Open Access Article
Open Access ArticleInsight into prognostics, diagnostics, and management strategies for SARS CoV-2
Umay Amara
ab,
Sidra Rashid
b,
Khalid Mahmood
 *a,
Mian Hasnain Nawaz
*a,
Mian Hasnain Nawaz
 b,
Akhtar Hayat
b and
Maria Hassana
b,
Akhtar Hayat
b and
Maria Hassana
aInstitute of Chemical Sciences, Bahauddin Zakariya University, Multan 608000, Pakistan. E-mail: khalidmahmood@bzu.edu.pk
bInterdisciplinary Research Centre in Biomedical Materials (IRCBM), COMSATS University Islamabad, Lahore Campus 54000, Pakistan
First published on 11th March 2022
Abstract
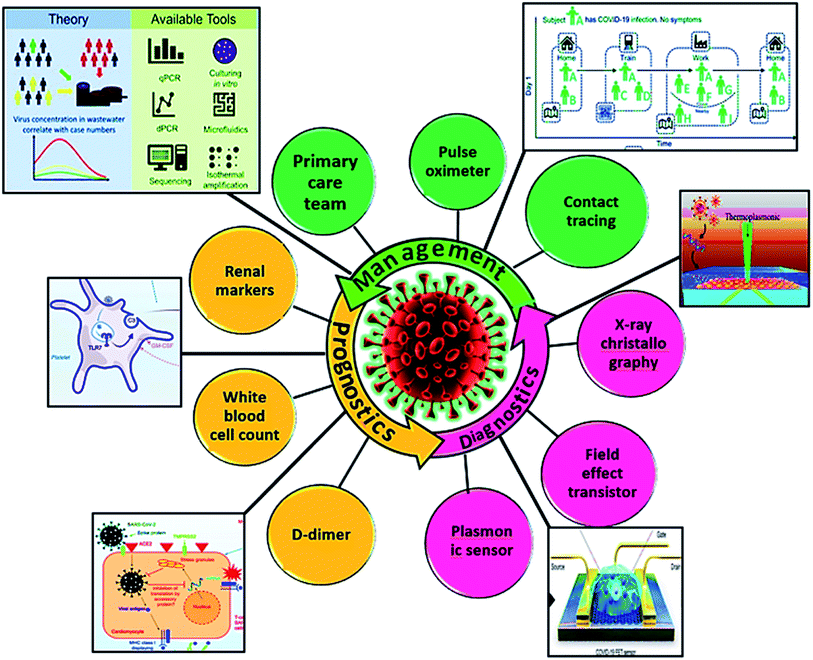
The foremost challenge in countering infectious diseases is the shortage of effective therapeutics. The emergence of coronavirus disease (COVID-19) outbreak has posed a great menace to the public health system globally, prompting unprecedented endeavors to contain the virus. Many countries have organized research programs for therapeutics and management development. However, the longstanding process has forced authorities to implement widespread infrastructures for detailed prognostic and diagnostics study of severe acute respiratory syndrome (SARS CoV-2). This review discussed nearly all the globally developed diagnostic methodologies reported for SARS CoV-2 detection. We have highlighted in detail the approaches for evaluating COVID-19 biomarkers along with the most employed nucleic acid- and protein-based detection methodologies and the causes of their severe downfall and rejection. As the variable variants of SARS CoV-2 came into the picture, we captured the breadth of newly integrated digital sensing prototypes comprised of plasmonic and field-effect transistor-based sensors along with commercially available food and drug administration (FDA) approved detection kits. However, more efforts are required to exploit the available resources to manufacture cheap and robust diagnostic methodologies. Likewise, the visualization and characterization tools along with the current challenges associated with waste-water surveillance, food security, contact tracing, and their role during this intense period of the pandemic have also been discussed. We expect that the integrated data will be supportive and aid in the evaluation of sensing technologies not only in current but also future pandemics.
1 Introduction
An initial insight into the current novel infectious ailment, namely, COVID-19, appeared on 31st December 2019 in Wuhan, China.1,2 A large number of individuals suffering from cough, shortness of breath, and fever were hospitalized at the start. A primary evaluation by compound tomography indicated divers as compared to healthy lungs.3 This finding directed to an early diagnosis of pneumonia. Further, nucleic acid investigations via multiplex real-time polymerase chain reaction (PCR) of already known pathogens led to negative outcomes, indicating that the actual cause of pneumonia was unfamiliar (1). A genetic sequence parallel to that of the beta coronavirus B was discovered from a sample of patients suffering from Bronchoalveolar Lavage (BAL) by 10th January 2020. This study also revealed a genomic similarity of 50%, 80%, and 96% to the Middle East Respiratory Syndrome virus (MERS-CoV), Severe Acute Respiratory Syndrome virus (SARS-CoV), and bat coronavirus (RaTG13), respectively.4,5Therefore, this unknown pathogen was named as SARS-CoV-2, which is responsible for COVID-19. By April 2, 2020, this infection had blown out to almost 202 countries, infecting approximately 1 million people with 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526 deaths worldwide. Consequently, on March 11, 2020, this novel COVID-19 outbreak was declared as a pandemic by the World Health Organization (WHO).6,7 However, the total number of coronavirus cases till December 29, 2021 were 283
526 deaths worldwide. Consequently, on March 11, 2020, this novel COVID-19 outbreak was declared as a pandemic by the World Health Organization (WHO).6,7 However, the total number of coronavirus cases till December 29, 2021 were 283![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 374
374![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 722 with 5
722 with 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434
434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 143 deaths by the Worldometer website (https://www.worldometers.info) (Scheme 1).
143 deaths by the Worldometer website (https://www.worldometers.info) (Scheme 1).
The percentage of individuals affected by SARS CoV-2 infection that remained asymptomatic has not yet been fully assessed. On the other hand, in symptomatic individuals, clinical manifestations of this infection start usually within a week including cough, fatigue, fever, and upper respiratory tract infection.8 It can also progress to several other diseases such as dyspnoea and other chest infections in almost 75% of the affected persons. It was also reported by one of the studies that the interaction with the laryngopharynx, nasal mucosa, and trachea of patients can develop innate-immune response for instance tissue damage in different parts of the body. It was also found that this infection may affect the brain via different passages also including peripheral nerves.9 More recently, the investigation of 425 confirmed infected cases demonstrate that the current pandemic may double the number of affected individuals every seven days and that each patient spreads the infection to 2.2 to 3.58 other individuals on an average.10 Standing against these current circumstances, scientists are actively probing for the important attributes accountable for this pandemic. However, examining the target sequences carried by exogenous nucleic acids along with CT scans11 suffers from serious limitations produced by nonspecific hybridization or amplification, as well as overpriced infrastructure and difficult work.12,13 To mitigate such challenges, protein-based sensing assays that principally depend on identifying antibodies released by individuals on contact with SARS CoV-2 have been introduced.2,14–16 Certain groups have employed plasmonic sensors as robust and economically viable substitute with much lower detection limits compared to the abovementioned techniques.17,18 In parallel, nanoscale visualization procedures such as electron microscopy (EM), atomic force microscopy (AFM), and X-ray diffraction (XRD) have also been exploited for timely analysis and enlightening prognostic topographies of a specific virion.19,20
Despite huge successes in vaccine development, the COVID-19 variants, especially the most recent one called Omicron, have raised severe concerns as it is significantly limiting antibody-based neutralization and elevated the chances of re-infection.21 Also, an increased number of cases reported have put the efficacy of functional vaccinations in question for emerging alternates.13,22 To solve such challenges, there is a need to improve the management strategies by moving toward tele-health care and tele-medicine23 to provide care from a distance.24 It would help to decrease the burden on the healthcare system and preventing exposure to highly vulnerable patients. Numerous clinics have implemented this practice globally. The current review aims to report the importance of the state-of-art instruments and technology employed so far to lessen the current and additional waves of COVID-19. The study also underlines the positive impact of contact tracing and quarantine in this critical pandemic situation.25–28
2 Sequential and molecular data analysis
Researchers have targeted the analysis of beta-coronavirus including SARS-CoV-1 as well as MERS to elaborate the real nature of the COVID-19 virus.29 This research shows that ab1 encodes the protein involved in the formation of envelope €, spike protein (S), as well as nucleocapsid (N) (Fig. 1).30 The sequential data of the viral genome was evaluated from Korean patients by Han et al.31 Almost >99.9% of the sequential homology was found for CoV-2 from various other corners of the world. Further, a similar kind of resemblance was also observed by Sah et al. in a 32 years old Nepalese patients32 that is similar to the previously reported viral genome obtained from Wuhan. However, seven new sequences were observed, which vary from MERS and SARS-CoV-1 on the basis of structural evaluation based on biochemical and structural experiments. Meanwhile, a comparative study between α and β coronaviruses revealed that SARS-CoV-2 may have the ability to bind effectively with the human-receptor gene called Angiotensin-Converting-Enzyme-2 (ACE-2).33 However, computational studies did not prove any linkage between the spike protein and ACE-2.34 It led the scientists to believe that a greater attraction between ACE-2 and the CoV-2 gene may be the result of mutation to ACE-2.35 It was also found that this mutation can easily alter the phenotype of SARS-CoV-2, which in turn can alter the diagnosis of the virus.36 In addition, the mutated portion of the respective genome can forecast the linkage between probe binding and the primer.37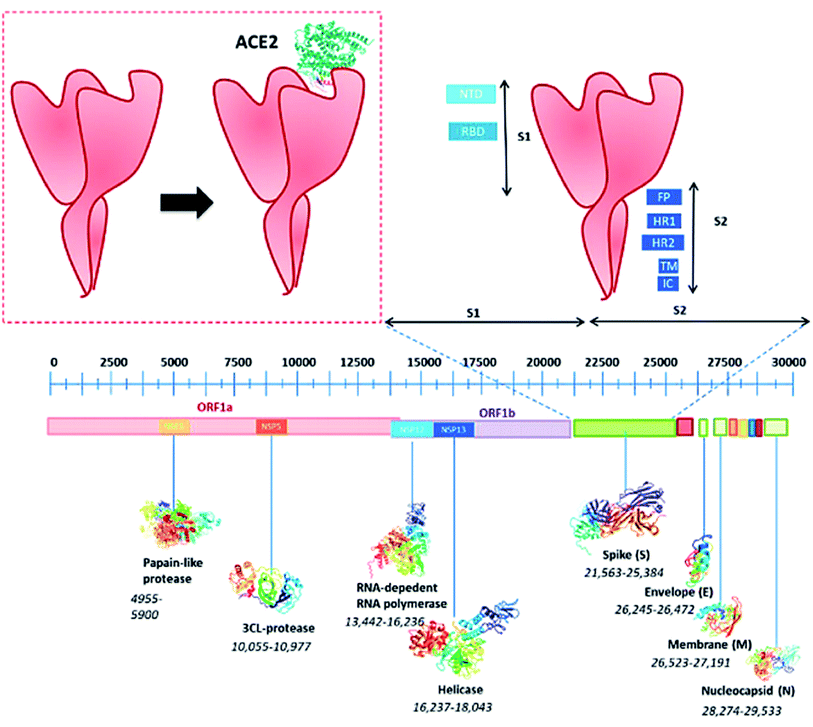 | ||
| Fig. 1 Structure of SARS-COV-2; prime structure of spike glycoprotein (S1) of SARS-COV-2; lateral and upper views of S1; ACE-2 and RDB of S1; binding sites of spike S1 protein. The corresponding amino acids marked as red for the positive charge, purple for the hydrophilic, and blue for the hydrophobic residues. The figure has been reproduced from ref. 30 with the permission from WILEY, copyright 2020. | ||
3 Prognostic strategies using biomarkers for COVID-19
The exploration of various risk factors caused by COVID-19 at different intervals is essential on an urgent basis due to its severity. It would be of great help to take timely actions and reasonable intervention to enhance the cure rate and the prognosis quality.38 Various biomolecules have been utilized for the effective prognosis evaluation of COVID-19 in laboratory tests. In the following section, we carefully evaluated the utilization of different prognostic strategies including D-dimer, IL-6, and some other biomarkers particularly during the early stages of infection.393.1 C-Reactive protein (CRP) for COVID-19
This protein is developed by liver and various inflammatory mediators such as IL-6. Its higher level is parallel to the disease intensity.40,41 CRP, as a biomarker in disease diagnostics, has been highlighted by a retrospective center in Wuhan. An increased level of CRP (57.9 mg L−1) was reported in the patients in Wuhan.42 Another study reported CRP levels >41.8 mg L−1 and found the likelihood of COVID-19 progression.43,44 Pathologically, CT scans have been employed to identify lung lesions during COVID-19 progression but are unable to differentiate between mild and severe cases, as we reported previously.45 The outstanding activity of CRP as a biomarker was revealed in the ‘area under the curve’ in the receiver operating examination of 0.87 (95% CI, 0.10–1.00), where 83% sensitivity and 91% specificity were reported. Henceforth, CT scans alone along with CRP values have been proved to be authentic for the earlier detection of cases. Many other studies also corroborated the abovementioned findings.46,473.2 Interleukin-6 (IL-6) for COVID-19 diagnostics
IL-6 is a primary trigger for cytokine storms. Wan et al.48 found that cytokine storm is crucial to the progression of COVID-19 and can lead to severe complications even death. The 5th edition of “Diagnosis and Treatment of COVID-19”49 suggests that observation of the cytokine level helps to improvise the treatment efficacy with reduced mortality. According to Yang et al., this inflammatory factor plays a crucial role in the progression of the disease from a mild to a severe level.50,51 Cox-proportional-hazard-model-analysis (CPHMA) showed that IL-6 could be utilized as an independent factor to predict the severity of SARS-COVID-19 hematological diseases.38 Thus, we can conclude that an increased level IL-6 can be a sign of a higher inflammatory cytokine storm. Thus, the role of IL-6 in this disease deserves special attention.52,53One meta-analysis recounted that the mean IL-6 concentrations were 2.9 times greater in complicated COVID-19 patients in comparison to non-complicated disease, in which (n = 1302; 95% CI 1.17–7.19).54 The findings of the study include ICU admittance, onset of acute respiratory distress syndrome (ARDS), and deaths. Groundbreaking conclusions suggested that the proportional upsurge of IL-6 is directly allied with disease severity. Another group proposed that SARS CoV-2 affects both the upper and lower respiratory tract and causes a high or mild acute respiratory syndrome with the consequent release of IL-6.55 This enhanced appearance of IL-6 in the serum possibly indicates the severity and prognosis of the disease in SARS COVID-19 patients.56 In line with the findings of many researchers, the dynamic changes in the level of IL-6 have proven to be an important biomarker for disease diagnostics in severe COVID-19 patients.57,58
3.3 White blood cell count
Leucocytes, also known as white blood cells (WBCs), are constituents of blood generated from lymphoid tissue and bone marrow. They are divided into two main categories, i.e., granulocytes and agranulocytes. Eosinophils, neutrophils, and (NC) basophils come under the granulocytes group, while monocytes and lymphocytes (LC) are agranulocytes. Their inconsistent number may cause infection and can be analyzed by blood tests, producing a white blood cell count. A survey was made and numerous differences in the WBC of severe and non-severe COVID-19 patients were noted.42 Patients of both cases have been reported to have increased leucocytes numbers, while the severe group experienced a comparatively large rise (5.6 vs. 4.9 × 109 L−1; P < 0.001). NCs were found to be primarily driving this increase. More interestingly, the level of monocytes, lymphocytes, eosinophils, and basophils were less, causing a larger neutrophil-to-lymphocyte ratio (NLR; 5.5 vs. 3.2; P < 0.001). Also, this NLR has been reported as a biomarker that reflects the disease severity.Another study conducted in China also corroborated the findings that higher NC and low LC count is associated with the severity of affected patients, signifying WBCs as a potent biomarker for the timely recognition of severe COVID-19.59 Nevertheless, glucocorticoid and other viral and bacterial infections disturb the accuracy of WBCs findings.60 A specific study made in China reported the depleted LC level in most of the COVID-19 affected patients.53 Another research group reported low blood lymphocyte percentage (LBL%) in critical cases, proposing that a lower LC count is indicative of a poor prognosis. Many other groups justified the same findings.61,62 However, the virus can target the mechanisms of IL-6 and lymphoid tissue. Meanwhile, additional reasons for low LC count need to be examined to determine the effect of WBC on COVID-19 severity.18
3.4 Lactate dehydrogenase (LDH)
In the glucose metabolic pathway, the enzyme LDH catalyzes the conversion of pyruvate to lactate.LDH is produced by cell membrane necrosis owing to lung damage or viral infection such as pneumonia caused by SARS CoV-2. Many evidences suggest the strong relationship between the LDH levels and the growth of COVID-19 disease.63 One study proposed enhanced serum LDH values as one of the unusual diagnostic parameters in SARS CoV-2 patients with severe or fatal development of the disease. Pulmonary injury along with pervasive organ damage are of potential biological and clinical significance at elevated LDH levels.64 One meta-study reported significantly greater levels of LDH in ICU patients, i.e., 248 U L−1 than in non-ICU patients 151 U L−1 (p = 0.002). Higher LDH level is sustained in ICU patients even after the several post-admission days (160 U L−1 vs. 218 U L−1, p = 0.002). Therefore, LDH may be a prognostic biomarker of severe disease. Multi-center research involving 1099 COVID-19 patients supported the evidence associating inflammation and tissue damage with enhanced LDH levels.6 Moreover, when LDH was at higher levels, as seen in the associated CT scans, the severity of pneumonia was revealed.65
Another study was carried out with 87 confirmed COVID-19 patients, assessed by the LDH level at diagnosis and follow-ups.66 The enhancement or reduction in the LDH level was noted to be representative of radiographic progress. A positive correlation was observed between the time to LDH normalization (5.67 ± 0.55, days) and the time to radiographic absorption (5.57 ± 0.65 days). The study validates the potential utilization of serum LDH as a biomarker for assessing the severity and observing treatment feedback in COVID-19 pneumonia.67 Many other studies by different research groups also supported the above findings.68,69
3.5 D-Dimer
D-Dimer released from the degradation of the cross-linked fibrin signifies the activation of fibrinolysis and coagulation.70 Early research has linked elevated levels of D-dimer and hemostatic abnormalities in non-survivors of COVID-19 compared to survivors.71 A survey performed by a group of researchers on 191 cases reported that a D-dimer level greater than 1.0 μg mL−1 (p = 0.0033) resulted in greater mortality among COVID-19 cases. Moreover, they optimized 2.0 μg mL−1 or more as a cut-off to predict the mortality in SARS CoV-2 patients in hospital.72 Another group reported a marked rise in the D-dimer level and enhanced coagulation in almost 90% of patients with pneumonia.73 The researchers also reported the elevated D-dimer level of 2.4 mg L−1 in ICU patients in contrast to non-ICU specimens 0.5 mg L−1 (p = 0.0042). Another study by Sakkat et al. with 1355 patients also justifies the previous findings.74 Yumeng et al. reported a D-dimer elevation of 74.6% (185/248) in patients.75 Furthermore, another research group suggested that the enhanced level of the D-dimer on admittance can be used as a marker to refer patients to critical care. This, along with the many reported studies, proposed that D-dimer levels should be employed as a prognostic marker and aid doctors to monitor and risk-stratify those who are probable to expire earlier.76–783.6 Platelet count
As we have discussed in the previous section, the COVID-19 virus leads to severe hematological variation, resulting in thrombocytopenia (Fig. 2). A survey of 1799 CoV-2 victims has revealed a fewer platelet count (WMD −31 × 109 L−1; 95% CI, −35 to −29 × 109 L−1) in patients with severe infection.79 Likewise, the mortality rate is higher in those with fewer platelets count (WMD −31 × 109 L−1; 95% CI, −35 to −29 × 109 L−1). Considering thrombocytopenia as an endpoint, a fivefold higher possibility of SARS CoV-2 (OR, 5.13; 95% CI, 1.81–14.58) has also been observed. Reconsidering research that castoff Cox proportional hazard regression analysis revealed the platelet count as an independent risk factor for deaths amongst CoV-2 patients, where a 50 × 109 L−1 rise is related to 40% mortality.80 Another study reports the 5.45% death rate (29 subjects died) and decreased platelet count among non-survivors compared to survivors after one week of admission. The difference was enhanced by 50 × 109 L−1, as reported by previous research.81 Another group endorsed the previously discussed work by suggesting that the lowest platelet count was directly related to death and lower the count, more the association.82 Hence, the literature and many findings by researchers suggested that the platelet count could be exploited feasibly to clinically analyze the severity of infection.83–85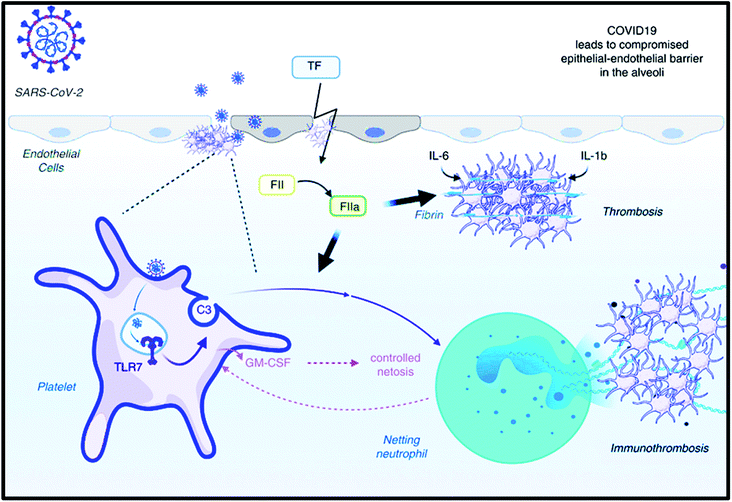 | ||
| Fig. 2 Potential role of platelets in SARS CoV-2: Implications for thrombosis. The figure has been reproduced from ref. 86 with the permission from WILEY, copyright 2020. | ||
3.7 Cardiac troponin
There are increasing shreds of evidence of higher mortality rates in COVID-19 infected patients with underlying cardiovascular disease.87,88 Few research groups have examined the utilization of high-sensitivity cardiac troponin I (hs-TnI) as a biomarker of infection development and transience.89–93 A study conducted on COVID-19 patients exploiting SARS CoV-2 RNA detection depicted an invariable odd ratio for death at 80.1 (95% CI 10.3–620.4, p < 0.0001) for hs-TnI.94 Another study on 416 hospitalized patients with SARS CoV-2 conveyed that hs-TnI was higher in 1 among 5 patients.95 These patients were more likely to develop infection (59% vs. 15%, p < 0.001) along with acute kidney injury (9% vs. 0%, p < 0.001) and needed non-invasive (46% vs. 4%, p < 0.001) or invasive (22% vs. 4%, p < 0.001) ventilation. Prompt analysis of myocardial damage identified by raised hs-TnI helps in suitable triage to enlighten the use of vasopressors and inotropes. In addition, the enhanced procoagulant, prothrombotic and inflammatory responses following COVID-19 infection enhanced the likelihood of acute myocardial infection and acute ischemic myocardial injury owing to respiratory failure with hemodynamic instability and hypoxia in severely sick patients. The use of hs-Tn, as discussed above, has the ability to aid in risk stratification and disease phenotyping in hospitalized SARS CoV-2 patients.96 The probable pathophysiology and anticipated guideline for identification and management has been reported by Bhurint et al., as shown in (Fig. 3).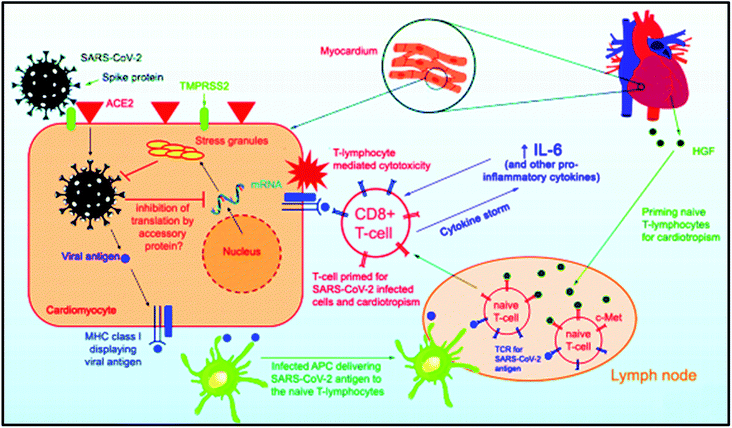 | ||
| Fig. 3 Proposed pathophysiology of COVID-19 myocarditis. The figure has been reproduced from ref. 97 with permission from Heart Rhythm Society, copyright 2020. | ||
3.8 Renal markers
Several pieces of evidence have also been reported on the association between the severity of COVID-19 infection and chronic kidney diseases.98 A study on 28 patients has confirmed suggestively that greater levels of renal biomarkers, for example, creatinine, serum urea, and markers of glomerular filtration have been associated with severe cases.99 A retrospective study on 701 patients exposed raised serum creatinine levels on admittance, associated with severity, owing to substantial anomalies in the coagulation pathway.100 The group reported that these cases require intensive care and mechanical ventilation. Univariate Cox regression investigation observing higher creatinine was also related to the in-hospital death rate (HR 2.99, 95% CI: 2.00, 4.47). Likewise, haematuria, proteinuria, and raised urea levels had parallel, if not superior, threat effects. Remarkably, another group of researchers reported a superior role for urinalysis over serum markers for kidney function and suggested the disease severity in case of any abnormalities in the urine test at the time of admission.101 Another meta-analysis by Michael et al. on 3615 patients reported that acute kidney injure is related to poor outcomes in patients with SARS CoV-2.102 Hence, renal abnormalities in COVID-19 patients at the time of admission might specify greater risks of deterioration, confirming suitable triaging.1033.9 Lymphopenia
The control of viral infection necessarily depends on natural killer cells and cytotoxic T lymphocytes. A functional collapse of antiviral lymphocytes has been noted in patients with COVID-19.104 It has been observed that the degree of pro-inflammatory cytokine storm lymphopenia is greater in severe SARS COV-2 patients than in mild cases, and is related to disease severity.105,106 The N8R, NLR, and lymphocyte percentage are considered to be useful reliable prognostic factors for the timely diagnosis and progress of severe COVID-19 cases. Literature reports the survey of 12 patients (death case) with an average age of 76 years, in which lymphocyte percentage decreased up to 5% in 2 weeks after disease inception. However, when a survey was performed for 7 patients with severe symptoms within a therapeutic window of 35 days and an average age of 35 years, the lymphocyte percentage was reduced at the start but raised to greater than 10% at the time of discharge. Conversely, another survey with 11 patients having a therapeutic time of 26 days and an average age of 49 years was performed in patients with moderate indications. The lymphocyte percentage was found to be persistent and greater than 20% at the time of discharge. The whole study elucidates the practical applicability of lymphopenia-based prognosis for early SARS COV-19 detection.944 Detection techniques for SARS-COVID
4.1 Computer tomography (CT)/chest CT scan-based SARS COVID-2 detection
Chest CT scan or computer tomography is an important technique employed for the detection of COVID-19. It is being utilized either alone or in combination with RT-PCR.32,107,108 The methodology involves a large number of chest X-rays from different dimensions and angles to obtain 3D images, which are later examined by some radiologists to validate the presence of the SARS-CoV2 virus. Consolidation of fluid in lungs along with ground glass opacification are observed in patients of COVID-19 associated with pneumonia. Other abnormalities observed in the CT-scan of COVID-19 patients are bilateral lung, diffused dissimilar airspace, or opaque cavity-like patch, involving lower lobes along with peripheral distribution. For instance, at the initial stages (0 to 2 days) of the infection, the CT scan bears a resemblance to a typical chest condition. However, as the infection develops more, the impermeability of the scan rises, and after 4 days, ground-glass opacity has been observed, which seems to show irregular patterns in the scan images. Fig. 4 illustrates the CT scan image of altered sufferers on different days. Likewise, oddity can be noticeably witnessed.109,110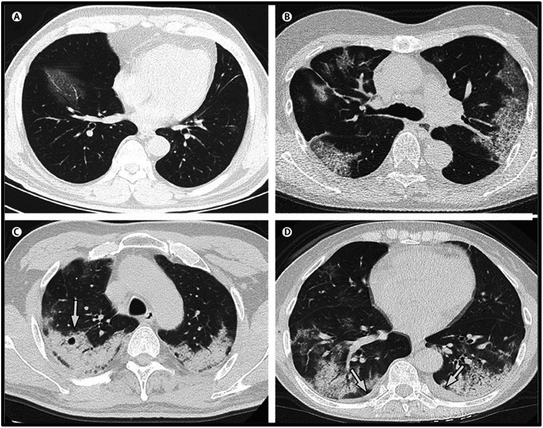 | ||
| Fig. 4 CT scans (transverse thin-section) in SARS COV-19 patients: (A) a 56 year-old man at 3rd day after symptom onset; (B) a 74 year-old woman at 10th day after symptom onset; (C) a 61 year-old woman at 20th day after symptom onset; (D) 63-year-old woman at 17th day after symptom onset. The figure has been reproduced from ref. 111 with permission from Elsevier, copyright 2020. | ||
Lately, a 3D framework built on chest CT scan images was constructed for CoV-2 analysis.112 The described model accomplished a high sensitivity and specificity of 90% and 96%, respectively. Therefore, CT scan images have been employed as a prognostic tool for SARS CoV-2 detection. However, high prices, the requirement of large medical instrumentation, and presence in very few hospitals have limited the use of CT scan technology. Meanwhile, this technology is also unable to distinguish different viruses and unable to distinguish between pre-symptomatic, asymptomatic, and some milder symptomatic patients without pneumonia.
4.2 Nucleic acid-based detection
After the recognition of SARS CoV-2 as the main agent for the pandemic, the genome of the virus was sequenced rapidly and distinctive sequences have been recognized for analysis, and different strategies have been proposed for CoVID-19 detection based on the nucleic acid.For the SARS-CoV-2 detection process, initially, DNA polymerase enzyme screened envelope gene (E gene) nucleotides, followed by the verification of the nucleocapsid gene (N gene) to the particular fragment of the virus cDNA. Polymerase enzyme encounters the double-stranded DNA and through exonuclease activity, it parts the quencher and fluorophore molecules for the real-time recognition of the viral cDNA using the RT-PCR approach. If the system is well-calibrated, a large number of cDNA copies along with fluorescence signals are formed after a few series of polymerase reactions. The fluorescence intensity and virus concentration are directly proportional. If negative results are obtained, further analysis will be performed by sequencing the RNA polymerase (RdRp) gene. After that analysis, if a positive outcome is achieved, it confirms the existence of SARS-CoV-2 in the suspected patients, and is labeled as COVID-19 positive clinically (Fig. 5). Currently, three regions of the cDNA, i.e., RdRP, E, and N genes have been recognized for the SARS-CoV2 virus detection. The high specificity and sensitivity of (500 or 1000 copies mL−1 of viral RNA) are the main causes of the commercialization of PCR-based diagnostic SARS CoV-2 kits.113 However, this technique has inherent challenges such as extended turnaround time and a sampling procedure that is affected by the viral load (VL). Meanwhile, SARS-CoV2 destroys the targeted RNA while opening a viral capsid that results in the discharge of small RNA fragments into the bloodstream that challenges detection by RT-PCR. Numerous testing kits have been established worldwide on the principle of RT-PCR.
 | ||
| Fig. 5 Steps involved in the qRT-PCR testing methodology. The figure has been reproduced from ref. 114 with permission from WILEY, copyright 2020. | ||
 | ||
| Fig. 6 Lateral flow immunoassay-based sensing of anti-SARS-CoV-2 antibodies. The figure has been reproduced from ref. 129 with permission from ACS, copyright 2020. | ||
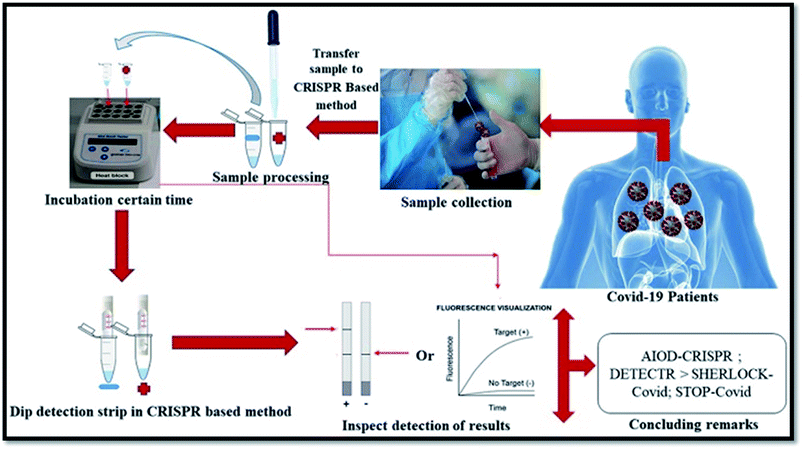 | ||
| Fig. 7 CRISP-based method for SARS CoV-2 detection. The figure has been reproduced from ref. 135 with permission from Elsevier, copyright 2021. | ||
4.3 Protein-based detection
The antibodies and antigens that are released in reaction to SARS CoV-2 have been exploited as a source to assess COVID-19. However, viral load variations during disease progression have limited the long scale application of methodology.A study was made on COVID-19 diagnosed cases, in which a group collected ELISA IgM and IgG antibodies from 63 samples. Meanwhile, certain researchers collected 91 plasma specimens for the colloidal gold-immuno-chromatographic assay (GICA). Meanwhile, the sensitivity of ELISA (IgM and IgG) was found to be 55/63 (87.3%). However, the sensitivities of the GICA IgM and IgG were found to be 75/91 (82.4%). Both methodologies showed negative results for healthy controls with 100% specificity.150 Another study was made for antibody screening to identify SARS-CoV-2 in which IgG and IgA ELISAs were evaluated in unification with the EURO-Lab workstation. The premediated specificities were 91.9% for IgG and 73.0% for IgA.151
 | ||
| Fig. 8 ELISA assays identify antibodies (A) or antigens (B). The figure has been reproduced from ref. 129 with permission from European Centre for Disease Prevention and Control, Copyright 2020. | ||
Another (LFICS) has been sanctioned in China for point-of-care disease COVID-19 diagnosis.153 The colloidal gold immunochromatographic assay (CGICA) based on gold nanoparticles (Au NPs) can identify IgM and IgG antibodies concurrently within 15 min, against the SARS COVID-2 virus in the blood. The gross sensitivity of the LFICS assay was found to be 88.66% with 90.63% specificity.99 The results demonstrate that immunoassay-based COVID-19 detection can also be used as a tool owing to its portable and economically viable infrastructure, along with good sensitivity and specificity. The inclusive process and data analysis of the abovementioned immunodiagnostic assay are revealed in Fig. 9. The immunoassay involves a few drops of blood to detect a virus qualitatively in a miniaturized set-up. Nonetheless, the analysis of the exact titer is not probable using this methodology.
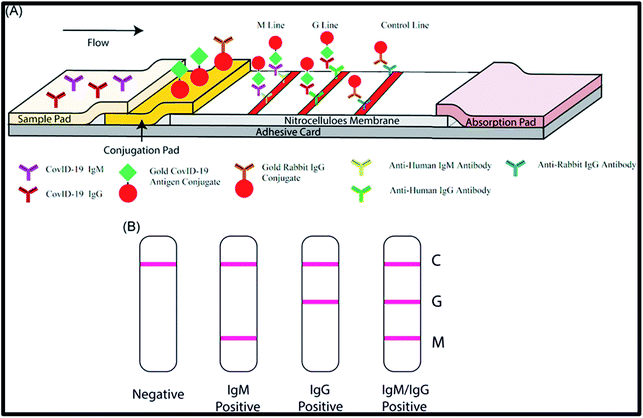 | ||
| Fig. 9 A systematic diagram of the detection device based on IgM-IgG combined antibody test. The figure has been reproduced from ref. 154 with permission from WILEY, copyright 2020. | ||
4.4 Plasmonic sensors
The ongoing SARS-COV-2 pandemic situation highlights the need for reliable and fast sensing strategies. Deep down, this pandemic is improving virus sensing to some extent, particularly plasmonic techniques, as portable, cheap, reliable, and fast sensors are in demand now. PCR is considered as the most effective and real time detection methodology for SARS-CoV-2 with almost 100% selectivity.155 However, it still has several drawbacks including cost and long response time along with a complex system.156 Another major drawback of RT-PCR-based detection has been associated with a large amount of false-positive and negative cases, thus limiting its practical applicability. In this regard, plasmonic detection techniques integrated with lateral flow processes have tremendously increased the detection of viruses in a limited amount of time.157 Apart from the fastest way with higher sensitivity and selectivity for different applications,158 plasmonic sensors in point-of-care (POC) devices for the early detection of viral diseases remain nascent.159 Efforts are being made for the improved functionality of such sensors by implementing various techniques, namely, interference lithography, nanoimprinting microsphere lithography, roll-to-roll pattering, and oblique angle deposition. These methods not only advance the accuracy of the sensors but also bringing the cost down.157,160,161 Besides, such modifications also simplify the detection of coronavirus and thus may help in disease control.162Recently, a plasmonic sensor with dual functionality combining the plasmonic photothermal (PPT) as well as surfaced plasmonic resonance effects has been employed as a robust and cost-effective substitute to RT-PCR. A selective sequence of CoV-2 has been analyzed via nucleic acid hybridization by exploiting complementary DNA-functionalized 2-D gold nanoislands (AuNIs). The two dissimilar incident angles were employed for the excitation of LSPR and plasmonic resonance of PPT, which desirably improved the sensing stability, reliability, and sensitivity, as shown in Fig. 10. A label-free and real-time recognition of viral sequences comprising ORF1ab, RdRp, and the E genes from the COVID-19 has been made owing to the attained ideal configuration. In addition, the enhanced specificity along with the hybridization kinetics about nucleic acid-based recognition have also been achieved by the utilization of PPT enhancement on AuNPs-chip. Such a biosensor exhibited a low detection limit of 0.22 pM along with multigene combination.163
 | ||
| Fig. 10 Graphical presentation of plasmonic sensing of SARS CoV-2 at lower concentrations. The figure has been reproduced from ref. 58 with permission from European Centre for Disease Prevention and Control, Copyright 2020. | ||
Another group developed a selective methodology for the N-gene detection of SARS CoV-2 by the naked eye. They reported a colorimetric assay using AuNPs integrated with thiolated antisense oligonucleotides (ASOs). This protocol was employed for confirming positive COVID-19 cases in a short period of 10 min. The capped AuNPs showed aggregation, while its targeted RNA sequence came into the picture. Furthermore, the existence of RNaseH (enzyme) enhances the splits of the RNA, particularly in RNA-DNA fusion, directing the additional accumulation of AuNPs, as shown in (Fig. 11). The assay worked selectively for MERS-CoV viral RNA having an LOD of 0.18 ng μL−1. Consequently, the current work reported an effective reproducible and reliable methodology for the naked eye detection of CoV-2 without the need for any advanced instrumental techniques.164
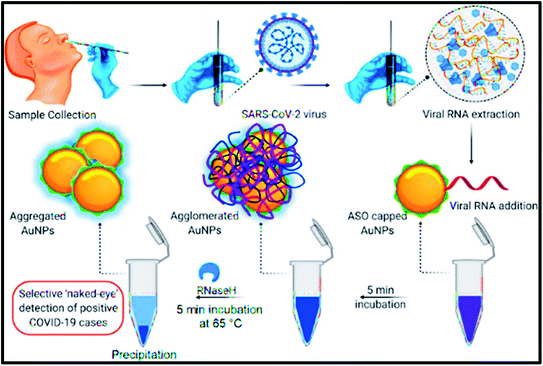 | ||
| Fig. 11 Schematic illustration of naked eye sensing of COVD-19 by AuNPs capped by ASO. The figure has been reproduced from ref. 164 with permission from ACS, copyright 2020. | ||
4.5 Surface-enhanced raman spectroscopy (SERS)
For the evaluation of organic and inorganic compounds, Raman spectroscopy (RS) is a well-known technique.165 It has been employed for the recognition of bacteria in food 22 as well as for HIV-1 detection.166–168 Recently, RS was applied for SARS-COVID-19 detection using Au nanoparticles.169 However, signal amplification was required due to the very low signal intensity, for which the plasmonic effects of different metallic nanoparticles can be applied. This plasmonic field is able to enhance the signal by ∼108 to 109 times, based on the chemical composition or the aspect ratio (shape).170 Further, various 2-D materials can easily provide an additional signal amplification of approximately 102 because of the effective transfer of electrons.171 This signal enhancement is known as Surface-Enhanced Raman Spectroscopy (SERS) for the detection of viruses as well as other pathogens172 with improved specificity and sensitivity.173According to Sanchez et al.,175 Raman-SERS spectral analysis of any viral particle is complex and can give broad peaks due to multiple chemical species (Fig. 12). Lee et al.176 reported a similar trend in the spectra of HIV-1 viral infection. An interesting fact regarding virion and S-protein spectra is that they lack the peaks characteristic (1640–1678 cm−1) of amide-I bonds. Kuroski et al. explained this with reference to the SERS studies of various homopeptides composed of Tyr-, Ala-, Trp-, and Gly-chains with a variety of investigational circumstances.177 In this study, it was found that the lack of some bands is due to amino side chains. This side chain increased the distance among the particles and the peptide bond. However, another possibility might be the deactivation that can disrupt amino acids. In another study, Zhang et al. employed SERS based on silver nanorods, which were functionalized by binding protein and cellular-receptor angiotensin converting enzyme 2 (ACE2) (Fig. 13).174 A very weak peak for amide I was found in this case, which was closer to the noise. They used the 1189 cm−1 peak in ACE-2 with a peak shift of 1182 cm−1 as the spike protein was attached. Then, they used a ratio of these two peaks in order to predict viral presence. Their SARS-CoV-2 spectra showed very few weak peaks.
 | ||
| Fig. 12 Application of SERS indicated by (a) the SEM image of Au–Cu nanostars, (b) optical absorption spectra (OAS) of these nanostars. The figure has been reproduced from ref. 174 with permission from ACS, copyright 2020. | ||
 | ||
| Fig. 13 Diagram illustration of the RT-LAMP-LFA scheme. (A) Workflow of the developed scheme for the detection of COVD-19, (B) principle involved in RT-LAMP amplification. (C) Principle of visual detection with LFA. The figure has been reproduced from ref. 174 with permission from ACS, copyright 2020. | ||
Besides, SERS amplification requires a molecule, which should be in contact with any suitable metallic nanoparticle. This method was used by Jhon et al. by combining SERS with a particular substrate, which is able to combine plasmonic amplification with excitation amplification and provides us with a greater number of peaks. A consistent spectrum with almost 5 peaks was obtained by this methodology. These peaks were connected to S and N proteins. However, fewer peaks were assigned to the particles deactivated due to the destruction of some viral parts. The range of 744–7 nM was obtained in the SERS-spectra of InBios-Spike-Protein. Afterward, for the calibration curve, PLSR (Partial Least Squares Regression) was employed with the variance of 99.9%. Finally, a limit of detection (LOD) of 8.89 × 10−9 M with a Root Mean Square Error (RMSE) of ≈2.27 × 10−9 was obtained, which is a good approximation for concentrations in the nM range. Meanwhile, we have summarized the pros and cons of all the discussed techniques as shown in (Table 1).
| Diagnostic techniques | Pros | Cons |
|---|---|---|
| Computer topography/Chest CT scan | 1. Provides high quality, detailed image of patient organs | 1. Higher price |
| 2. Images are problem-solving for cases that do not show up in initial PCR tests | 2. Requirement of large medical instrumentation | |
| 3. Presence in very few hospitals | ||
| 4. Can result in a fast negative result, especially at early infection | ||
| 5. Unable to distinguish different viruses and unable to detect pre-symptomatic, asymptomatic, and some milder symptomatic patients without pneumonia | ||
| Nucleic acid-based detection | 1. High specificity | 1. Extended turnaround time |
| 2. High sensitivity | 2. The sampling procedure is affected by the viral load (VL) | |
| 3. Promising LOD | 3. SARS-CoV2 destroys the targeted RNA while opening viral capsid that results in the discharge of small RNA fragments into the bloodstream that challenges detection by RT-PCR. | |
| 4. Less assay time | 4. A large amount of false negative and positive cases | |
| Protein-based detection | 1. Good specificity | 1. Viral load variations during disease progression |
| 2. Promising method to obtain the pervasiveness of COVID-19 in a larger population | 2. Sometimes false-positive results | |
| 3. Economic and portable infrastructure along with good sensitivity | 3. Detection of low concentrations of viral protein | |
| Plasmonic sensor based detection | 1. Take a few to tens of minutes or even less | 1. Early diagnostics of viral diseases remains nascent |
| 2. Rapid sampling | ||
| 3. Broad linear range | ||
| 4. Lower LOD. | ||
| 5. High sensitivity | ||
| 6. Higher selectivity | ||
| Signal enhanced Raman spectroscopy (SERS) | 1. High selectivity | 1. Low signal amplification |
| 2. High specificity | ||
| A field-effect transistor (FET) | 1. Miniaturization | 1. Less sensitive |
| 2. Robust | ||
| 3. Real-time | ||
| 4. Selective | ||
| 4. Label-free probing | ||
| 5. Use of ultralow concentration |
4.6 Field-effect transistor (FET)
The FETs and amperometric systems are important electronic and digital devices that are broadly employed for the sensing of biomolecules and pathogens. The remunerations of low cost, mass manufacturing, and miniaturization make these systems most desirable and applicable. FET biosensors have also been applied for the clinical diagnosis of COVD-19. The sensor was established by integrating the COVD-19 spike antibody with the graphene sheets of FET. The developed transducing interface detected the COVID-19 protein in phosphate buffer saline (PBS) in the concentration range of 1–100 fg mL−1. Meanwhile, the OFET was also apt to analyze COVID-19 in clinical trials with an LOD of 2.42 × 102 copies mL−1 and a detection bound (DB) of 1.6 × 101 pfu mL−1.178 Thus, the developed FET-based interfaces can be employed for the production of compact POC devices and provide the advantages of robust, real-time, selective, and label-free probing of SARS Cov-2 at an ultralow concentration. The working methodology of FET-based COVD-19 detection has been shown in Fig. 14. | ||
| Fig. 14 Representation of the operational setup of FET-based COVID-19 sensor. The figure has been reproduced from ref. 179 with permission from ACS, copyright 2020. | ||
4.7 Visualization and characterization tools for COVID-19 detection
These basic techniques are usually employed separately or with the combination of other detection techniques for the viral structure. The outstanding proficiencies of these techniques provide a deep examination of the viral structure and function along with a detailed examination of viral influence on the extracellular environment and host cells, which are beneficial in drug discovery applications.180–190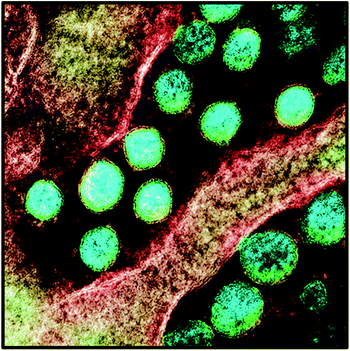 | ||
| Fig. 15 SARS CoV-2 virus particles envisioned by TEM. Viral particles are revealed in blue-green with yellow viral envelopes. This TEM was captured and color-magnified. The figure has been reproduced from ref. 199 under Creative Commons license agreement, copyright 2020. | ||
Recently, cryo-EM was utilized for the identification of multiple mono-clonal antibodies, aiming at the coronavirus, particularly using S-protein from memory B-cells of the infected person. It was found that an antibody called S309 has the potential to neutralize CoV-2 and SARS-CoV.196
The methodology has been utilized for the investigation of RNA-dependent RNA polymerase nsp12 of the COVID-19 structure, which catalyzes the synthesis of viral RNA in complexation with 2 cofactors, namely, nsp7 and nsp8.197 In ref. 198, detailed information about the 3D structure and the morphological surface of the causative agent in SARS-CoV was achieved. These results suggest that such techniques can easily be exploited for the detailed analysis of SARS COVID-19.
Nonetheless, this cryo technique could be an analogous process for viral protein crystallography.204 The details provided by 3D cryo-EM could be assimilated with the obtained X-ray data to ameliorate the constructed model of the virus particles.205 Serial femtosecond X-ray crystallography (SFX) also uses X-ray free-electron lasers (XFELs). It has been exploited to scrutinize the viruses as single particles and crystals owing to its unmatched brilliance of XFEL beams and the pulse duration in a femtosecond. The variations that occurs in the life cycles of viruses, such as response to the changes in pH and interaction of the viral protein with the receptors, could be sensed using time-resolved SFX.206–208 Recently, a 3D crystal structure of unliganded CoV-2 was determined by a resolution power of 1.75 Å, as shown in Fig. 16 (ref. 209) and was utilized to determine the optimization of α-ketoamide inhibitor series.
 | ||
| Fig. 16 The structure of SARS CoV-2 Mpro from two altered 3D viewpoints. The figure has been reproduced from ref. 209 with permission from AAAS, copyright 2020. | ||
However, EM and XRD methodologies are established on the average of countless particles that exist in the crystal or electron micrographs. Consequently, it generates inadequacy in the data acquired from the structural differences between the discrete particles present in a large population. Moreover, the abovementioned methodologies require an atmosphere that is far from the physiological conditions of viral functioning and prevent the characterization of vibrant properties in real-time. Thus, it obscures the study of the viruses that lack distinct structural symmetry, such as SARS CoV-2, which is an enveloped virus. Lastly, AFM is also a direct imaging technology that offers noteworthy impression of the virus study. It offers prospects for studying the COV-2 virus particles.
Furthermore, to examine the chemical, physical, and viscoelastic traits of the viruses, the force–distance curve-based AFM (FD-based AFM) and currently, multifrequency methods, are the frequently employed approaches. In the latest FD curve-based AFM, while imaging the biological sample, several FD curves are recorded.232 Meanwhile, chemical and biological properties mapping methodology has been developed on the idea of FD-based imaging.233,234 In the above methodology, the tips are modified by particular ligands. Furthermore, based on the mechanical strength and adhesion of bonds developed among the modified tips and the receptors of the specimen, the biological traits can be investigated during imaging.235 The time of data acquisition and high volume are the major challenges associated with FD-based AFM. Numerous multifrequency AFM approaches have been projected to decrease the amount of data and increase the imaging speed.236,237 However, their complicated physical principle, mechanism of data interpretation, and theoretical development require more research.
The former research on coronaviruses exploiting AFM techniques238–241 indicates the substantial potential of AFM to visualize, image, and investigate the morphological topographies of the SARS-CoV virus, as shown in Fig. 17. Generally, characterization/visualization tools are valuable as they deliver surface and subsurface information of COVID-19 affected cells, perceive multiple versatile interactions amid host cell and SARS CoV-2 virus, explore the mechanism of the COVID-19 virus to pass the cellular membrane and transport its genome into the host cell, study its replication into the cell along with SARS-CoV2 virus nucleic acids characterization, and recognize the way it is packaged and condensed inside capsids. Recently, anticipated techniques at the nanoscale map that the directional flow patterns can also be castoff to examine the effects of ion-specific sieving traits of the host cell and different pH on the binding of the SARS CoV-2 S protein with the receptor at the interface of the cell.242 Currently, Yang et al. characterized molecular binding and studied the inhibition relation of SARS CoV-2 with ACE2 receptor by exploiting AFM.243
 | ||
| Fig. 17 (A) 2D, (B) 3D AFM images, and contour map (C) of a single SARS-CoV virion. The scale bar = 100 nm in (A) and (C). The resultant cursor profiles (middle and bottom row) give quantitative measurements of the dimensions for the spike proteins (1–15) revealed in (C). The figure has been reproduced from ref. 238 with permission from WILEY, copyright 2005. | ||
4.8 Detection of COVID-19 from common sources
The systematic study publicized 96 records after the removal of duplicates and among those, 5 records were included for quantitative and 26 records for qualitative synthesis. The findings proposed 91% (CI 80–99%) sensitivity of saliva-based diagnostics and 98% (CI 89–100%) sensitivity for nasopharyngeal-based diagnostics for CoV-2 patients with moderate heterogeneity in the studies. The same group also recognized 18 registered patients with ongoing clinical trials of saliva-centered prognosis for COVID-19 diagnosis.245 In Hong Kong, a study was made, in which patients were considered to be diseased if SARS CoV-2 was identified in their sputum or nasopharyngeal specimen. Saliva of a total of 12 patients was obtained and subjected to nucleic acid extraction and RT-PCR for COVID-19 prognosis. Saliva samples were collected at an average of almost 2 days after hospitalization (range, 0–7 days) and initially, SARS CoV-2 was found to be present in saliva samples of 110 patients (91.7%). Meanwhile, there were 33 cases whose nasopharyngeal samples were found to be negative for COVID-19. Their saliva specimen also showed the same negative response. Recently, a group of scientists has effectually endorsed saliva as a viable source compared to oropharyngeal swabs for COVID-19 detection. They reported the utilization of saliva as a robust source to extract viral RNA for COVID-19 detection. Saliva-based tests will help with the global scarcity of swab-based sampling for COVID-19 analysis.246 Likewise, many other groups reported the early detection of COVID-19 based on saliva specimen findings.247,248 However, the low concentration of the analyte in the saliva compared to blood limits its practical application, which is circumvented by the development of precise molecular methodologies and nanotechnology.
RT-qPCR has been utilized to test N(N1–N3) and gene E, and they all were identified at different sites and validated the presence of SARS CoV-2. In the United States, the SARS-CoV2 at high titers was also detected in wastewater specimens by RT-PCR.260 Their work further revealed that the longitudinal examination of wastewater can give an approximation of the population level without onsite analysis. The existence of SARS CoV-2 RNA was also confirmed in 6 WWTP wastewater specimens from a lower frequency area in Spain.261 Environmental surveillance outcomes elucidated that SARS CoV-2 had been transmitted among the population before initial reporting by the municipality. Therefore, WBE can be employed as a surveillance tool for the detection and pervasiveness of COVID-19 and gives a detailed insight into the development magnitude of COVID-19 than clinical analysis. RT-PCR assays have been executed for SARS CoV-2 assessment in many disease control and research centers, as we have discussed previously.58,262 However, the requirement of skilled technicians, longstanding data processing, and examination limits its practical application globally. Digital PCR has also been utilized and its sustainability for certain samples has been evaluated.263
Therefore, it is of paramount importance to manufacture efficient robust and transportable analytical tools to analyze low-level SARS CoV-2 specimens through WBE to validate doubtful cases and monitor asymptomatic patients without the need for centralized laboratories. Thus, exploiting the WBE approach for timely warning and intervention needs a prompt biosensing approach for the on-site recognition of all viruses. The latest progress in sensing devices makes field analysis possible so that the system can deliver real-time monitoring and public health information, as shown in Fig. 18. Recently, it has been proposed by a few scientists that paper-based biosensors can be employed for WBE.262 Owing to the merits such as budget-friendliness, robustness, accuracy, specificity, and sensitivity, these biosensors have been employed previously in the clinical analysis of different analytes,264 food protection, and environmental monitoring systems. Recently, biosensors have been exploited for electrochemical, acoustic, thermal, electrical, and piezoelectric analysis of contagious diseases. The indicators of these contagious diseases are typically recognized in the nasal mucosa, plasma, sputum, serum, blood, urine, feces, and saliva. Exploiting high-affinity probes for sensing wastewater specimens can also decrease the matrix effects.265 In addition, biosensors can also be employed for the multiplex analysis of contagious ailments.
 | ||
| Fig. 18 Design of the established sensor for the recognition of multiplex infectious disease pathogens. (a) Components of the sensor, (b) sketch of the whole sample processing methodology ranging from sample addition to pathogen detection. After amplification, the signal is read out with a UV flashlight (365 nm). (c–f) The figure has been reproduced from ref. 266 with permission from ACS, copyright 2018. | ||
Moreover, biosensors as medical tools can be established for the POC recognition of contagious ailments in resource-limited areas. For instance, Yang et al. introduced a fast “sample-to-answer” detection process, which can offer quantitative analysis of nucleic acids along with the genetic data by the exploration of sewage waste,267 and these findings were further validated by agarose gel image assay and robust electrophoresis, displaying good consistency for wastewater examination. The anticipated biosensors will display advantages such as excellent sensitivity, affordability, superior specificity, rapid sensing time, and low sample consumption for the user-friendly recognition of SARS CoV-2 in sewage water despite conventional detection methods. Under this standpoint, the advanced study on the environment for the examination of disease biomarkers is needed to efficiently exploit WBE sensors for the updated status of COVID-19 in a community (Fig. 19).
 | ||
| Fig. 19 Wastewater surveillance potential for COVID-19 monitoring. The figure has been reproduced from ref. 268 with permission from Elsevier, copyright 2020. | ||
4.9 Role of nanotechnology for SARS-CoV-2 detection
As mentioned previously, the first technique employed for the detection of SARS-CoV-2 and to gain information regarding primers, genetic biomarkers, molecular probes, and different concentration of antibodies in samples was rRT-PCR. But the complex infrastructure, longstanding time requirements,282,283 and excessive use of reagents for the diagnostic test are the major drawbacks of the technique. Therefore, simplistic diagnostic tools with robust response are the immediate requirement. In this regard, nanotechnology has played an important role to counter the aforementioned challenges. For instance, nanodiagnostics works on the principle of the binding capacity of the nanomaterials and the biomolecules under consideration to generate a quantifiable signal for pathogen detection.284 Meanwhile, nanotechnology has also aided in providing effectual results with prompt and timely diagnosis of the disease.285 In addition, the role of nanomaterials in bioengineering can also be of significant importance for SARS-CoV-2 detection as this virus itself has a core shell nanostructure with the size ranging from 60 nm to 140 nm.286,287 Thus, it allows the bioengineered nanomaterials to specifically bind with the virus,288 permitting the evaluation, engineering, and development of procedures for diagnosis, treatment, as well as the preventive measures of SARS-COV-2.289,290 These traits of nanotechnology also lead to the development of handheld devices/tools that can be commercialized with additional benefits of stability, sensitivity, and high accuracy (Tables 2–8).| Product | Detail | Sponsor |
|---|---|---|
| CDC 2019-nCoV real-time RT-PCR diagnostic panel | Manufactured by CDC and firstly distributed to public health laboratories but testing is limited to complex labs | Center for Disease Control and Prevention |
| New York SARS-CoV-2 real-time reverse transcriptase (RT)-PCR diagnostic panel | Developed on CDC's principal and works incapable labs. However, testing is restricted to complex laboratories | Wadsworth – New York State Public Health |
| Panther fusion SARS-CoV-2 assay | Reagents are disseminated as a kit. However, testing is restricted to complex laboratories only | Hologic, Inc. |
| Lyra® SARS-CoV-2 assay | Kits are commercialized but restricted to complex labs only | Quidel Corporation |
| Abbott RealTime SARS-CoV-2 assay | Reagents are commercialized as kits but testing is confined to extraordinary complex labs | Abbott Molecular, Inc. |
| Xpert Xpress SARS-CoV-2 test | Reagents are commercialized and circulated as kits to laboratories. It can run 2000 specimens in a day and tests are confined to moderately to highly complex labs along with Point of Care (POC) settings | Cepheid |
| Actual SARS-CoV-2 test | Reagents commercialized and disseminated as a kit to laboratories, run only a single sample at a time, and tests are restricted to moderately to highly complex laboratories along with Point of Care (POC) infrastructure working under a CLIA Certificate | Mesa Biotech inc |
| BioFire COVID-19 test | Reagents distributed commercially to laboratories, test 264 samples a day, and can perform in both moderate or complex labs | BioFire Defense, LLC |
| AvellinoCoV2 test | Tests are restricted to high complexity Avellino laboratories USA, Inc. | Avellino Labs USA |
| NxTAG CoV extended panel assay | Reagents circulated to labs and tests are restricted to highly complex laboratories only | Luminex Molecular Diagnostics, Inc. |
| ID NOW™ COVID-19 | Reagents commercialized and disseminated as a kit, need a specific platform, only one sample runs at a time, and every specimen takes greater than 13 min. The test is restricted to moderately as well as highly complex labs along with Point of Care (POC) sets working under a CLIA Certificate of Waiver | Abbott Diagnostics Scarborough, Inc. |
| NeuMoDx SARS-CoV-2 assay | The reagents are commercialized, 288 or 96 samples run at a single time, and sometimes takes 80 samples depending on the instrument. However, tests are restricted to moderate as well as complex laboratories | NeuMoDx Molecular, Inc. |
| QIAstat-Dx respiratory SARS-CoV-2 panel | Identifies many other respiratory viruses and bacterial organisms, and reagents are distributed commercially as kits to laboratories. It takes one hour and runs one sample at a time | QIAGEN GmbH |
| COVID-19 IDx assay | Utilizes already commercialized reagents and the test is restricted to complex laboratories by Ipsum | Ipsum Diagnostics |
| BioGX SARS-CoV-2 reagents for BD MAX system | Commercialized reagents are distributed as a kit to moderate to high complexity fully automated laboratories that run 8 samples per hour | Becton, Dickinson & Company (BD) |
| ARIES SARS-CoV-2 assay | Reagents distributed commercially as a kit to labs and tests are conducted at moderate and complex labs | Luminex Corporation |
| Logix Smart Coronavirus Disease 2019 (COVID-19) kit | Reagents are commercialized and distributed as a kit to laboratories and testing is bound to highly complex labs | Co-Diagnostics, Inc. 4-3-2020 |
| Gnomegen COVID-19 RT-Digital PCR detection kit | Reagents are spread commercially to labs as kits. Tests are restricted to very complex labs | Gnomegen LLC |
| Smart detect SARS-CoV-2 rRT-PCR kit | Reagents are distributed and commercialized as a kit to laboratories and testing is restricted to complex laboratories | InBios International, Inc |
| QuantiVirus SARS-CoV-2 test kit | Reagents are commercialized and testing is confined to highly complex labs only | DiaCarta, Inc. |
| iAMP COVID-19 detection kit | Reagents are circulated commercially as kits and tests are restricted to extraordinary complex laboratories only | Atila BioSystems, Inc. |
| SARS-CoV-2 fluorescent PCR kit | Reagents are distributed commercially to laboratories as a kit and are restricted to very complex laboratories for testing | Macura Biotechnology (USA) LLC |
| Curative-Korva SARS-cov-2 assay | Tests are restricted to KorvaLabs as that is developed specifically for laboratories | KorvaLabs inc |
| GeneFinder COVID-19 plus RealAmp kit | Reagents are commercialized to the lab and restricted to complex labs for testing | OSANG Healthcare |
| PhoenixDx 2019-CoV | Reagents are distributed to labs and tests are confined to highly complex labs | Trax Management Services Inc. |
| Allplex 2019-nCoV assay | Reagents are commercialized and distributed to labs and are restricted to highly complex laboratories for tests | Seegene, Inc. |
| Rheonix COVID-19 MDx assay, COVID-19 | Reagents are commercialized and distributed to laboratories as kits and testing is restricted to complex labs | Rheonix, Inc. |
| LabGunCOVID-19, RT-PCR kit | Reagents are commercially divided into laboratories and testing is restricted to complex laboratories | LabGenomics Co., Ltd. |
| Bio-Rad, SARS-CoV-2 ddPCR test | Reagents are commercialized and distributed to laboratories as kits; the test is restricted to highly complex laboratories | Bio-Rad Laboratories, Inc. |
| Sherlock CRISPR SARS-CoV-2 kit | Commercialized reagents are circulated to highly complex laboratories as kits | Sherlock Biosciences, Inc. |
| Gnomegen COVID-19-RT-qPCR detection kit | Reagents are commercialized and distributed to laboratories and are limited to highly complex labs for testing | Gnomegen LLC |
| NeoPlex COVID-19 detection kit, COVID-19 | Reagents are distributed commercially to labs as kits and testing is restricted to highly complex labs | GeneMatrix, Inc. |
| Optima SARS-CoV-2 assay | Reagents are commercialized and disseminated to labs; tests are restricted to complex labs | Hologic, Inc. |
| Lyra direct SARS-CoV-2 assay | Reagents are commercialized and distributed to labs; testing is confined to complex labs | Quidel Corporation |
| AQ-TOP COVID-19 rapid detection kit | Reagents are commercially disseminated to complex laboratories | Season Biomaterials, Inc. |
| DiaPlexQ novel coronavirus detection kit | Reagents are circulated commercially as a kit to highly complex laboratories for testing | SolGent Co., Ltd |
| BioCore 2019-nCoV real time PCR kit | Reagents are commercialized and distributed to highly complex labs only for testing | BioCore Co., Ltd |
| FRL SARS CoV-2 test | Laboratory-based testing restricted to UAB Fungal Reference Laboratory, a high complexity lab. | The University of Alabama at Birmingham Fungal Reference Lab |
| LifeHope 2019-nCoV real- | Laboratory manufactured test, Time RT-PCR Diagnostic Panel. Tests are narrowed to complex Life Hope Laboratory in plainview | LifeHope Labs |
| COVID-19 RT-PCR peptide nucleic acid (PNA) kit | Reagents are distributed commercially in the laboratory as a kit; testing is restricted to highly complex laboratories | TNS Co., Ltd (Bio TNS) |
| Psoma COVID-19 RT test | Tests are confined to the complex laboratory of Psomagen, Inc. in RockvilleMD. | Psomagen, Inc. |
| Solaris multiplex SARS-CoV-2 assay | Laboratory manufactured to test and is restricted to Solaris Diagnostics, a highly complex lab situated in NicholasvilleKY. | Solaris diagnostics |
| SARS CoV-2 RNA STAR | Reagents distributed commercially to labs as kits and limited to highly complex laboratories | LumiraDx UK Ltd. |
| SARS CoV-2 Real-Time RT-PCR Test | Reagents are commercially distributed to labs as kits and tests are confined to highly complex laboratories | Biomeme, Inc. |
| Pro-Amp RT SARS-CoV-2 Test | Lab-based test and restricted to Pro-Lab Diagnostics, a highly complex lab in Round Rock, TX | Pro-Lab Diagnostics |
| SalivaDirect | Recognition of SARS CoV-2 in specimens of saliva obtained without preserving. Commands for utilization is spread to laboratories and all constituents are available commercially | Yale School of Public Health, Department of Epidemiology of Microbial Diseases |
| Advanta Dx SARS-CoV-2 RT-PCR assay | Sensing of COVID-19 in saliva and reagents are disseminated commercially as kits to labs. Testing is restricted to highly complex laboratories | Fluidigm Corporation |
| Product | Detail | Sponsor |
|---|---|---|
| COVID-19 RT-PCR Test | Manufactured and functions in LabCorp labs; not for larger laboratories | Laboratory Corporation of America |
| COVID-19 RT-PCR Amendment | Amendment allows utilization of the Pixel via LabCorp COVID-19 testing and home collection kit permitting suspected victims to self-collect nasal swab specimens. It also offers the transfer of the collected sample materials safely from homes to an authorized laboratory | Amended |
| SARS-CoV-2 RNA, Qualitative Real-Time RT-PCR | Manufactured to function in Quest labs only and not for distribution, and tests are restricted to highly complex laboratories | Quest Diagnostics Infectious Disease, Inc. |
| P23 Labs TaqPath SARS CoV-2 Assay | Laboratory manufactured test, confined to P23 Labs, situated in Little Rock, a high complexity lab and utilized in-home for collecting saliva samples using OMNIgene ORAL Collection Device and then referred to the laboratory for further tests | P23 Labs, LLC. |
| LetsGetChecked Coronavirus (COVID-19) Test | Laboratory-based test restricted to PrivaPath Labs, high complexity labs and utilized only for nasal swabs comprised of COVID-19 LetsGetChecked Home-based Kit and directed to the laboratory for tests | PrivaPath Diagnostics, Inc. |
| KPMAS COVID-19 Test | Lab-based testing confined to KPMAS Regional Lab and can be utilized with nasal swab samples collected via KPMAS Health Plan members | Kaiser Permanente Mid-Atlantic States |
| Compass Laboratory Services SARS-CoV2 Assay | Laboratory-based testing restricted to Compass Lab Services, a high complexity laboratory. It can be utilized for nasal swab samples obtained at home also via a video witnessed by a healthcare worker | Compass Laboratory Services, LLC |
| Quest Diagnostics PF SARS-CoV-2 Assay | Laboratory-based test, confined to labs selected through Quest Diagnostics that meet the necessities to conduct complex testing. It could be cast-off for nasal swab samples at home under the supervision of a healthcare worker by a COVID-19 Self-Collection Quest Diagnostics Kit | Quest Diagnostics Infectious Disease, Inc. |
| CRL Rapid Response | Lab manufactured test, restricted to the Clinical Reference Library laboratory placed in Lenexa, high complexity laboratory. It could be exploited for saliva samples obtained at home or under the supervision of a healthcare provider | Clinical Reference Laboratory, Inc |
| QDX SARS-CoV-2 Assay | Laboratory-based test, restricted to QDx Pathology Services situated in Cranford a giant complex laboratory. It could be applied for samples that are collected at home by utilizing the Qdetect collection kit or any other approved home-based collection kit | QDx Pathology Services |
| 8-25-2020 |
| Product | Detail | Sponsor |
|---|---|---|
| BinaxNOW COVID-19 Ag Card | Moderate to highly complex laboratories are exploited for tests and POC sets working below a CLIA Certificate. It visually recites assays that do not need an instrument | Abbott Diagnostics Scarborough, Inc. |
| LumiraDx SARS-CoV-2 Ag Test | Tests are confined from moderately to highly complex laboratories and POC settings work under a waiver of CLIA Certificate | LumiraDx UK Ltd. |
| Sofia SARS Antigen FIA | Tests are confined from moderately to highly complex laboratories and POC settings functioning under a waiver of CLIA Certificate | Quidel Corporation |
| Product | Detail | Sponsor |
|---|---|---|
| Serology Test qSARS | The very first authorized serological test by EUA, spots CoV-2 antibodies in the blood, distinguishes between IgM and IgG antibodies. It is a robust test and gives an outcome in 15–20 min and is restricted to adequate to high complexity labs | Cellex Inc. |
| CoV-2 IgG/IgM Rapid Test | ||
| VITROS immunodiagnostic products anti-SARS-CoV-2 IgG reagent pack | Identifies IgG antibodies in human serum in moderate to high complexity labs | Ortho-Clinical Diagnostics, Inc. |
| COVID-19 ELISA IgG Antibody Test | Used to identify IgG antibodies in human plasma and serum. Tests are restricted to the high complexity Mount Sinai Lab | Mount Sinai Laboratory |
| LIAISON SARS-CoV-2 S1/S2 IgG | Identifies IgG antibodies in serum and human plasma and tests are restricted from moderately to largely complex labs | DiaSorin Inc. |
| SARS-CoV-2 IgG assay | Perceives IgG antibodies in human plasma, as well as serum and tests, and are restricted from moderate to large complexity laboratories | Abbott Laboratories Inc. |
| Elecsys Anti-SARS CoV-2 | Senses Anti-SARS Cov-2 antibodies in human serum and plasma, and tests are restricted to moderate as well as highly complex laboratories | Wadsworth Center, NY State Department of Health |
| Vibrant COVID-19 Ab Assay | Detects IgM and IgG in dried blood and human serum, functional in the highly complex Vibrant America Clinical Laboratories | Siemens Healthcare Diagnostics Inc. |
| Biohit SARS-CoV-2 IgM/IgG Antibody Test Kit | Senses IgM and IgG in human plasma, serum, and venipuncture whole blood; tests are constrained to medium and high complexity laboratories | Biohit Healthcare (Hefei) Co. Ltd. |
| DZ-Lite SARS-CoV-2 IgM CLIA Kit | Identifies IgM in serum along with plasma | Diazyme Laboratories, Inc. |
| SARS-CoV-2 IgG and IgM Combo Test | Identifies and distinguishes between IgM and IgG in human serum | BioCheck, Inc. |
| Tell Me Fast Novel Coronavirus (COVID-19) IgG/IgM Antibody Test | Senses and differentiates between IgM and IgG in venous whole blood, plasma, and serum | Biocan Diagnostics, Inc |
| Product | Detail | Sponsor |
|---|---|---|
| Elecsys IL-6 | To help in recognizing severe inflammatory response in COVID-19 confirmed cases and for analyzing the risk of intubation with mechanical ventilators | Roche Diagnostics |
| Product | Detail | Sponsor |
|---|---|---|
| Battelle Decontamination System | A well-suited respirator was reused and recycled up to twenty times utilizing the Battelle Decontamination System. It has been approved to ameliorate their actions in order to purify approximately 120,000 respirators every day through 12 satellite amenities that transfer data to the FDA | Battelle |
| Duke Decontamination System | Purifies compatible N95 or analogous respirators | Duke University Health System |
| STERIS STEAM Decon Cycle in AMSCO Medium Steam Sterilizers | Disinfects N95 respirators for single-user reprocess by HCP to stop acquaintance to pathogenic airborne particulates in shortage of face-filtering respirators (FFRs) owing to COVID-19 | STERIS |
| Stryker Sustainability Solutions VHP Decontamination System | Purifies N95 respirators for numerous user reuses via healthcare workers to avoid acquaintance to infectious airborne particles in shortage of face-filtering respirators as consequences of the pandemic | Stryker |
| Nova2200 | Disinfects N95 respirators for one person reuse for healthcare personnel (HCP) to stop contact to airborne pathogenic species in insufficient supply of face-filtering respirators (FFRs) owing to the SARS COV-19 | NovaSterilis, Inc. |
| Product | Detail | Sponsor |
|---|---|---|
| Space and Outlook Pumps (Infusion Pump) | A pump (infusion) for the transport of medicines into a nebulizer for the treatment of SARS CoV-2 patients of all ages | B. Braun |
The below table contains the data regarding the confirmed in vitro detection tests, which can be exploited for COVID-19 patients' management.
5 Management of Covid-19
Still various rational and distinguishing management strategies are critical even after the development of intensive detection methodologies to overcome the after effects from this pandemic situation.291 For intense cases, survivors of SARS COV-19 could have a higher risk of long-term lung infection. Moreover, some other serious problems such as serious interstitial, upper, and lower respiratory body infection can be possible.2925.1 Contact tracing-based SARS COV-19 detection
The isolation of symptomatic COVID-19 patients and tracing of contacts along with social distancing have been employed as an early measure to mitigate COVID-19 spread as outbreaks have grown. Reducing the disruption to the population while controlling infection, there is a dire need to recognize what combination of measures are required to decrease the transmission. Thus, in this area, the use of smartphone-based digital technologies have been executed effectively to cope with the spread of COVID-19.293 Recently, the application of contact tracing has been developed and employed as shown in Fig. 20. The main idea of this application is to substitute manual contact tracing with an immediate broadcast of signal toward central/main server. Principally, the information of virus patients has been transferred to the server that recommends risk-stratified quarantine along with robust social distancing to possible contacts. The tests of symptomatic patients have been requested through developed applications, while maintaining the anonymity of the infected patients. This developed software-based technology can be tuned to more informative infrastructure owing to its accessibility to almost every community member.294 Nevertheless, to achieve the complete exploitation of this technology, the tests number also needs to be increased. Meanwhile, privacy consideration also limits the application of this software-based technology.26 However, as the virus responsible for COVID-19 is continuously emerging, thus, such an improved developed system of contact tracing could play an important role in lowering the intensive impact generated by COVID-19.295 | ||
| Fig. 20 Schematic representation of contact tracing application. The figure has been reproduced from ref. 296 with permission from AAAS, copyright 2020. | ||
5.2 Use of pulse oximetry
Intense shallow breathing has been commonly observed after severe SARS-COVID-19 infection. The Pulse Oximeter (PO) can play an important role in monitoring different respiratory symptoms after SARS-COVID-19 without any extreme anxiety or stress.297 Infected persons should be given an observation diary with a pulse oximeter and proper guideline to self-monitor. Normally, in this practice, a daily reading would be taken by a neat and warm finger after an interval of 20 min. However, the important thing to keep in mind while taking the reading is that the applied device should be allowed to stabilize first and then the highest reading should be considered.298 Another major consideration is that commercially available O-saturated probes with a normal range of 92% should be used instead of smartphone apps.2995.3 Repercussions of the primary care Team
Most of the COVID-19 patients suffered from prolonged recovery without any special input through a holistic approach. Much of it could be achieved via community rehabilitation strategies and inter-professional services, which improve a person's self-management and harness the possible video and related technologies. Also, an informative as well as rehabilitation platforms should be established at the community level.300 In this time of uncertainty, the major role of these platforms should be the witness of “honoring the story” of a patient whose recovery was found to be unexpected.3016 Recent trends and future perspective
To date, no accurate treatment/therapy has been proved to circumvent the mortality from this pandemic completely. Researchers are continuously struggling to invent a universal treatment to reduce the severity of disease. Various COVID-19 vaccines across the world have been developed, as listed in Table 9. Experts consider Remdesivir as a significant and clear-cut vaccine for reducing the recovery time by blocking this virus. It could be considered as a magic bullet. Further, Favilavir has been considered as an antiviral drug by China. However, its sample size is still very small. Experts also consider that any new compound could take a decade from its initial discovery to reach the final marketplace; many compounds would never be able to make it that far.| No | Vaccine | Manufacturer | Brand Name |
|---|---|---|---|
| a NA = not applicable. | |||
| 1 | Janssen | Janssen Pharmaceutica | Johnson & Johnson |
| 2 | Sinopharm BIBP | China National Pharmaceutical Group | NA |
| 3 | Sputnik V | Russian Gamaleya Research Institute of Epidemiology and Microbiology | NA |
| 4 | CoronaVac | Sinovac Biotech | NA |
| 5 | Novavax | Novavax and the Coalition for Epidemic Preparedness Innovations | Covavax |
| 6 | Covaxin | Bharat Biotech in collaboration with the Indian Council of Medical Research-National Institute of Virology | Covaxin |
| 7 | Sputnik Light | Russian Gamaleya Research Institute of Epidemiology | NA |
| 8 | Convidecia | Chinese company CanSino Biologics and the Beijing Institute of Biotechnology of the Academy of Military Medical Sciences | NA |
| 9 | Sinopharm WIBP | China National Pharmaceutical Group | Sinopharm |
| 10 | Abdala | Center for Genetic Engineering and Biotechnology in Cuba | NA |
| 11 | EpiVacCorona | State Research Center of Virology and Biotechnology | NA |
| 12 | Zifivax | Chinese company Anhui Zhifei Longcom Biopharmaceutical | NA |
| 13 | Soberana O2 | Finlay Institute in Cuba | NA |
| 14 | QazCovid-in | Research Institute for Biological Safety Problems in Kazakhstan | QazVac |
| 15 | Minhai | Minhai Biotechnology Co. and Shenzhen Kangtai Biological Products Co. Ltd. in China | NA |
| 16 | CoviVac | Chumakov Centre at the![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Russian Academy of Sciences Russian Academy of Sciences |
NA |
| 17 | COVIran Barekat | Shifa Pharmed Industrial Co. in Iran | NA |
| 18 | Medigen | Taiwan's Medigen Vaccine Biologics and Dynavax Technologies | NA |
| 19 | FAKHRAVAC | Organization of Defensive Innovation and Research | MIVAC |
| 20 | COVAX-19 | Australian-based company Vaxine and Iran-based company CinnaGen | NA |
| 21 | Razi Cov Pars | Razi Vaccine and Serum Research Institute | NA |
| 22 | Turkovac | Health Institutes of Turkey and Erciyes University | NA |
| 23 | Corbevax | Indian biopharmacutical firm Biological E. Limited (BioE), the Baylor College of Medicine in Houston, United States, and American company Dynavax Technologies (DVAX) | NA |
This pandemic situation needs intensive actions in various areas from prevention to detection and then to final treatment. A cheap, simple, and fast testing methodology is required to detect the antibodies able to neutralize COVID-19. In this regard, some of the opportunities have to intervene in long as well as short terms to save lives. But before that, we need a guideline from science. Besides all these scientific efforts, everyone has to contribute to control this uncertain situation to lessen the risk assessment. At this stage, food, shelter, and healthcare for the needy sections of the community are massive challenges. Everyone including migrants and refugees need special attention.
Shaping the future as a global nation is anticipated. The education sector is one of the highly affected areas by SARS-COV-2. All the nations must have strategies to protect children's education. Teenagers are the major group affected by this pandemic-isolation. As the situation prolongs, the need for healthcare is increasing, which also includes the counseling of young generation missing out on school meals amid school closures.
7 Conclusion
The SARS-CoV-2 pandemic offers an incomparable global humanitarian and medical trial. Although this has impelled unprecedented growth in the development of therapeutics and vaccines, it has also highlighted the vulnerability of limited resources. The limited testing capacity along with the infrastructure to produce therapeutic drugs was largely absent to combat COVID-19 in the beginning. Efforts were made to exploit available resources to manufacture cheap and robust diagnostic methodologies. The review discusses in detail the standard diagnostic methods along with their pros and cons for the detection of COVID-19 as well as to avoid consequent secondary blow-out. Nucleic acid plus protein-based diagnosis along with biomarkers to evaluate the disease severity have been employed at the start to sense SARS CoV-2. Furthermore, as research proceeded, non-invasive sensing protocols were explored for wastewater surveillance for early detection. Similarly, nanoscale tools have also been discussed to examine viral morphology. Although various nanobiosystems have clearly revealed the viral inhibitory properties of COVID-19, still, most studies have shown the significance of initial findings of the virus. The perseverance of nanosystems has not fully been evaluated, regardless of their endpoint applications including therapeutics, surface protection, and water disinfection. Further, the administration of investigative nanobiosystems to animal models should be validated along with its development. This validation would lead to subsequent antiviral coatings for medical devices. The review was an effort to provides insights into the diagnostics and prognostic approaches for current and future pandemics.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.References
- W. H. Organization, https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en 2020.
- G. Das, S. Ghosh, S. Garg, S. Ghosh, A. Jana, R. Samat, N. Mukherjee, R. Roy and S. Ghosh, RSC Adv., 2020, 10, 28243–28266 RSC.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, H.-D. Chen, J. Chen, Y. Luo, H. Guo, R.-D. Jiang, M.-Q. Liu, Y. Chen, X.-R. Shen, X. Wang, X.-S. Zheng, K. Zhao, Q.-J. Chen, F. Deng, L.-L. Liu, B. Yan, F.-X. Zhan, Y.-Y. Wang, G.-F. Xiao and Z.-L. Shi, Nature, 2020, 579, 270–273 CrossRef CAS PubMed.
- A. Linet, M. M. Joseph, M. Haritha, K. Shamna, S. Varughese, P. S. Devi, C. H. Suresh, K. K. Maiti and I. Neogi, New J. Chem., 2021, 45, 17777–17781 RSC.
- W.-j. Guan, Z.-y. Ni, Y. Hu, W.-h. Liang, C.-q. Ou, J.-x. He, L. Liu, H. Shan, C.-l. Lei and D. S. Hui, N. Engl. J. Med., 2020, 382, 1708–1720 CrossRef CAS PubMed.
- Z. Wu and J. M. McGoogan, JAMA, 2020, 323, 1239–1242 CrossRef CAS PubMed.
- K. Payne, P. Kenny, J. M. Scovell, K. Khodamoradi and R. Ramasamy, Sexual Medicine Reviews, 2020, 8, 518–530 CrossRef PubMed.
- C. Wang, P. W. Horby, F. G. Hayden and G. F. Gao, Lancet, 2020, 395, 470–473 CrossRef CAS.
- T. P. Velavan and C. G. Meyer, Trop. Med. Int. Health, 2020, 25(3), 278 CrossRef CAS PubMed.
- Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying, P. Pang and W. Ji, Radiology, 2020, 200432 Search PubMed.
- G. Das, N. Mukherjee and S. Ghosh, ACS Chem. Neurosci., 2020, 11, 1206–1209 CrossRef CAS PubMed.
- A. Tahamtan and A. Ardebili, Expert Rev. Mol. Diagn., 2020, 20(5), 453–454 CrossRef CAS PubMed.
- C. Bundschuh, M. Egger, K. Wiesinger, C. Gabriel, M. Clodi, T. Mueller and B. Dieplinger, Clin. Chim. Acta, 2020, 509, 18–21 CrossRef PubMed.
- R. Zhao, M. Li, H. Song, J. Chen, W. Ren, Y. Feng, G. F. Gao, J. Song, Y. Peng and B. Su, Clin. Infect. Dis., 2020, 71(16), 2066–2072 CrossRef CAS PubMed.
- M. Yuan, N. C. Wu, X. Zhu, C. D. Lee, R. T. Y. So, H. Lv, C. K. P. Mok and I. A. Wilson, Science, 2020, 368, 630–633 CrossRef CAS PubMed.
- G. Ponti, M. Maccaferri, C. Ruini, A. Tomasi and T. Ozben, Crit. Rev. Clin. Lab. Sci., 2020, 1–11 Search PubMed.
- L. Tan, Q. Wang, D. Zhang, J. Ding, Q. Huang, Y.-Q. Tang, Q. Wang and H. Miao, Signal Transduction Targeted Ther., 2020, 5, 1–3 CrossRef PubMed.
- C. Mao, A. Liu and B. Cao, Angew. Chem., Int. Ed., 2009, 48, 6790–6810 CrossRef CAS PubMed.
- Y. Saylan, Ö. Erdem, S. Ünal and A. Denizli, Biosensors, 2019, 9, 65 CrossRef CAS PubMed.
- A. Wilhelm, M. Widera, K. Grikscheit, T. Toptan, B. Schenk, C. Pallas, M. Metzler, N. Kohmer, S. Hoehl, F. A. Helfritz, T. Wolf, U. Goetsch and S. Ciesek, medRxiv, 2021 DOI:10.1101/2021.12.07.21267432.
- L. Zhang, Q. Li, Z. Liang, T. Li, S. Liu, Q. Cui, J. Nie, Q. Wu, X. Qu and W. Huang, Emerging Microbes Infect., 2021, 1–11 Search PubMed.
- J. E. Hollander and B. G. Carr, N. Engl. J. Med., 2020, 382, 1679–1681 CrossRef CAS PubMed.
- S. Assari, Hosp Pract Res, 2020, 5, 81–86 CrossRef PubMed.
- A. J. Kucharski, P. Klepac, A. Conlan, S. M. Kissler, M. Tang, H. Fry, J. Gog, J. Edmunds and C. C.-W. Group, MedRxiv, 2020 DOI:10.1101/2020.06.05.20123141.
- H. Cho, D. Ippolito and Y. W. Yu, arXiv preprint arXiv: 2003.11511, 2020.
- N. Ahmed, R. A. Michelin, W. Xue, S. Ruj, R. Malaney, S. S. Kanhere, A. Seneviratne, W. Hu, H. Janicke and S. K. Jha, IEEE Access, 2020, 8, 134577–134601 Search PubMed.
- M. M. Joseph, A. N. Ramya, V. M. Vijayan, J. B. Nair, B. T. Bastian, R. K. Pillai, S. T. Therakathinal and K. K. Maiti, Small, 2020, 16, 2003309 CrossRef CAS PubMed.
- C. C.-Y. Yip, C.-C. Ho, J. F.-W. Chan, K. K.-W. To, H. S.-Y. Chan, S. C.-Y. Wong, K.-H. Leung, A. Y.-F. Fung, A. C.-K. Ng and Z. Zou, Int. J. Mol. Sci., 2020, 21, 2574 CrossRef CAS PubMed.
- C. Wang, Z. Liu, Z. Chen, X. Huang, M. Xu, T. He and Z. Zhang, J. Med. Virol., 2020, 92, 667–674 CrossRef CAS PubMed.
- J.-M. Kim, Y.-S. Chung, H. J. Jo, N.-J. Lee, M. S. Kim, S. H. Woo, S. Park, J. W. Kim, H. M. Kim and M.-G. Han, Osong Public Health and Research Perspectives, 2020, 11, 3 CrossRef PubMed.
- R. Sah, A. J. Rodriguez-Morales, R. Jha, D. K. Chu, H. Gu, M. Peiris, A. Bastola, B. K. Lal, H. C. Ojha and A. A. Rabaan, Microbiol. Resour. Announce., 2020, 9(11), e00169-20 CrossRef PubMed.
- D. Batlle, J. Wysocki and K. Satchell, Clin. Sci., 2020, 134, 543–545 CrossRef CAS PubMed.
- Y. Zhou, Y. Hou, J. Shen, Y. Huang, W. Martin and F. Cheng, Cell Discovery, 2020, 6, 1–18 Search PubMed.
- Winners and Losers of EU Integration, Policy Issues for Central and Eastern Europe, ed. H. Tang, World Bank Publications, 2000 Search PubMed.
- J. Zou, J. Yin, L. Fang, M. Yang, T. Wang, W. Wu, M. A. Bellucci and P. Zhang, J. Chem. Inf. Model., 2020, 60(12), 5794–5802 CrossRef CAS PubMed.
- N. Rabiee, M. Bagherzadeh, A. Ghasemi, H. Zare, S. Ahmadi, Y. Fatahi, R. Dinarvand, M. Rabiee, S. Ramakrishna and M. Shokouhimehr, Int. J. Mol. Sci., 2020, 21, 5126 CrossRef CAS PubMed.
- L. Zhang and H. Guo, Advances in Biomarker Sciences and Technology, 2020, 2, 1–23 CrossRef CAS PubMed.
- W. Ye, G. Chen, X. Li, X. Lan, C. Ji, M. Hou, D. Zhang, G. Zeng, Y. Wang, C. Xu, W. Lu, R. Cui, Y. Cai, H. Huang and L. Yang, Respir. Res., 2020, 21, 169 CrossRef CAS PubMed.
- J. Gong, H. Dong, S. Q. Xia, Y. Z. Huang, D. Wang, Y. Zhao, W. Liu, S. Tu, M. Zhang and Q. Wang, medRxiv, 2020 DOI:10.1101/2020.02.25.20025643.
- W. Ling, Medecine et maladies infectieuses, 2020, 50(4), 332–334 CrossRef PubMed.
- C. Qin, L. Zhou, Z. Hu, S. Zhang, S. Yang, Y. Tao, C. Xie, K. Ma, K. Shang and W. Wang, Clin. Infect. Dis., 2020, 71(15), 762–768 CrossRef CAS PubMed.
- L. Liu, Q. Huang, Z. I. Tanveer, K. Jiang, J. Zhang, H. Pan, L. Luan, X. Liu, Z. Han and Y. Wu, Sens. Actuators, B, 2020, 302, 127212 CrossRef CAS.
- W. Ji, G. Bishnu, Z. Cai and X. Shen, medRxiv, 2020 DOI:10.1101/2020.03.10.20033613.
- C. Tan, Y. Huang, F. Shi, K. Tan, Q. Ma, Y. Chen, X. Jiang and X. Li, J. Med. Virol., 2020, 92(7), 856–862 CrossRef CAS PubMed.
- G. Wang, C. Wu, Q. Zhang, F. Wu, B. Yu, J. Lv, Y. Li, T. Li, S. Zhang, C. Wu, G. Wu and Y. Zhong, Open Forum Infect. Dis., 2020, 7(5), ofaa153 CrossRef PubMed.
- L. A. Potempa, I. M. Rajab, P. C. Hart, J. Bordon and R. Fernandez-Botran, Am. J. Trop. Med. Hyg., 2020, 103, 561–563 CrossRef PubMed.
- S. Wan, Q. Yi, S. Fan, J. Lv, X. Zhang, L. Guo, C. Lang, Q. Xiao, K. Xiao and Z. Yi, MedRxiv, 2020 DOI:10.1101/2020.02.10.20021832.
- Y. He, Infect. Microbes Dis., 2020 Search PubMed.
- M. Kermali, R. K. Khalsa, K. Pillai, Z. Ismail and A. Harky, Life Sci., 2020, 254, 117788 CrossRef CAS PubMed.
- P. Yang, Y. Ding, Z. Xu, R. Pu, P. Li, J. Yan, J. Liu, F. Meng, L. Huang and L. Shi, Medrxiv, 2020 DOI:10.1101/2020.02.28.20028068.
- R. Antiochia, Biosens. Bioelectron., 2021, 173, 112777 CrossRef CAS PubMed.
- N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu and Y. Wei, Lancet, 2020, 395, 507–513 CrossRef CAS.
- E. A. Coomes and H. Haghbayan, Rev. Med. Virol., 2020, 30(6), 1–9 CrossRef CAS PubMed.
- P. Conti, G. Ronconi, A. Caraffa, C. Gallenga, R. Ross, I. Frydas and S. Kritas, J. Biol. Regul. Homeostatic Agents, 2020, 34, 1 Search PubMed.
- L. Chen, H. Liu, W. Liu, J. Liu, K. Liu, J. Shang, Y. Deng and S. Wei, Chin. J. Tuberc. Respir. Dis., 2020, 43, E005 CAS.
- Z. S. Ulhaq and G. V. Soraya, Medecine et maladies infectieuses, 2020, 50(4), 382 CrossRef PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brünink, J. Schneider and M. L. Schmidt, Eurosurveillance, 2020, 25, 2000045 Search PubMed.
- D. Wang, B. Hu, C. Hu, F. Zhu, X. Liu, J. Zhang, B. Wang, H. Xiang, Z. Cheng and Y. Xiong, Jama, 2020, 323, 1061–1069 CrossRef CAS PubMed.
- T. T. Yip, J. W. Chan, W. C. Cho, T.-T. Yip, Z. Wang, T.-L. Kwan, S. C. Law, D. N. Tsang, J. K. Chan and K.-C. Lee, Clin. Chem., 2005, 51, 47–55 CrossRef CAS PubMed.
- J. Osman, J. Lambert, M. Templé, F. Devaux, R. Favre, C. Flaujac, D. Bridoux, S. Marque-Juillet, F. Bruneel and F. Mignon, Br. J. Haematol., 2020, 190(5), 718–722 CrossRef CAS PubMed.
- T. Yang, Y.-C. Wang, C.-F. Shen and C.-M. Cheng, Diagnostics, 2020, 10(3), 165 CrossRef CAS PubMed.
- D. Ferrari, A. Motta, M. Strollo, G. Banfi and M. Locatelli, Clin. Chem. Lab. Med., 2020, 58(7), 1095–1099 CrossRef CAS PubMed.
- M. Eller and D. Williams, FESCC, 2011, 49, 403 CAS.
- Y. Xiong, D. Sun, Y. Liu, Y. Fan, L. Zhao, X. Li and W. Zhu, Invest. Radiol., 2020, 55(6), 332–339 CrossRef CAS PubMed.
- B. M. Henry, M. H. S. De Oliveira, S. Benoit, M. Plebani and G. Lippi, Clin. Chem. Lab. Med., 2020, 58, 1021–1028 CrossRef CAS PubMed.
- Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K. S. Leung, E. H. Lau and J. Y. Wong, N. Engl. J. Med., 2020 Search PubMed.
- M. H. Mahnashi, Microchim. Acta, 2020, 187, 1–8 CrossRef PubMed.
- Y. Han, H. Zhang, S. Mu, W. Wei, C. Jin, C. Tong, Z. Song, Y. Zha, Y. Xue and G. Gu, Aging, 2020, 12, 11245 CrossRef CAS PubMed.
- L. Zhang, Y. Long, H. Xiao, J. Yang, P. Toulon and Z. Zhang, International Journal of Laboratory Hematology, 2018, 40, 503–507 CrossRef CAS PubMed.
- N. Tang, D. Li, X. Wang and Z. Sun, J. Thromb. Haemostasis, 2020, 18, 844–847 CrossRef CAS PubMed.
- L. Zhang, X. Yan, Q. Fan, H. Liu, X. Liu, Z. Liu and Z. Zhang, J. Thromb. Haemostasis, 2020, 18, 1324–1329 CrossRef CAS PubMed.
- E. B. Milbrandt, M. C. Reade, M. Lee, S. L. Shook, D. C. Angus, L. Kong, M. Carter, D. M. Yealy, J. A. Kellum and G. Investigators, Mol. Med., 2009, 15, 438–445 CAS.
- M. Sakka, J. M. Connors, G. Hékimian, I. Martin-Toutain, B. Crichi, I. Colmegna, D. Bonnefont-Rousselot, D. Farge and C. Frere, Journal de Médecine Vasculaire, 2020, 45(5), 268–274 CrossRef CAS PubMed.
- Y. Yao, J. Cao, Q. Wang, Q. Shi, K. Liu, Z. Luo, X. Chen, S. Chen, K. Yu and Z. Huang, J. Intensive Care, 2020, 8, 1–11 CrossRef CAS PubMed.
- B. Yu, X. Li, J. Chen, M. Ouyang, H. Zhang, X. Zhao, L. Tang, Q. Luo, M. Xu and L. Yang, J. Thromb. Thrombolysis, 2020, 1–10 Search PubMed.
- E. J. Favaloro and J. Thachil, Clin. Chem. Lab. Med., 2020, 58(8), 1191–1199 CrossRef CAS PubMed.
- D. Liao, F. Zhou, L. Luo, M. Xu, H. Wang, J. Xia, Y. Gao, L. Cai, Z. Wang and P. Yin, Lancet Haematology, 2020, 7(9), e671–e678 CrossRef PubMed.
- G. Lippi, M. Plebani and B. M. Henry, Clin. Chim. Acta, 2020, 506, 145–148 CrossRef CAS PubMed.
- Y. Liu, W. Sun, Y. Guo, L. Chen, L. Zhang, S. Zhao, D. Long and L. Yu, Platelets, 2020, 31, 490–496 CrossRef CAS PubMed.
- X. Zhao, K. Wang, P. Zuo, Y. Liu, M. Zhang, S. Xie, H. Zhang, X. Chen and C. Liu, EPMA J., 2020, 1 Search PubMed.
- X. Yang, Q. Yang, Y. Wang, Y. Wu, J. Xu, Y. Yu and Y. Shang, J. Thromb. Haemostasis, 2020, 18, 1469–1472 CrossRef CAS PubMed.
- S. Q. Jiang, Q. F. Huang, W. M. Xie, C. Lv and X. Q. Quan, Br. J. Haematol., 2020, 190(1), e29–e33 CrossRef CAS PubMed.
- Y. Zhang, X. Zeng, Y. Jiao, Z. Li, Q. Liu, J. Ye and M. Yang, Thromb. Res., 2020, 193, 110–115 CrossRef CAS PubMed.
- J. Bennett, C. Brown, M. Rouse, M. Hoffmann and Z. Ye, Cureus, 2020, 12(7) DOI:10.7759/cureus.9083.
- M. Koupenova, Res. Pract. Thromb. Haemostasis, 2020, 4(5), 737–740 CrossRef CAS PubMed.
- I. H. Khan, S. A. Zahra, S. Zaim and A. Harky, J. Card. Surg., 2020, 35(6), 1287–1294 CrossRef PubMed.
- T. R. Khashkhusha, J. S. K. Chan and A. Harky, J. Card. Surg., 2020, 1172–1173 CrossRef PubMed.
- R. O. Bonow, G. C. Fonarow, P. T. O'Gara and C. W. Yancy, JAMA Cardiol., 2020, 5(7), 751–753 CrossRef PubMed.
- G. Tersalvi, M. Vicenzi, D. Calabretta, L. Biasco, G. Pedrazzini and D. Winterton, J. Card. Failure, 2020, 26(6), 470–475 CrossRef PubMed.
- R. M. Inciardi, L. Lupi, G. Zaccone, L. Italia, M. Raffo, D. Tomasoni, D. S. Cani, M. Cerini, D. Farina and E. Gavazzi, JAMA Cardiol., 2020, 5(7), 819–824 CrossRef PubMed.
- Y.-Y. Zheng, Y.-T. Ma, J.-Y. Zhang and X. Xie, Nat. Rev. Cardiol., 2020, 17, 259–260 CrossRef CAS PubMed.
- A. R. Chapman, A. Bularga and N. L. Mills, Circulation, 2020, 141, 1733–1735 CrossRef CAS PubMed.
- F. Zhou, T. Yu, R. Du, G. Fan, Y. Liu, Z. Liu, J. Xiang, Y. Wang, B. Song and X. Gu, Lancet, 2020, 395(10229), 1054–1062 CrossRef CAS.
- C. A. Labarrere, J. Woods, J. Hardin, G. Campana, M. Ortiz, B. Jaeger, B. Reichart, J. Bonnin, A. Currin and S. Cosgrove, Am. J. Transplant., 2011, 11, 528–535 CrossRef CAS PubMed.
- Y. Sandoval, J. L. Januzzi Jr and A. S. Jaffe, J. Am. Coll. Cardiol., 2020, 76(10), 1244–1258 CrossRef CAS PubMed.
- B. Siripanthong, S. Nazarian, D. Muser, R. Deo, P. Santangeli, M. Y. Khanji, L. T. Cooper Jr and C. A. A. Chahal, Heart Rhythm, 2020, 17(9), 1463–1471 CrossRef PubMed.
- B. M. Henry and G. Lippi, Int. Urol. Nephrol., 2020, 1–2 CAS.
- J. Xiang, J. Wen, X. Yuan, S. Xiong, X. Zhou, C. Liu and X. Min, medRxiv, 2020 DOI:10.1101/2020.03.19.20034447.
- Y. Cheng, R. Luo, K. Wang, M. Zhang, Z. Wang, L. Dong, J. Li, Y. Yao, S. Ge and G. Xu, Kidney Int., 2020, 97(5), 829–838 CrossRef CAS PubMed.
- H. Zhou, Z. Zhang, H. Fan, J. Li, M. Li, Y. Dong, W. Guo, L. Lin, Z. Kang and T. Yu, Diagnostics, 2022, 12, 602 CrossRef.
- M. A. Lim, R. Pranata, I. Huang, E. Yonas, A. Y. Soeroto and R. Supriyadi, Can. J. Kidney Health Dis., 2020, 7 DOI:10.1177/2054358120938573.
- C. Sardu, J. Gambardella, M. B. Morelli, X. Wang, R. Marfella and G. Santulli, J. Clin. Med., 2020, 9, 1417 CrossRef CAS PubMed.
- Q. Zhao, M. Meng, R. Kumar, Y. Wu, J. Huang, Y. Deng, Z. Weng and L. Yang, Int. J. Infect. Dis., 2020, 96, 131–135 CrossRef CAS PubMed.
- J. Liu, S. Li, J. Liu, B. Liang, X. Wang, H. Wang, W. Li, Q. Tong, J. Yi and L. Zhao, EBioMedicine, 2020, 102763 CrossRef PubMed.
- B. Zhang, X. Zhou, C. Zhu, Y. Song, F. Feng, Y. Qiu, J. Feng, Q. Jia, Q. Song and B. Zhu, Front. Mol. Biosci., 2020, 7, 157 CrossRef CAS PubMed.
- W. Yang and F. Yan, Radiology, 2020, 295, E3 CrossRef PubMed.
- A. T. Xiao, Y. X. Tong and S. Zhang, J. Med. Virol., 2020, 92(10), 1755–1756 CrossRef CAS PubMed.
- F. Chua, D. Armstrong-James, S. R. Desai, J. Barnett, V. Kouranos, O. M. Kon, R. José, R. Vancheeswaran, M. R. Loebinger and J. Wong, Lancet Respir. Med., 2020, 8, 438–440 CrossRef CAS PubMed.
- T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun and L. Xia, Radiology, 2020, 200642 Search PubMed.
- H. Shi, X. Han, N. Jiang, Y. Cao, O. Alwalid, J. Gu, Y. Fan and C. Zheng, Lancet Infect. Dis., 2020, 20(4), 425–434 CrossRef CAS PubMed.
- Y. Zheng, X. Li, C. Pi, H. Song, B. Gao, P. K. Chu and K. Huo, FlatChem, 2020, 19, 100149 CrossRef CAS.
- L. Peñarrubia, M. Ruiz, R. Porco, S. N. Rao, M. Juanola-Falgarona, D. Manissero, M. López-Fontanals and J. Pareja, Int. J. Infect. Dis., 2020, 97, 225–229 CrossRef PubMed.
- I. Santiago, ChemBioChem, 2020, 21(20), 2880–2889 CrossRef CAS PubMed.
- Y. Zhou, Z. Wan, S. Yang, Y. Li, M. Li, B. Wang, Y. Hu, X. Xia, X. Jin and N. Yu, Front. Microbiol., 2019, 10, 1056 CrossRef PubMed.
- X. Li, L. Wang, S. Yan, F. Yang, L. Xiang, J. Zhu, B. Shen and Z. Gong, Int. J. Infect. Dis., 2020, 94, 128–132 CrossRef CAS PubMed.
- R. Lu, X. Wu, Z. Wan, Y. Li, X. Jin and C. Zhang, Int. J. Mol. Sci., 2020, 21, 2826 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- Y. Mori, K. Nagamine, N. Tomita and T. Notomi, Biochem. Biophys. Res. Commun., 2001, 289, 150–154 CrossRef CAS PubMed.
- P. Yu, J. Zhu, Z. Zhang and Y. Han, J. Infect. Dis., 2020, 221, 1757–1761 CrossRef CAS PubMed.
- G.-S. Park, K. Ku, S.-H. Baek, S.-J. Kim, S. I. Kim, B.-T. Kim and J.-S. Maeng, J. Mol. Diagn., 2020, 22(6), 729–735 CrossRef CAS PubMed.
- L. Yu, S. Wu, X. Hao, X. Li, X. Liu, S. Ye, H. Han, X. Dong, X. Li and J. Li, medRxiv, 2020 DOI:10.1101/2020.02.20.20025874.
- Y. H. Baek, J. Um, K. J. C. Antigua, J.-H. Park, Y. Kim, S. Oh, Y.-i. Kim, W.-S. Choi, S. G. Kim and J. H. Jeong, Emerging Microbes Infect., 2020, 9, 998–1007 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C.-Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- L. Yu, et al., Clin. Chem., 2020, 66(7), 975–977 CrossRef PubMed.
- L. E. Lamb, S. N. Bartolone, E. Ward and M. B. Chancellor, PLoS One, 2020, 15, e0234682 CrossRef CAS PubMed.
- T. Nguyen, D. Duong Bang and A. Wolff, Micromachines, 2020, 11, 306 CrossRef PubMed.
- L. J. Carter, L. V. Garner, J. W. Smoot, Y. Li, Q. Zhou, C. J. Saveson, J. M. Sasso, A. C. Gregg, D. J. Soares and T. R. Beskid, ACS Cent. Sci., 2020, 591–605 CrossRef CAS PubMed.
- S. Chekani-Azar, E. Gharib Mombeni, M. Birhan and M. Yousefi, J. Life Sci. Biomed., 2020, 10, 01–09 CrossRef.
- M. Dara and M. Talebzadeh, Avicenna J. Med. Biotechnol., 2020, 12, 201–202 Search PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados and A. Sotomayor-Gonzalez, Nat. Biotechnol., 2020, 1–5 Search PubMed.
- A. J. Rodriguez-Morales, J. A. Cardona-Ospina, E. Gutiérrez-Ocampo, R. Villamizar-Peña, Y. Holguin-Rivera, J. P. Escalera-Antezana, L. E. Alvarado-Arnez, D. K. Bonilla-Aldana, C. Franco-Paredes and A. F. Henao-Martinez, Travel Medicine and Infectious Disease, 2020, 101623 CrossRef PubMed.
- M. Azhar, R. Phutela, A. H. Ansari, D. Sinha, N. Sharma, M. Kumar, M. Aich, S. Sharma, K. Singhal and H. Lad, Biosens. Bioelectron., 2021, 183, 113207 CrossRef CAS PubMed.
- M. R. Rahman, M. A. Hossain, M. Mozibullah, F. Al Mujib, A. Afrose, M. Shahed-Al-Mahmud and M. A. I. Apu, Biomed. Pharmacother., 2021, 140, 111772 CrossRef CAS PubMed.
- A. L. Marca, et al., Reprod. BioMed. Online, 2020, 41(3), 483–499 CrossRef PubMed.
- J. Xiang, M. Yan, H. Li, T. Liu, C. Lin, S. Huang and C. Shen, medRxiv, 2020 DOI:10.1101/2020.02.27.20028787.
- Y. Bai, L. Yao, T. Wei, F. Tian, D.-Y. Jin, L. Chen and M. Wang, JAMA, 2020, 323, 1406–1407 CrossRef CAS PubMed.
- Q. X. Long, et al., medRxiv, 2020 DOI:10.1101/2020.03.18.20038018.
- N. M. Okba, M. A. Müller, W. Li, C. Wang, C. H. GeurtsvanKessel, V. M. Corman, M. M. Lamers, R. S. Sikkema, E. de Bruin and F. D. Chandler, Emerging Infect. Dis., 2020, 26, 1478 CrossRef CAS PubMed.
- L. Zhang, R. Pang, X. Xue, J. Bao, S. Ye, Y. Dai, Y. Zheng, Q. Fu, Z. Hu and Y. Yi, Aging, 2020, 12, 6536 CrossRef CAS PubMed.
- Z. Chen, Z. Zhang, X. Zhai, Y. Li, L. Lin, H. Zhao, L. Bian, P. Li, L. Yu and Y. Wu, Anal. Chem., 2020, 92, 7226–7231 CrossRef CAS PubMed.
- R. A. Perera, C. K. Mok, O. T. Tsang, H. Lv, R. L. Ko, N. C. Wu, M. Yuan, W. S. Leung, J. M. Chan and T. S. Chik, Eurosurveillance, 2020, 25, 2000421 CrossRef PubMed.
- Q. Wang, Q. Du, B. Guo, D. Mu, X. Lu, Q. Ma, Y. Guo, L. Fang, B. Zhang and G. Zhang, J. Clin. Microbiol., 2020, 58(6), e00375-20 CrossRef PubMed.
- B. Tang, F. Xia, S. Tang, N. L. Bragazzi, Q. Li, X. Sun, J. Liang, Y. Xiao and J. Wu, Int. J. Infect. Dis., 2020, 96, 636–647 CrossRef CAS PubMed.
- L. Guo, L. Ren, S. Yang, M. Xiao, D. Chang, F. Yang, C. S. Dela Cruz, Y. Wang, C. Wu, Y. Xiao, L. Zhang, L. Han, S. Dang, Y. Xu, Q.-W. Yang, S.-Y. Xu, H.-D. Zhu, Y.-C. Xu, Q. Jin, L. Sharma, L. Wang and J. Wang, Clin. Infect. Dis., 2020, 71(15), 778–785 CrossRef CAS PubMed.
- I. Cassaniti, F. Novazzi, F. Giardina, F. Salinaro, M. Sachs, S. Perlini, R. Bruno, F. Mojoli and F. Baldanti, J. Med. Virol., 2020, 92(10), 1724–1727 CrossRef CAS PubMed.
- A. F. S. Laureano and M. Riboldi, JBRA assisted reproduction, 2020, 24, 340 Search PubMed.
- G. Eick, S. S. Urlacher, T. W. McDade, P. Kowal and J. J. Snodgrass, Biodemography and social biology, 2016, 62, 222–233 CrossRef PubMed.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu and W. Chen, J. Med. Virol., 2020, 92(9), 1518–1524 CrossRef CAS PubMed.
- A. J. Jääskeläinen, E. Kekäläinen, H. Kallio-Kokko, L. Mannonen, E. Kortela, O. Vapalahti, S. Kurkela and M. Lappalainen, Eurosurveillance, 2020, 25, 2000603 CrossRef PubMed.
- M. Feng, J. Chen, J. Xun, R. Dai, W. Zhao, H. Lu, J. Xu, L. Chen, G. Sui and X. Cheng, ACS Sens., 2020, 5(8), 2331–2337 CrossRef CAS PubMed.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Analyst, 2020, 145, 2828–2840 RSC.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu, S. Li, R. Sun, Y. Wang, B. Hu and W. Chen, J. Med. Virol., 2020, 92, 1518–1524 CrossRef CAS PubMed.
- R. M. Pallares and R. J. Abergel, ACS Pharmacol. Transl. Sci., 2021, 4(1), 1–7 CrossRef CAS PubMed.
- Y. Orooji, H. Sohrabi, N. Hemmat, F. Oroojalian, B. Baradaran, A. Mokhtarzadeh, M. Mohaghegh and H. Karimi-Maleh, Nano-Micro Lett., 2021, 13, 1–30 CrossRef CAS PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, Y. M. Kamil, W. M. E. M. M. Daniyal, A. R. Sadrolhosseini and M. A. Mahdi, Sci. Rep., 2020, 10, 1–15 CrossRef PubMed.
- W. M. E. M. M. Daniyal, Y. W. Fen, S. Saleviter, N. Chanlek, H. Nakajima, J. Abdullah and N. A. Yusof, Polymers, 2021, 13, 478 CrossRef CAS PubMed.
- J. Lukose, S. Chidangil and S. D. George, Biosens. Bioelectron., 2021, 113004 CrossRef CAS PubMed.
- V. Shvalya, G. Filipič, D. Vengust, J. Zavašnik, M. Modic, I. Abdulhalim and U. Cvelbar, Appl. Surf. Sci., 2020, 517, 146205 CrossRef CAS.
- B. Mondal and S. Zeng, Nanophotonics, 2021, 21–48 Search PubMed.
- A. M. Shrivastav, U. Cvelbar and I. Abdulhalim, Commun. Biol., 2021, 4, 70 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- P. Moitra, M. Alafeef, K. Dighe, M. Frieman and D. Pan, ACS Nano, 2020, 14(6), 7617–7627 CrossRef CAS PubMed.
- H. Ma, X. X. Han and B. Zhao, TrAC, Trends Anal. Chem., 2020, 131, 116019 CrossRef CAS.
- Y. Guo, M. Girmatsion, H.-W. Li, Y. Xie, W. Yao, H. Qian, B. Abraha and A. Mahmud, Crit. Rev. Food Sci. Nutr., 2021, 61, 3555–3568 CrossRef CAS PubMed.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, Talanta, 2020, 206, 120201 CrossRef CAS PubMed.
- S. Yadav, S. Senapati, D. Desai, S. Gahlaut, S. Kulkarni and J. P. Singh, Colloids Surf., B, 2021, 198, 111477 CrossRef CAS PubMed.
- C. Carlomagno, D. Bertazioli, A. Gualerzi, S. Picciolini, P. I. Banfi, A. Lax, E. Messina, J. Navarro, L. Bianchi, A. Caronni, F. Marenco, S. Monteleone, C. Arienti and M. Bedoni, Sci. Rep., 2021, 11, 4943 CrossRef CAS PubMed.
- J. Reguera, J. Langer, D. Jiménez de Aberasturi and L. M. Liz-Marzán, Chem. Soc. Rev., 2017, 46, 3866–3885 RSC.
- X. Zhou, D. Wu, Z. Jin, X. Song, X. Wang and S. L. Suib, J. Mater. Sci., 2020, 55, 16374–16384 CrossRef CAS.
- A. C. Hernández-Arteaga, J. de Jesús Zermeño-Nava, M. U. Martínez-Martínez, A. Hernández-Cedillo, H. J. Ojeda-Galván, M. José-Yacamán and H. R. Navarro-Contreras, Arch. Med. Res., 2019, 50, 105–110 CrossRef PubMed.
- M. M. Joseph, N. Narayanan, J. B. Nair, V. Karunakaran, A. N. Ramya, P. T. Sujai, G. Saranya, J. S. Arya, V. M. Vijayan and K. K. Maiti, Biomaterials, 2018, 181, 140–181 CrossRef CAS PubMed.
- C. Zhang, T. Zheng, H. Wang, W. Chen, X. Huang, J. Liang, L. Qiu, D. Han and W. Tan, Anal. Chem., 2021, 93, 3325–3330 CrossRef CAS PubMed.
- J. E. Sanchez, S. A. Jaramillo, E. Settles, J. J. Velazquez Salazar, A. Lehr, J. Gonzalez, C. Rodríguez Aranda, H. R. Navarro-Contreras, M. O. Raniere, M. Harvey, D. M. Wagner, A. Koppisch, R. Kellar, P. Keimb and M. J. Yacaman, RSC Adv., 2021, 11, 25788–25794 RSC.
- J.-H. Lee, B.-K. Oh and J.-W. Choi, Biosens. Bioelectron., 2013, 49, 531–535 CrossRef CAS PubMed.
- D. Kurouski, T. Postiglione, T. Deckert-Gaudig, V. Deckert and I. K. Lednev, Analyst, 2013, 138, 1665–1673 RSC.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park and H. G. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- T. H. V. Nguyen, J. Lichiere, B. Canard, N. Papageorgiou, S. Attoumani, F. Ferron and B. Coutard, Acta Crystallogr., Sect. D: Struct. Biol., 2019, 75, 8–15 CrossRef CAS PubMed.
- S. Ricagno, B. Coutard, S. Grisel, N. Brémond, K. Dalle, F. Tocque, V. Campanacci, J. Lichière, V. Lantez and C. Debarnot, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2006, 62, 409–411 CrossRef CAS PubMed.
- M. Luo, Crystallogr. Rev., 2015, 21, 103–121 CrossRef.
- C. S. Goldsmith and S. E. Miller, Clin. Microbiol. Rev., 2009, 22, 552–563 CrossRef PubMed.
- F. Ohnesorge, J. Hörber, W. Häberle, C. Czerny, D. Smith and G. Binnig, Biophys. J., 1997, 73, 2183–2194 CrossRef CAS PubMed.
- Y. G. Kuznetsov and A. McPherson, Microbiol. Mol. Biol. Rev., 2011, 75, 268–285 CrossRef CAS PubMed.
- K. R. Richert-Pöggeler, K. Franzke, K. Hipp and R. G. Kleespies, Front. Microbiol., 2019, 9, 3255 CrossRef PubMed.
- Y. G. Kuznetsov, J. Victoria, W. Robinson and A. McPherson, J. Virol., 2003, 77, 11896–11909 CrossRef CAS PubMed.
- M. Laue, in Methods in cell biology, Elsevier, 2010, vol. 96, pp. 1–20 Search PubMed.
- C. E. Conrad, S. Basu, D. James, D. Wang, A. Schaffer, S. Roy-Chowdhury, N. A. Zatsepin, A. Aquila, J. Coe and C. Gati, IUCrJ, 2015, 2, 421–430 CrossRef CAS PubMed.
- J. E. Pak, C. Sharon, M. Satkunarajah, T. C. Auperin, C. M. Cameron, D. J. Kelvin, J. Seetharaman, A. Cochrane, F. A. Plummer and J. D. Berry, J. Mol. Biol., 2009, 388, 815–823 CrossRef CAS PubMed.
- A. Handisurya, S. Gilch, D. Winter, S. Shafti-Keramat, D. Maurer, H. M. Schätzl and R. Kirnbauer, FEBS J., 2007, 274, 1747–1758 CrossRef CAS PubMed.
- R. Sougrat, A. Bartesaghi, J. D. Lifson, A. E. Bennett, J. W. Bess, D. J. Zabransky and S. Subramaniam, PLoS Pathog., 2007, 3, e63 CrossRef PubMed.
- S. Subramaniam, A. Bartesaghi, J. Liu, A. E. Bennett and R. Sougrat, Curr. Opin. Struct. Biol., 2007, 17, 596–602 CrossRef CAS PubMed.
- J. Liu, A. Bartesaghi, M. J. Borgnia, G. Sapiro and S. Subramaniam, Nature, 2008, 455, 109–113 CrossRef CAS PubMed.
- Y. Chen, Y. Guo, Y. Pan and Z. J. Zhao, Biochem. Biophys. Res. Commun., 2020, 525, 135–140 CrossRef PubMed.
- D. Pinto, Y.-J. Park, M. Beltramello, A. C. Walls, M. A. Tortorici, S. Bianchi, S. Jaconi, K. Culap, F. Zatta and A. De Marco, Nature, 2020, 1–6 Search PubMed.
- D. Cao, Y. Gao, C. Roesler, S. Rice, P. D'Cunha, L. Zhuang, J. Slack, M. Domke, A. Antonova and S. Romanelli, Nat. Commun., 2020, 11, 1–9 CrossRef PubMed.
- Y. Lin, X. Yan, W. Cao, C. Wang, J. Feng, J. Duan and S. Xie, Antiviral Ther., 2004, 9, 287–289 CAS.
- J. R. Greenland, M. D. Michelow, L. Wang and M. J. London, Anesthesiology: The Journal of the American Society of Anesthesiologists, 2020, 132, 1346–1361 CrossRef CAS PubMed.
- W. Chiu, R. L. Garcea and R. Burnette, Structural biology of viruses, Oxford University Press Oxford, UK, 1997 Search PubMed.
- C. Kannicht and B. Fuchs, in Molecular Biomethods Handbook, Springer, 2008, pp. 427–449 Search PubMed.
- N. Papageorgiou, J. Lichiere, A. Baklouti, F. Ferron, M. Sévajol, B. Canard and B. Coutard, Acta Crystallogr., Sect. D: Struct. Biol., 2016, 72, 192–202 CrossRef CAS PubMed.
- A. McPherson and S. B. Larson, Crystallogr. Rev., 2015, 21, 3–56 CrossRef CAS.
- L. A. Earl and S. Subramaniam, Proc. Natl. Acad. Sci., 2016, 113, 8903–8905 CrossRef CAS PubMed.
- S. C. Shoemaker and N. Ando, Biochemistry, 2018, 57, 277–285 CrossRef CAS PubMed.
- A. Barty, C. Caleman, A. Aquila, N. Timneanu, L. Lomb, T. White, J. Andreasson, D. Arnlund, S. Bajt and T. Barends, Nat. Photonics, 2011, 6, 35–40 CrossRef PubMed.
- J. S. Harrison, C. D. Higgins, M. J. O'Meara, J. F. Koellhoffer, B. A. Kuhlman and J. R. Lai, Structure, 2013, 21, 1085–1096 CrossRef CAS PubMed.
- R. Perera, M. Khaliq and R. J. Kuhn, Antiviral Res., 2008, 80, 11–22 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- E. J. Miller, W. Trewby, A. F. Payam, L. Piantanida, C. Cafolla and K. Voïtchovsky, JoVE, 2016, e54924 Search PubMed.
- K. S. Novoselov, D. Jiang, F. Schedin, T. Booth, V. Khotkevich, S. Morozov and A. K. Geim, Proc. Natl. Acad. Sci., 2005, 102, 10451–10453 CrossRef CAS PubMed.
- R. S. McLean, M. Doyle and B. B. Sauer, Macromolecules, 2000, 33, 6541–6550 CrossRef CAS.
- S. C. Warren, K. Voïtchovsky, H. Dotan, C. M. Leroy, M. Cornuz, F. Stellacci, C. Hébert, A. Rothschild and M. Grätzel, Nat. Mater., 2013, 12, 842–849 CrossRef CAS PubMed.
- C. A. Amo, A. P. Perrino, A. F. Payam and R. Garcia, ACS Nano, 2017, 11, 8650–8659 CrossRef CAS PubMed.
- A. F. Payam, D. Martin-Jimenez and R. Garcia, Nanotechnology, 2015, 26, 185706 CrossRef PubMed.
- A. Belianinov, R. Vasudevan, E. Strelcov, C. Steed, S. M. Yang, A. Tselev, S. Jesse, M. Biegalski, G. Shipman and C. Symons, Adv. Struct. Chem. Imaging, 2015, 1, 1–25 CrossRef.
- A. F. Payam, J. R. Ramos and R. Garcia, ACS Nano, 2012, 6, 4663–4670 CrossRef CAS PubMed.
- H. Huang, I. Dobryden, P.-A. Thorén, L. Ejenstam, J. Pan, M. Fielden, D. Haviland and P. M. Claesson, Compos. Sci. Technol., 2017, 150, 111–119 CrossRef CAS.
- G. Rosso, I. Liashkovich and V. Shahin, Adv. Sci., 2019, 6, 1801638 CrossRef PubMed.
- C. Martinez-Torres, A. Arneodo, L. Streppa, P. Argoul and F. Argoul, Appl. Phys. Lett., 2016, 108, 034102 CrossRef.
- A. F. Payam, O. Payton, L. Picco, S. Moore, T. Martin, A. Warren, M. Mostafavi and D. Knowles, Int. J. Fatigue, 2019, 127, 1–9 CrossRef CAS.
- B. Laperrousaz, L. Berguiga, F. Nicolini, C. Martinez-Torres, A. Arneodo, V. M. Satta and F. Argoul, Phys. Biol., 2016, 13, 03LT01 CrossRef CAS PubMed.
- K. N. Baumann, L. Piantanida, J. García-Nafría, D. Sobota, K. Voïtchovsky, T. P. Knowles and S. Hernández-Ainsa, ACS Nano, 2020, 14, 2316–2323 CrossRef CAS PubMed.
- A. Engel and D. J. Müller, Nat. Struct. Biol., 2000, 7, 715–718 CrossRef CAS PubMed.
- R. Garcıa and R. Perez, Surf. Sci. Rep., 2002, 47, 197–301 CrossRef.
- Y. F. Dufrêne, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin, A. Engel, C. Gerber and D. J. Müller, Nat. Nanotechnol., 2017, 12, 295–307 CrossRef PubMed.
- C. A. Putman, K. O. Van der Werf, B. G. De Grooth, N. F. Van Hulst and J. Greve, Appl. Phys. Lett., 1994, 64, 2454–2456 CrossRef CAS.
- D. J. Müller and Y. F. Dufrene, in Nanoscience and Technology: A Collection of Reviews from Nature Journals, World Scientific, 2010, pp. 269–277 Search PubMed.
- F. Oesterhelt, D. Oesterhelt, M. Pfeiffer, A. Engel, H. Gaub and D. Müller, Science, 2000, 288, 143–146 CrossRef CAS PubMed.
- S. Kufer, E. Puchner, H. Gumpp, T. Liedl and H. Gaub, Science, 2008, 319, 594–596 CrossRef CAS PubMed.
- W. Roos, R. Bruinsma and G. Wuite, Nat. Phys., 2010, 6, 733–743 Search PubMed.
- Y. F. Dufrêne, D. Martínez-Martín, I. Medalsy, D. Alsteens and D. J. Müller, Nat. Methods, 2013, 10, 847–854 CrossRef PubMed.
- P. Hinterdorfer, W. Baumgartner, H. J. Gruber, K. Schilcher and H. Schindler, Proc. Natl. Acad. Sci., 1996, 93, 3477–3481 CrossRef CAS PubMed.
- M. Grandbois, W. Dettmann, M. Benoit and H. E. Gaub, J. Histochem. Cytochem., 2000, 48, 719–724 CrossRef CAS PubMed.
- P. Hinterdorfer and Y. F. Dufrêne, Nat. Methods, 2006, 3, 347–355 CrossRef CAS PubMed.
- R. Garcia and E. T. Herruzo, Nat. Nanotechnol., 2012, 7, 217 CrossRef CAS PubMed.
- T. Ando, Biophys. Rev., 2018, 10, 285–292 CrossRef CAS PubMed.
- S. Lin, C. K. Lee, S. Y. Lee, C. L. Kao, C. W. Lin, A. B. Wang, S. M. Hsu and L. S. Huang, Cell. Microbiol., 2005, 7, 1763–1770 CrossRef CAS PubMed.
- M.-L. Ng, J. Lee, M. Leong, A.-E. Ling, H.-C. Tan and E. Ooi, Emerging Infect. Dis., 2004, 10, 1907 CrossRef CAS PubMed.
- M. Aznar, S. Roca-Bonet and D. Reguera, J. Phys.: Condens. Matter, 2018, 30, 264001 CrossRef PubMed.
- O. Y. Limanskaya, Biopolym. Cell, 2009, 25, 307 CAS.
- L. Piantanida, A. F. Payam, J. Zhong and K. Voïtchovsky, Phys. Rev. Appl., 2020, 13, 064003 CrossRef CAS.
- J. Yang, S. Petitjean, S. Derclaye, M. Koehler, Q. Zhang, A. C. Dumitru, P. Soumillion and D. Alsteens, Nat. Commun., 2020, 11(1), 1–10 CrossRef PubMed.
- K. K.-W. To, C. Yip, C. Lai, C. Wong, D. Ho, P. Pang, A. Ng, K.-H. Leung, R. Poon and K.-H. Chan, Clin. Microbiol. Infect., 2019, 25, 372–378 CrossRef CAS PubMed.
- L. M. Czumbel, S. Kiss, N. Farkas, I. Mandel, A. E. Hegyi, A. K. Nagy, Z. Lohinai, Z. Szakacs, P. Hegyi, M. C. Steward and G. Varga, medRxiv, 2020 DOI:10.1101/2020.05.26.20112565.
- P. Harikrishnan, J. Craniofacial Surg., 2020, e656–e658 CrossRef PubMed.
- K. K.-W. To, O. T.-Y. Tsang, C. C.-Y. Yip, K.-H. Chan, T.-C. Wu, J. M.-C. Chan, W.-S. Leung, T. S.-H. Chik, C. Y.-C. Choi and D. H. Kandamby, Clin. Infect. Dis., 2020, 71(15), 841–843 CrossRef CAS PubMed.
- R. Anjum, Biomedica, 2020, 36, 97–99 CrossRef.
- D. J. O'Bannon, Women in Water Quality: Investigations by Prominent Female Engineers, Springer, 2019 Search PubMed.
- J. Y. Kim, J.-H. Ko, Y. Kim, Y.-J. Kim, J.-M. Kim, Y.-S. Chung, H. M. Kim, M.-G. Han, S. Y. Kim and B. S. Chin, J. Korean Med. Sci., 2020, 35(7) DOI:10.3346/jkms.2020.35.e86.
- M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman, H. Bruce, C. Spitters, K. Ericson, S. Wilkerson and A. Tural, N. Engl. J. Med., 2020, 382(10), 929–936 CrossRef CAS PubMed.
- B. E. Young, S. W. X. Ong, S. Kalimuddin, J. G. Low, S. Y. Tan, J. Loh, O.-T. Ng, K. Marimuthu, L. W. Ang and T. M. Mak, Jama, 2020, 323, 1488–1494 CrossRef CAS PubMed.
- R. Woelfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Mueller, D. Niemeyer, P. Vollmar, C. Rothe and M. Hoelscher, Nature, 2020, 581, 465–469 CrossRef CAS PubMed.
- Y. Xu, X. Li, B. Zhu, H. Liang, C. Fang, Y. Gong, Q. Guo, X. Sun, D. Zhao and J. Shen, Nat. Med., 2020, 26, 502–505 CrossRef CAS PubMed.
- G. D. Bhowmick, D. Dhar, D. Nath, M. M. Ghangrekar, R. Banerjee, S. Das and J. Chatterjee, npj Clean Water, 2020, 3, 1–8 CrossRef.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Preprint here, 2020.
- F. Wu, A. Xiao, J. Zhang, X. Gu, W. L. Lee, K. Kauffman, W. Hanage, M. Matus, N. Ghaeli and N. Endo, mSystems, 2020, 5(4), e00614-20 CrossRef PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins, J. W. O'Brien, P. M. Choi, M. Kitajima, S. L. Simpson and J. Li, Sci. Total Environ., 2020, 138764 CrossRef CAS PubMed.
- S. Wurtzer, V. Marechal, J.-M. Mouchel and L. Moulin, MedRxiv, 2020 DOI:10.1101/2020.04.12.20062679.
- K. Mao, K. Zhang, W. Du, W. Ali, X. Feng and H. Zhang, Current Opinion in Environmental Science & Health, 2020, 17, 1–7 Search PubMed.
- W. Randazzo, P. Truchado, E. C. Ferrando, P. Simon, A. Allende and G. Sanchez, MedRxiv, 2020 DOI:10.1101/2020.04.22.20075200.
- K. Mao, H. Zhang and Z. Yang, Biosens. Bioelectron., 2020, 169, 112617 CrossRef CAS PubMed.
- N. Sims and B. Kasprzyk-Hordern, Environment international, 2020, 105689 CrossRef PubMed.
- L. Liu, D. Yang and G. Liu, Biosens. Bioelectron., 2019, 136, 60–75 CrossRef CAS PubMed.
- Z. Yang, B. Kasprzyk-Hordern, C. G. Frost, P. Estrela and K. V. Thomas, Community sewage sensors for monitoring public health, 2015, pp. 5845–5846 Search PubMed.
- Z. Yang, G. Xu, J. Reboud, S. A. Ali, G. Kaur, J. McGiven, N. Boby, P. K. Gupta, P. Chaudhuri and J. M. Cooper, ACS Sens., 2018, 3, 403–409 CrossRef CAS PubMed.
- Z. Yang, G. Xu, J. Reboud, B. Kasprzyk-Hordern and J. M. Cooper, Anal. Chem., 2017, 89, 9941–9945 CrossRef CAS PubMed.
- K. Farkas, L. S. Hillary, S. K. Malham, J. E. McDonald and D. L. Jones, Curr. Opin. Environ. Sci. Health, 2020, 17, 14–20 CrossRef PubMed.
- A. Bosch, E. Gkogka, F. S. Le Guyader, F. Loisy-Hamon, A. Lee, L. Van Lieshout, B. Marthi, M. Myrmel, A. Sansom and A. C. Schultz, Int. J. Food Microbiol., 2018, 285, 110–128 CrossRef PubMed.
- L. Wu, W. Yin, K. Tang, K. Shao, Q. Li, P. Wang, Y. Zuo, X. Lei, Z. Lu and H. Han, Biosens. Bioelectron., 2016, 82, 177–184 CrossRef CAS PubMed.
- L. Wu, G. Li, X. Xu, L. Zhu, R. Huang and X. Chen, TrAC, Trends Anal. Chem., 2019, 113, 140–156 CrossRef CAS.
- Y. Chen and L. Li, Lancet Infect. Dis., 2020, 20, 515–516 CrossRef CAS PubMed.
- J. Zhao, Q. Yuan, H. Wang, W. Liu, X. Liao, Y. Su, X. Wang, J. Yuan, T. Li and J. Li, Clin. Infect. Dis., 2020, 71(16), 2027–2034 CrossRef CAS PubMed.
- J. Cai, W. Sun, J. Huang, M. Gamber, J. Wu and G. He, Emerging Infect. Dis., 2020, 26(6), 1343 CrossRef CAS PubMed.
- C. F. Yung, K.-q. Kam, M. S. Wong, M. Maiwald, Y. K. Tan, B. H. Tan and K. C. Thoon, Ann. Intern. Med., 2020, 173(3), 240–242 CrossRef PubMed.
- Z.-D. Guo, Z.-Y. Wang, S.-F. Zhang, X. Li, L. Li, C. Li, Y. Cui, R.-B. Fu, Y.-Z. Dong and X.-Y. Chi, Emerging Infect. Dis., 2020, 26, 10.3201 Search PubMed.
- Q. Sun, X. Xu, J. Xie, J. Li and X. Huang, Korean J. Radiol., 2020, 21, 614–619 CrossRef PubMed.
- F.-C. Jiang, X.-L. Jiang, Z.-G. Wang, Z.-H. Meng, S.-F. Shao, B. D. Anderson and M.-J. Ma, Emerging Infect. Dis., 2020, 26(9), 2162 CrossRef CAS PubMed.
- M. Suzuki, et al., MedRxiv, 2020 DOI:10.1101/2020.05.02.20088567.
- K. Razzini, M. Castrica, L. Menchetti, L. Maggi, L. Negroni, N. V. Orfeo, A. Pizzoccheri, M. Stocco, S. Muttini and C. M. Balzaretti, Sci. Total Environ., 2020, 140540 CrossRef CAS PubMed.
- G. Ye, H. Lin, L. Chen, S. Wang, Z. Zeng, W. Wang, S. Zhang, T. Rebmann, Y. Li and Z. Pan, J. Infect., 2020, 81(2), e1–e5 CrossRef CAS PubMed.
- D. K. Chu, Y. Pan, S. M. Cheng, K. P. Hui, P. Krishnan, Y. Liu, D. Y. Ng, C. K. Wan, P. Yang and Q. Wang, Clin. Chem., 2020, 66, 549–555 CrossRef PubMed.
- P. Poizot, J. Gaubicher, S. Renault, L. Dubois, Y. Liang and Y. Yao, Chem. Rev., 2020, 120, 6490–6557 CrossRef CAS PubMed.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu and X. Gu, Lancet, 2020, 395, 497–506 CrossRef CAS.
- M. Ezhilan, I. Suresh and N. Nesakumar, Measurement, 2021, 168, 108335 CrossRef PubMed.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang and B. Cao, Lancet, 2020, 395, 497–506 CrossRef CAS.
- H. Zhong, et al., ACS Nano, 2020, 14(5), 6213–6221 CrossRef CAS PubMed.
- D. Lauster, S. Klenk, K. Ludwig, S. Nojoumi, S. Behren, L. Adam, M. Stadtmüller, S. Saenger, S. Zimmler and K. Hönzke, Nat. Nanotechnol., 2020, 15, 373–379 CrossRef CAS PubMed.
- R. Vinodh, R. S. Babu, C. V. M. Gopi, C. Deviprasath, R. Atchudan, L. M. Samyn, A. L. F. de Barros, H.-J. Kim and M. Yi, Journal of Energy Storage, 2020, 28, 101196 CrossRef.
- A. Lopez, Nanotechnol. Nanomater. Res., 2020, 1, 1 Search PubMed.
- A. Padmanaban, N. Padmanathan, T. Dhanasekaran, R. Manigandan, S. Srinandhini, P. Sivaprakash, S. Arumugam and V. Narayanan, J. Electroanal. Chem., 2020, 877, 114658 CrossRef CAS.
- H. Weerahandi, K. A. Hochman, E. Simon, C. Blaum, J. Chodosh, E. Duan, K. Garry, T. Kahan, S. L. Karmen-Tuohy and H. C. Karpel, J. Gen. Intern. Med., 2021, 1–8 Search PubMed.
- J. Hellewell, S. Abbott, A. Gimma, N. I. Bosse, C. I. Jarvis, T. W. Russell, J. D. Munday, A. J. Kucharski, W. J. Edmunds and F. Sun, Lancet Global Health, 2020, 8(4), e488–e496 CrossRef PubMed.
- M. E. Kretzschmar, G. Rozhnova, M. C. Bootsma, M. van Boven, J. H. van de Wijgert and M. J. Bonten, Lancet Public Health, 2020, 5(8), e452–e459 CrossRef PubMed.
- A. Aleta, D. Martín-Corral, A. P. y Piontti, M. Ajelli, M. Litvinova, M. Chinazzi, N. E. Dean, M. E. Halloran, I. M. Longini Jr and S. Merler, Nature Human Behaviour, 2020, 1–8 CAS.
- L. Ferretti, C. Wymant, M. Kendall, L. Zhao, A. Nurtay, L. Abeler-Dörner, M. Parker, D. Bonsall and C. Fraser, Science, 2020, 368(6491), eabb6936 CrossRef CAS PubMed.
- K. J. Monahan, A. Lincoln, J. E. East, S. Benton, J. Burn, B. DeSouza, H. Hanson, F. Lalloo, T. McVeigh and M. D. Rutter, Gut, 2021, 70, 624–626 CrossRef CAS PubMed.
- E. O. Tingaz, IJERI: International Journal of Educational Research and Innovation, 2021, 73–81 Search PubMed.
- G. A. COVID and P.-A. C. S. Group, Aging: Clin. Exp. Res., 2020, 1 Search PubMed.
- T. Greenhalgh, M. Knight, C. A'Court, M. Buxton and L. Husain, BMJ, 2020, 370, m3026 CrossRef PubMed.
- S. H. Ahmedzai, A. Dickman and A. C. Nwosu, BMJ, 2020, 369, m1461 Search PubMed.
| This journal is © The Royal Society of Chemistry 2022 |