Zeng, Y. Ming and S. Wu, Microchim. Acta, 2013, 180, 453–460 CrossRef CAS.
- Z. S. Stojanovi, A. D. Urovi, A. M. Ashrafi and Z. L. Richter, Sens. Actuators, B, 2020, 318, 128141 CrossRef.
- G. Z. Wang, Y. Wang, H. Liu, S. Dong and J. Hao, New J. Chem., 2019, 43, 4965–4974 RSC.
- Y. F. K. Du, F. Zeng, Y. H. Ming and S. Z. Wu, Chem. Mater., 2009, 21, 3042–3044 CrossRef.
- B. Rawat, K. K. Mishra, U. Barman, L. Arora, D. Pal and R. P. Paily, IEEE Sens. J., 2020, 20, 6937–6944 CAS.
- J. H. Yuan, A. Q. Li, T. G. Chen, J. Du, A. D. Ma and J. L. Pan, Anal. Chim. Acta, 2020, 1102, 24–35 CrossRef CAS PubMed.
- A. Tsiasioti, A. S. Zotou and P. D. Tzanavaras, Food Chem., 2021, 361, 130173 CrossRef CAS.
- L. Han, S. G. Liu, J. Y. Liang, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2019, 288, 195–201 CrossRef CAS.
- A. Simindokht, A. Mehdinia, R. Niroumand and A. Jabbari, Anal. Chim. Acta, 2020, 1120, 11–23 CrossRef.
- M. H. Lin, X. Ma, S. J. Lin, X. J. Zhang, Y. Dai and F. Xia, RSC Adv., 2020, 10, 33635–33641 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08107a |
| This journal is © The Royal Society of Chemistry 2022 |