Open Access Article
Open Access ArticleA rapid water bath PCR combined with lateral flow assay for the simultaneous detection of SARS-CoV-2 and influenza B virus†
Hong Chen‡
ab,
Yunxiang Wang‡ab,
Hongjuan Weiab,
Zhen Rong *ab and
Shengqi Wang*ab
*ab and
Shengqi Wang*ab
aBeijing Institute of Radiation Medicine, Beijing 100850, P. R. China. E-mail: sqwang@bmi.ac.cn; rongzhen0525@sina.com
bBeijing Key Laboratory of New Molecular Diagnosis Technologies for Infectious Diseases, Beijing 100850, P. R. China
First published on 25th January 2022
Abstract
The outbreak of the coronavirus disease 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in significant global health and economic threats to the human society. Thus, a rapid and accurate detection method for early testing and diagnosis should be established. In this study, a rapid water bath polymerase chain reaction (PCR) combined with lateral flow assay was developed to detect SARS-CoV-2 and influenza B virus simultaneously. A homemade automated transfer device equipped with reaction tube shuttled rapidly between two water baths at 98 °C and 53 °C to realize rapid PCR. After amplification, two-ended labeled PCR products were detected using the lateral flow strip with two test lines and streptavidin-conjugated quantum dot nanobeads. The fluorescence value was read using a handheld instrument. The established assay could complete reverse-transcription PCR amplification and lateral flow detection in 45 minutes. The detection limits were 8.44 copies per μL and 14.23 copies per μL for SARS-CoV-2 and influenza B virus, respectively. The coefficients of variation of the test strip were 10.10% for the SARS-CoV-2 and 4.94% for the influenza B virus, demonstrating the excellent repeatability of the experiment. These results indicated that the rapid PCR combined with lateral flow assay could detect SARS-CoV-2 and influenza B virus simultaneously at a short assay time and low cost, thereby showing the remarkable potential for the rapid and multiplex detection of respiratory viruses in resource-limited settings.
1 Introduction
The coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global health problem and resulted in significant economic losses throughout the world.1 Until now, the cumulative number of COVID-19 cases has been more than 200 million, including more than 4 million deaths (https://covid19.who.int/). The global epidemic prevention is grim. The common symptoms in people infected with SARS-COV-2 are cough, sore throat, high fever, headache, and difficulty in breathing and are similar to an influenza-like illness.1 Severe cases may manifest as acute respiratory distress syndrome within one week after infection.1 Given the similar symptoms between SARS-CoV-2 and influenza, the detection method is critical to distinguish between patients infected with SARS-CoV-2 and influenza viruses. The rapid and accurate detection is the key to early diagnosis, medical treatment, and effective halting of the global epidemic.The main detection methods for SARS-CoV-2 include nucleic acid amplification testing, antigen–antibody testing, sequencing, clustered regularly interspaced short palindromic (CRISPR), and other tests.2 Coronaviruses have many molecular targets in their positive single-stranded RNA genomes that can be detected by polymerase chain reaction (PCR). The real-time reverse-transcription PCR (RT-qPCR) assay is considered the “gold standard” for confirming clinical cases of COVID-19 due to its high sensitivity and specificity. The highly conservative gene regions of SARS-CoV-2, such as ORF1ab, nucleocapsid (N) and envelope (E), spike (S), and RNA-dependent RNA polymerase (RdRp) sequences, are utilized as single or multiple target fragments in current nucleic acid detection protocols.3,4 However, these protocols require at least 4 hours of operation performed by skilled technicians in professional laboratory facilities with precise PCR thermocycler and real-time fluorescence reader.5 The turnaround time for issuing results from RT-PCR is typically a few hours.6 Hence, methodological simplification, such as circumventing RNA extraction in SARS-CoV-2 detection by performing RT-PCR directly on heat-inactivated or lysed samples, can be crucial to benefit patient care and control infection.7 Droplet digital PCR is a more sensitive and accurate diagnostic tool compared with RT-qPCR. For example, 63 samples with negative RT-qPCR results for SARS-CoV-2, 55% of which have COVID-19-related symptoms, have shown positive droplet digital PCR results at low copy number level.8 However, droplet digital detection requires high-quality equipment, complex operation procedure, and long detection time, which limit the clinical application of droplet digital PCR. The lateral flow assay (LFA) is a widely used paper-based point-of-care testing biosensor that can detect target analytes in minutes and does not require a trained person to operate expensive and sophisticated instrument. The LFA has the advantages of low cost, easy operation, and rapid detection for point-of-care testing applications.9,10 The LFA is widely applied to the detection of cancer marker,11 infectious diseases,9,12 microorganisms, and mycotoxins.13 A common LFA strip is composed of sample pad, conjugation pad, nitrocellulose (NC) membrane with test (T) and control (C) lines, and absorbent and backing pads.14 The rapid antigen detection for SARS-CoV-2 by LFA has shown promising performance for mass population testing and can be applied to identify infected individuals.6
Since the COVID-19 outbreak, numerous RT-PCR kits have been developed to detect SARS-CoV-2. However, in cases where clinical symptoms and imaging findings raise strong suspicion, false-negative results are found to be as high as 20% to 40%.4,15 Thus, a test that can improve the detection sensitivity and specificity for SARS-CoV-2 should be developed. The novel lateral flow strips membrane assay combined with RT-PCR can simultaneously detect RdRp, ORF3a, and N genes with a detection limit of 10 copies per test for each gene.4 The gold magnetic nanoparticle-based LFA combined with amplification refractory mutation system PCR can be used to detect single-nucleotide polymorphism (SNP) and multiple SNPs.16,17 The RT-RPA with the CRISPR-Cas12a assay is an effective method for the screening of influenza viruses and SARS-CoV-2 with high specificity and sensitivity.1 The CRISPR combined with RT-LAMP for molecular diagnosis of COVID-19 is reported to improve specificity and sensitivity.18–20 These isothermal nucleic acid analysis techniques avoided time-consuming and precisely repeated thermocycling usually based on the Peltier effect. However, their shortcomings, such as complicated primer design, contamination from amplicons, and high expense, still limit their wide applications. Thus, a fast, easy-to-use, and highly sensitive PCR-based analysis approach that can be widely deployed in resource-limited settings with no professional equipment and technicians to contain the COVID-19 pandemic effectively is yet to be established.
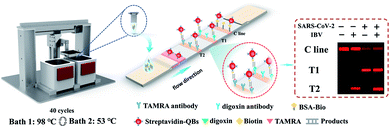
Here, we have developed a rapid nucleic acid amplification testing method integrating water bath PCR and fluorescent LFA for the simultaneous detection of SARS-CoV-2 and influenza B virus (IBV). Water bath PCR, as the first generation of PCR, can omit the process of temperature rise and fall and quickly achieve the suitable temperature for thermal cycles. Rapid water bath PCR combined with a molecular beacon can specifically detect two kinds of shrimp pathogens in one tube within 15 minutes and observe the results through a simple device with naked eyes.21 A water bath setup using sous vide immersion heaters, a Raspberry Pi computer, and a single servo motor can achieve rapid RT-PCR with thin-welled PCR tube to detect SARS-CoV-2.22 Our water bath PCR system is composed of two water baths, transfer equipment, a hot cover, and motor motion control circuit. Driven by the X- and Z-axis motors, the transfer equipment equipped with reaction systems shuttle rapidly between the two water baths to perform the PCR amplification. It can fit with PCR reaction tubes of varied sizes and quantities by simply adapting its tube holder, thus demonstrating its great potential for high-throughput analysis applications in a low cost and flexible manner. After amplification, the biotin and digoxin-labeled SARS-CoV-2 amplicon and the biotin and TAMRA-labeled IBV amplicon are tested using a lateral flow strip with two test lines and streptavidin-conjugated quantum dot nanobeads (SA-QBs). Immune complexes form on the test line in the presence of the target SARS-CoV-2 or IBV gene, and the fluorescence intensity is determined by our reported smartphone-based fluorescent analysis device under UV light illumination.23 The scheme aims to provide a rapid, low-cost, and sensitive multiplex method for the detection of SARS-CoV-2 and IBV in low resource settings.
2 Experimental section
2.1 Materials
The TIANamp virus DNA/RNA kit, DL500 DNA marker, and TaqPath™ 1-step Multiplex Master Mix were obtained from Tiangen Biotech (Beijing, China), Takara (Dalian, China), and Thermo Fisher Scientific (Shanghai, China), respectively. Goat anti-mouse IgG antibody was purchased from Sigma (Shanghai, China). TAMRA and digoxin antibodies were acquired from Biocare Biotech (Zhuhai, China). Bovine serum albumin–biotin conjugate (BSA-Bio) was provided by Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). Streptavidin-conjugated quantum dot nanobeads (SA-QBs, 615 nm emission wavelength) were obtained from Najing Technology Corp. (Beijing, China). UniSart CN140 nitrocellulose (NC) membrane was provided by Sartorius (Göttingen, Germany). Primers were synthesized, modified, and purified by Sangon Biotech Co., Ltd. (Shanghai, China). The IBV-NS1, SARS-CoV-2 N gene, SARS-CoV, and middle east respiratory syndrome (MERS)-CoV pseudoviruses were purchased from Fubio Biotech (Suzhou, China). IBV, influenza A virus (IAV), adenovirus (ADV), parainfluenza virus (PIV) were provided by HyTest Ltd. (Shanghai, China).2.2 Nucleic acid extraction
In accordance with the instruction manual, RNA templates were extracted from the IBV-NS1 (105 copies per μL) and SARS-CoV-2 N gene (105 copies per μL) pseudoviruses by the TIANamp virus DNA/RNA kit. The RNA template (5 μL) was used for each testing.2.3 Primer design
A 354 bp RNA fragment of IBV and a 1260 bp RNA fragment of SARS-CoV-2 were chosen as template sequences. Primer sequences were designed using the Primer Premier 5 or reported in recent publications.24 Three pairs of primers were designed for each SARS-CoV-2 and IBV, arranged, and combined. Finally, a combination of primers that could simultaneously detect the two viruses was selected. The optimal primers are shown in Table 1.| Primer | Length (bp) | 5′ to 3′ sequence |
|---|---|---|
| Flu B-NS1-F | 20 | 5′-TAMRA-CCTTCCTCAAGCACCCTAAT-3′ |
| Flu B-NS1-R | 19 | 5′-Biotin-ACTCCCACCGCAGTTTCAG-3′ |
| SARS-CoV-2-N-F | 22 | 5′-Digoxin-GATTACAAACATTGGCCGCAAA-3′ |
| SARS-CoV-2-N-R | 18 | 5′-Biotin-GCGCGACATTCCGAAGAA-3′ |
2.4 Duplex rapid RT-PCR assay
The duplex rapid PCR system contained 5 μL 4× TaqPath™ 1-Step Multiplex Master Mix, 30 nM of each primer for SARS-CoV-2, 20 nM of each primer for influenza B, 2.5 μL RNA template each for influenza B and SARS-CoV-2, and 8 μL double-distilled water to obtain a total volume of 20 μL. The thermocycling program of rapid water bath PCR was as follows: 53 °C for 5 minutes, 98 °C for 40 seconds, and 40 cycles of 98 °C for 16 seconds and 53 °C for 12 seconds. The nucleic acid amplification with rapid water bath PCR cycles was completed in 30 minutes.2.5 Preparation of lateral flow strips
QBs-based immune test strips consisted of five components: sample pad, conjugate pad, detection line, quality control line, and absorbent pad. The IBV amplicon capture antibody (TAMRA antibody, 1 mg mL−1), SARS-CoV-2 amplicon capture antibody (digoxin antibody, 1 mg mL−1), and BSA-Bio (1 mg mL−1) were spotted onto NC membranes to form two T lines and one C line. SA-QBs (10 nM) were dispensed into a glass fiber paper and freeze-dried to prepare the conjugate pad. Eventually, the sample pad, conjugate pad, absorbent pad, plastic backing card, and NC membrane were assembled and cut into strips of 3.5 mm width.2.6 Detection of RT-PCR products by using LFA
Fluorescent lateral flow strips were used to quantitate the products of duplex RT-PCR. Amplification products (20 μL) were mixed with 70 μL running buffer solution (10 mM PBS, pH 7.4, 0.05% Tween-20, 1% BSA), and the mixture was dripped onto the strip sample pad. After 15 minutes, the corresponding T line turned into a bright red line under the UV light illumination in the presence of SARS-CoV-2 or IBV in the sample. Results were photographed using a handheld fluorescence analysis device,23 and the fluorescence intensity of T lines were measured to quantify the target virus load.2.7 Development of rapid water bath PCR equipment
As shown in Scheme 1, the rapid water bath PCR equipment consisted of the following components: two water baths, transfer equipment, a hot cover, and circuits. Gantry and large bottom plate constituted the main frame of the instrument. The length, width, and height of the instrument were 780, 670, and 268 mm, respectively. A 96-well PCR reaction plate was fixed in a holder that was shuttled rapidly between two water baths at 98 °C and 53 °C to realize rapid PCR by two stepper motors and linear guide rails. A separate heating plate was designed in the hot cover to prevent condensation on the wall or top of the tube after the evaporation of reaction mixture. At the same time, the cover opening button and lock tongue were designed to make the hot cover open and close under the drive of the torsion spring, which was easy to fix and take out the PCR reaction tube. For this 96-well plate holder, the shape could be changed in accordance with the different types of reaction tubes.The detection system was an enclosed portable device that could take a picture and read fluorescence values. The smartphone-based fluorescent measurement and analysis device was based on an equipment developed in a previous report.23
3 Results and discussion
3.1 Principle of rapid PCR-combined LFA
We have developed a rapid detection method for SARS-CoV-2 and IBV. The RNA template was extracted from the pseudoviruses of SARS-CoV-2 and IBV and amplified by rapid water bath PCR with specifically modified primers. As shown in Scheme 1, the fast water bath PCR consisted of two water baths, transfer equipment, and a hot cover. The water bath enabled rapid thermal cycling could be applied for the assembly of DNA fragments in vitro or PCR amplification. The assembly experiment was carried out in the reaction tube, and experimental results were verified by agarose gel electrophoresis, which indicated that DNA fragments with length of 1 kb could be assembled in vitro by water bath PCR. The results of the assembly are shown in the Fig. S1.† The rapid water bath PCR uses two temperature cycles and can complete 40 cycles of amplification in 30 minutes due to the reduced time of temperature rise and fall. The water bath PCR was utilized to amplify virus template because of its advantages, including short amplification time, low cost, easy accessibility, and high-throughput analysis capability. The traditional real-time fluorescence amplification instrument is expensive and takes about 2 hours to complete the experiment. In this assay, the two-ended labeled amplicons were quantitatively detected by a fluorescent lateral flow strip with two T lines and SA-QBs as the detection probe. Once added to the sample pad, the resultant mixture migrated along the strip due to capillarity effect. During this process, digoxin/biotin-labeled SARS-CoV-2 and TAMRA/biotin-labeled IBV amplicons in the amplified PCR products were bound to SA-QBs and were captured by digoxin antibody-coated T line 1 and TAMRA antibody-coated T line 2, respectively, leading to the appearance of bright red T lines in 15 minutes under the excitation of UV light. LFA results were recorded using a mobile phone camera, and the fluorescence value of the LFA was determined. The images plotted in Scheme 1 show the results for negative control, IBV-positive, SARS-CoV-2-positive, and SARS-CoV-2- and IBV-positive samples. Agarose gel electrophoresis was used to detect the amplified product and compared with the LFA.3.2 Optimization of rapid RT-PCR and LFA
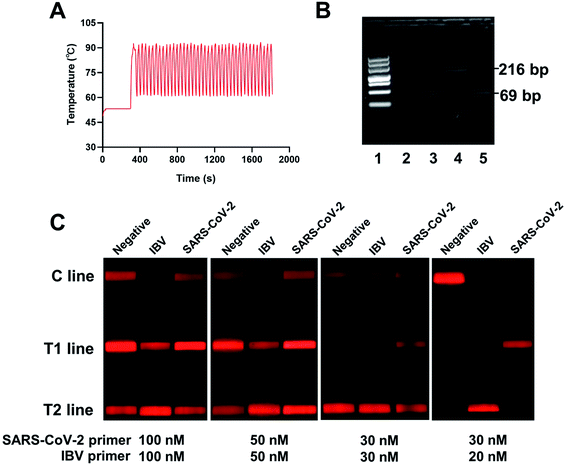
Several system parameters were tuned to shorten the assay time required for RT-PCR. The 96-well PCR reaction plate fixed in a holder was driven by the movement parts in the horizontal and vertical directions to achieve the frequent reciprocating movement between the two water baths. The stepping motors in the X and Z directions were selected as the moving driver, and the load and length of the guide rail were reasonably designed in accordance with force and stroke. The X-axis traveled a distance of 400 mm at speed of 72.5 mm s−1, and the Z-axis traveled a distance of 268 mm at speed of 72.5 mm s−1. Two water baths provided the rapid thermal cycling for PCR between ∼92 °C for denaturation, and ∼60 °C for annealing and extension. It should be noted that the temperatures of the two water baths were set as 98 °C and 53 °C to decrease the ramp time. The incubation time for each water bath was also adjusted to guarantee the shortest possible amplification time. As shown in Fig. S2,† the reaction tube was incubated for 16 s in 98 °C water bath and 12 s in 53 °C water bath in each cycle, which can achieve the shortest possible incubation time. The temperature diagram of an entire reaction is shown in Fig. 1A. It required about 30 minutes to perform the water bath RT-PCR procedure, including reverse transcription, 40 cycles of denaturation, and annealing/extension, and shuttle movement. Setting the same temperature program for nucleic acid amplification, the time required by the conventional instrument to complete the cycles was about 1 hour. The amplified PCR products were obtained and analyzed using agarose electrophoresis gel (Fig. 1B). Clear bands appeared at the corresponding positions for target fragments, indicating that the target sequences could be specifically amplified under the set conditions.The RNA samples of IBV and SARS-CoV-2 virus containing 104 copies per μL were used as template to assess the effect of primer concentration on the PCR amplification. In accordance with the instruction manual of the TaqPath™ 1-step Multiplex Master Mix, primers with a concentration of 200 nM readily produced nonspecific products, leading to false-positive results in the test strips. Therefore, the primer concentration should be optimized. Several groups of primers with different concentrations were used for PCR amplification and tested using test strips. As shown in Fig. 1C, false-positive results were observed at high concentrations of primers by forming primer dimers. Thus, the concentrations of forward and reverse primers were determined as 20 nM for IBV and 30 nM for SARS-CoV-2.
The used SA-QBs were diluted by 100, 200, and 300 times for strip preparation to optimize its concentration. Then, 104 copies per μL of SARS-CoV-2 as the positive sample along with the blank control were tested using the proposed method. As shown in Fig. S3,† the resultant images indicated that SA-QBs diluted 100 times could achieve the brightest T line for the positive sample with negligible signal for the blank control. Thus, the 100 times diluted SA-QBs was applied to the following experiment. The concentrations of digoxin antibody and TAMRA antibody coated on the two test lines were both determined as 1 mg mL−1 on the basis of signal-to-noise ratios (Fig. S4†).
3.3 Specificity of the duplex RT-PCR-combined LFA
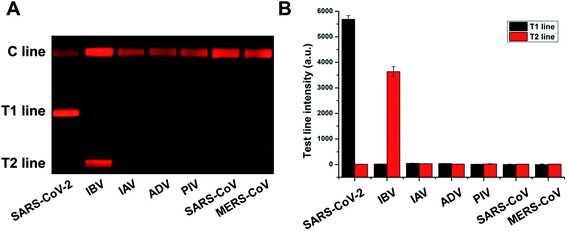
SARS-CoV-2 pseudovirus, IBV, IAV, ADV, PIV, SARS-CoV pseudovirus, and MERS-CoV pseudovirus were amplified in accordance with the above procedure to verify the assay specificity. The amplified product (20 μL) was mixed with the buffer solution (70 μL) and tested using the prepared test strips. Each sample was independently repeated thrice, and the image was recorded and processed to calculate its signal value. Fig. 2A shows that the RT-PCR product of SARS-CoV-2 resulted in a bright band on T line 1, whereas that of IBV exhibited a bright band on T line 2. The results for other respiratory viruses showed no positive band. Their corresponding fluorescence intensities were determined and plotted (Fig. 2B). Results showed no cross-reactivity of this designed system with other respiratory virus, indicating the excellent specificity of this assay.3.4 Sensitivity of the duplex RT-PCR-combined LFA
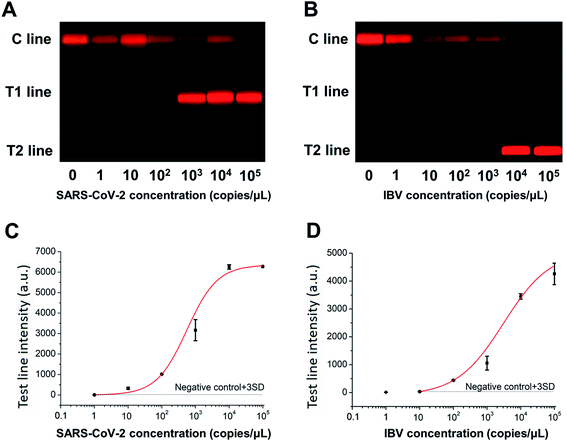
The nucleic acid templates of SARS-CoV-2 and IBV were serially diluted and subjected to RT-PCR-combined LFA. As shown in Fig. 3A and B, the red-colored T lines became brighter with increasing concentration of their corresponding target analyte. The calibration curves of the resultant fluorescence intensities were well fitted by a 4-parameter logistic model in Fig. 3C and D. The limits of detection (LODs), calculated as the analyte concentration of which the result signal was three times standard deviation higher than the blank control signal, were 8.44 copies per μL and 14.23 copies per μL, namely 42.20 copies/test and 71.15 copies/test, for SARS-CoV-2 and IBV, respectively. As a comparison, the RT-PCR product was also detected by agarose gel electrophoresis and had LODs of 5000 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies/test for SARS-CoV-2 and IBV, respectively (Fig. S5†). Assay performances of recently reported methods for SARS-CoV-2 nucleic acid detection were summarized in Table 3. It was reported that the RT-qPCR has an LOD of 5 copies/test for detecting SARS-CoV-2.25 The LOD of both RT-LAMP based on LFA and RT-PCR combined LFA assay were 10 copies/test of SARS-CoV-2,4,26 which were slightly superior to the detection method established in this experiment. A strand exchange amplification-based fluorescence LFA can detect 70 copies per μL of SARS-CoV-2 N gene.27 The sensitivity of CRISPR/Cas9-mediated LFA was 100 copies/test.28 It should be noted that the primer concentrations used in this work for duplex detection of SARS-CoV-2 and IBV were decreased to avoid the formation of primer dimers, which would directly affect the detection sensitivity. The assay performance targeting one analyte is expected to be improved. Thus, the proposed rapid RT-PCR-combined LFA has acceptable assay sensitivity as compared with these assays.
000 copies/test for SARS-CoV-2 and IBV, respectively (Fig. S5†). Assay performances of recently reported methods for SARS-CoV-2 nucleic acid detection were summarized in Table 3. It was reported that the RT-qPCR has an LOD of 5 copies/test for detecting SARS-CoV-2.25 The LOD of both RT-LAMP based on LFA and RT-PCR combined LFA assay were 10 copies/test of SARS-CoV-2,4,26 which were slightly superior to the detection method established in this experiment. A strand exchange amplification-based fluorescence LFA can detect 70 copies per μL of SARS-CoV-2 N gene.27 The sensitivity of CRISPR/Cas9-mediated LFA was 100 copies/test.28 It should be noted that the primer concentrations used in this work for duplex detection of SARS-CoV-2 and IBV were decreased to avoid the formation of primer dimers, which would directly affect the detection sensitivity. The assay performance targeting one analyte is expected to be improved. Thus, the proposed rapid RT-PCR-combined LFA has acceptable assay sensitivity as compared with these assays.
3.5 Repeatability of the duplex RT-PCR-combined LFA
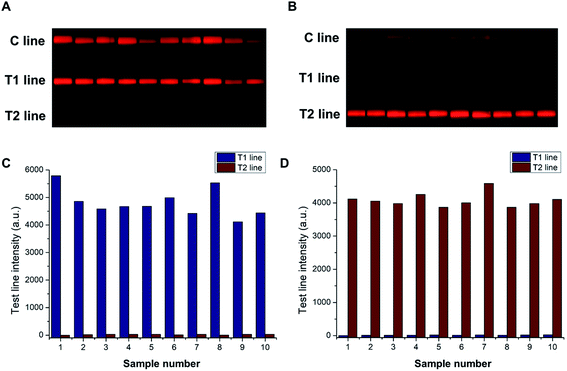
The widely used conventional Peltier-based metal bath often suffers from the edge effect due to the varied heat dissipation at its different positions, which can lead to the inhomogeneous amplification of target fragments. By contrast, the water bath heating method can achieve uniform thermal distribution in a low-cost and simple way. About 104 copies per μL of SARS-CoV-2 and IBV were independently amplified and tested using the strips to evaluate the repeatability of this approach. The resulting fluorescent images were captured using a smartphone-based device. As shown in Fig. 4A and B, the T lines for SARS-CoV-2 and IBV exhibited uniform bands under the excitation of UV light. Their corresponding fluorescent signals were obtained and plotted (Fig. 4C and D). The coefficients of variation (CV) of the test strips were 10.10% for SARS-CoV-2 and 4.94% for IBV, which indicated the good repeatability of this method.3.6 Analysis of the virus-spiked saliva
The clinical applicability of the proposed detection system was further confirmed by testing SARS-CoV-2- and IBV-spiked saliva. Different concentrations (102, 103, and 104 copies per μL) of SARS-CoV-2 and IBV pseudoviruses were added into saliva samples. RNA templates were extracted for RT-PCR amplification in accordance with the above procedure, and the sensitivity and quantitative analysis ability were verified using test strips. The quantitative analysis ability of the pseudovirus added into saliva was accurate. Results are displayed in Table 2. The measured concentration of the three repeated experiments was in acceptable agreement with the added concentration. The relative standard deviation of SARS-CoV-2 ranged from 13.08% to 19.23%, and that of the IBV ranged from 13.98% to 19.77%.| Virus | Added concentration (copies per μL) | Measured concentration (copies per μL) | SD | RSD |
|---|---|---|---|---|
| SARS-CoV-2 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636 636 |
2044.96 | 19.23% |
| 1000 | 808 | 105.71 | 13.08% | |
| 100 | 122 | 20.85 | 17.09% | |
| IBV | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 104 |
2393.58 | 19.77% |
| 1000 | 1610 | 287.36 | 17.84% | |
| 100 | 132 | 18.46 | 13.98% |
| Method | Target | Limit of detection | Assay time | Ref. |
|---|---|---|---|---|
| RT-qPCR | SARS-CoV-2 | 5 copies/reaction | ∼90 min | 25 |
| RT-LAMP based LFA | SARS-CoV-2 | 10 copies/reaction | ∼40 min | 26 |
| RT-PCR based on LFA | SARS-CoV-2 | 10 copies/reaction | ∼120 min | 4 |
| Strand exchange amplification-based fluorescence LFA | SARS-CoV-2 | 70 copies per μL | ∼60 min | 27 |
| CRISPR/Cas9-mediated LFA | SARS-CoV-2 | 100 copies/reaction | ∼60 min | 28 |
| Rapid water bath PCR combined LFA | SARS-CoV-2 | 42.2 copies/reaction | ∼45 min | This work |
4 Conclusion
We developed a rapid RT-PCR combined with LFA for the simultaneous detection of SARS-CoV-2 and IBV. The nucleic acid PCR amplification was accomplished in a simple, low-cost, and high-throughput manner by rapidly transferring reaction tubes between two water baths set at 98 °C and 53 °C. Two-ended labeled PCR products were detected by fluorescent lateral flow strip with two T lines for SARS-CoV-2 and IBV. The analysis procedure could be performed within 45 minutes. The LODs were 8.44 copies per μL of SARS-COV-2 and 14.23 copies per μL of IBV, and the CVs of the test strip was 10.10% for the SARS-CoV-2 and 4.94% for the IBV. This method had superior features, including short assay time, high-throughput analysis capability, and low cost. We believe that this method can provide a rapid and accurate approach for the multiplex quantification of virus nucleic acid in low resource settings.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Key R&D Program of China (grant no. 2018YFA0902300), and the National Natural Science Foundation of China (grant no. 81902159).References
- O. Mayuramart, P. Nimsamer, S. Rattanaburi, N. Chantaravisoot, K. Khongnomnan, J. Chansaenroj, J. Puenpa, N. Suntronwong, P. Vichaiwattana, Y. Poovorawan and S. Payungporn, Exp. Biol. Med., 2020, 246, 400–405 CrossRef PubMed.
- H. M. Yoo, I.-H. Kim and S. Kim, Int. J. Mol. Sci., 2021, 22, 6150 CrossRef CAS PubMed.
- Y. W. Tang, J. E. Schmitz, D. H. Persing and C. W. Stratton, J. Clin. Microbiol., 2020, 58, e00512–e00520 CrossRef PubMed.
- S. Yu, S. B. Nimse, J. Kim, K. S. Song and T. Kim, Anal. Chem., 2020, 92, 14139–14144 CrossRef CAS PubMed.
- H. Abbasi, A. Tabaraei, S. M. Hosseini, A. Khosravi and H. R. Nikoo, Infection, 2021 DOI:10.1007/s15010-021-01674-x.
- T. Peto and U. C.-L. F. O. Team, EClinicalMedicine, 2021, 36, 100924 CrossRef PubMed.
- I. Smyrlaki, M. Ekman, A. Lentini, N. Rufino de Sousa, N. Papanicolaou, M. Vondracek, J. Aarum, H. Safari, S. Muradrasoli, A. G. Rothfuchs, J. Albert, B. Hogberg and B. Reinius, Nat. Commun., 2020, 11, 4812 CrossRef CAS PubMed.
- K. B. Kim, H. Choi, G. D. Lee, J. Lee, S. Lee, Y. Kim, S. Y. Cho, D. G. Lee and M. Kim, Mol. Diagn. Ther., 2021, 25, 617–628 CrossRef CAS PubMed.
- H. Wei, Y. Peng, Z. Bai, Z. Rong and S. Wang, Analyst, 2021, 146, 558–564 RSC.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Biosens. Bioelectron., 2021, 186, 113279 CrossRef CAS PubMed.
- C. C. Fang, C. C. Chou, Y. Q. Yang, T. Wei-Kai, Y. T. Wang and Y. H. Chan, Anal. Chem., 2018, 90, 2134–2140 CrossRef CAS PubMed.
- R. Banerjee and A. Jaiswal, Analyst, 2018, 143, 1970–1996 RSC.
- E. B. Bahadır and M. K. Sezgintürk, TrAC, Trends Anal. Chem., 2016, 82, 286–306 CrossRef.
- X. Gong, J. Cai, B. Zhang, Q. Zhao, J. Piao, W. Peng, W. Gao, D. Zhou, M. Zhao and J. Chang, J. Mater. Chem. B, 2017, 5, 5079–5091 RSC.
- T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen, W. Lv, Q. Tao, Z. Sun and L. Xia, Radiology, 2020, 296, E32–E40 CrossRef PubMed.
- W. Hui, S. Zhang, C. Zhang, Y. Wan, J. Zhu, G. Zhao, S. Wu, D. Xi, Q. Zhang, N. Li and Y. Cui, Nanoscale, 2016, 8, 3579–3587 RSC.
- X. Liu, C. Zhang, K. Liu, H. Wang, C. Lu, H. Li, K. Hua, J. Zhu, W. Hui, Y. Cui and X. Zhang, Anal. Chem., 2018, 90, 3430–3436 CrossRef CAS PubMed.
- Y. Zhang, M. Chen, C. Liu, J. Chen, X. Luo, Y. Xue, Q. Liang, L. Zhou, Y. Tao, M. Li, D. Wang, J. Zhou and J. Wang, Sens. Actuators, B, 2021, 345, 130411 CrossRef CAS PubMed.
- B. Pang, J. Xu, Y. Liu, H. Peng, W. Feng, Y. Cao, J. Wu, H. Xiao, K. Pabbaraju, G. Tipples, M. A. Joyce, H. A. Saffran, D. L. Tyrrell, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 16204–16212 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- C. Qian, R. Wang, C. Wu, L. Wang, Z. Ye, J. Wu and F. Ji, Anal. Chim. Acta, 2018, 1040, 105–111 CrossRef CAS PubMed.
- A. Arumugam, M. L. Faron, P. Yu, C. Markham, M. Wu and S. Wong, Diagnostics, 2020, 10, 739 CrossRef CAS PubMed.
- Z. Bai, H. Wei, X. Yang, Y. Zhu, Y. Peng, J. Yang, C. Wang, Z. Rong and S. Wang, Sens. Actuators, B, 2020, 325, 128780 CrossRef CAS PubMed.
- J. Cheong, H. Yu, C. Y. Lee, J. U. Lee, H. J. Choi, J. H. Lee, H. Lee and J. Cheon, Nat. Biomed. Eng., 2020, 4, 1159–1167 CrossRef CAS PubMed.
- C. B. F. Vogels, A. F. Brito, A. L. Wyllie, J. R. Fauver, I. M. Ott, C. C. Kalinich, M. E. Petrone, A. Casanovas-Massana, M. Catherine Muenker, A. J. Moore, J. Klein, P. Lu, A. Lu-Culligan, X. Jiang, D. J. Kim, E. Kudo, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takahashi, M. Taura, M. Tokuyama, A. Venkataraman, O. E. Weizman, P. Wong, Y. Yang, N. R. Cheemarla, E. B. White, S. Lapidus, R. Earnest, B. Geng, P. Vijayakumar, C. Odio, J. Fournier, S. Bermejo, S. Farhadian, C. S. Dela Cruz, A. Iwasaki, A. I. Ko, M. L. Landry, E. F. Foxman and N. D. Grubaugh, Nat. Microbiol., 2020, 5, 1299–1305 CrossRef CAS PubMed.
- C. Zhang, T. Zheng, H. Wang, W. Chen, X. Huang, J. Liang, L. Qiu, D. Han and W. Tan, Anal. Chem., 2021, 93, 3325–3330 CrossRef CAS PubMed.
- L. Zhuang, J. Gong, M. Ma, Y. Ji, P. Tian, X. Mei, N. Gu and Y. Zhang, Analyst, 2021, 146, 6650–6664 RSC.
- E. Xiong, L. Jiang, T. Tian, M. Hu, H. Yue, M. Huang, W. Lin, Y. Jiang, D. Zhu and X. Zhou, Angew. Chem., Int. Ed., 2021, 60, 5307–5315 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07756b |
| ‡ H. Chen and Y.-X. Wang contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |