Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical constituents from Carica papaya Linn. leaves as potential cytotoxic, EGFRwt and aromatase (CYP19A) inhibitors; a study supported by molecular docking†
Ashraf N. E. Hameda,
Mohamed E. Abouelela b,
Ahmed E. El Zowalaty
b,
Ahmed E. El Zowalaty cde,
Mohamed M. Badrf and
Mohamed S. A. Abdelkader
cde,
Mohamed M. Badrf and
Mohamed S. A. Abdelkader *g
*g
aDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia 61519, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Al-Azhar University, Assiut-Branch, Assiut, 71524, Egypt
cSahlgrenska Center for Cancer Research, Department of Surgery, Institute of Clinical Sciences, University of Gothenburg, 40530 Gothenburg, Sweden
dWallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, 40530 Gothenburg, Sweden
eDepartment of Pharmacology and Toxicology, Faculty of Pharmacy, Zagazig University, 44519, Egypt
fDepartment of Biochemistry, Faculty of Pharmacy, Menoufia University, Menoufia, 32511, Egypt
gDepartment of Pharmacognosy, Faculty of Pharmacy, Sohag University, Sohag, 82524, Egypt. E-mail: m.salaheldin@pharm.sohag.edu.eg
First published on 23rd March 2022
Abstract
The phytochemical investigation of the hydromethanolic extract of Carica papaya Linn. leaves (Caricaceae) resulted in the isolation and characterization of ten compounds, namely; carpaine (1), methyl gallate (2), loliolide (3), rutin (4), clitorin (5), kaempferol-3-O-neohesperidoside (6), isoquercetin (7), nicotiflorin (8) and isorhamnetin-3-O-β-D-glucopyranoside (9). The compounds 2, 3, 5–7 and 9 were isolated for the first time from the genus Carica. An in vitro breast cancer cytotoxicity study was evaluated with an MCF-7 cell line using the MTT assay. Methyl gallate and clitorin demonstrated the most potent cytotoxic activities with an IC50 of 1.11 ± 0.06 and 2.47 ± 0.14 μM, respectively. Moreover, methyl gallate and nicotiflorin exhibited potential EGFRwt kinase inhibition activities with an IC50 of 37.3 ± 1.9 and 41.08 ± 2.1 nM, respectively, compared with the positive control erlotinib (IC50 = 35.94 ± 1.8 nM). On the other hand, clitorin and nicotiflorin displayed the strongest aromatase kinase inhibition activities with an IC50 of 77.41 ± 4.53 and 92.84 ± 5.44 nM, respectively. Clitorin was comparable to the efficacy of the standard drug letrozole (IC50 = 77.72 ± 4.55). Additionally, molecular docking simulations of the isolated compounds to EGFR and human placental aromatase cytochrome P450 (CYP19A1) were evaluated. Methyl gallate linked with the EGFR receptor through hydrogen bonding with a pose score of −4.5287 kcal mol−1 and RMSD value of 1.69 Å. Clitorin showed the strongest interaction with aromatase (CYP19A1) for the breast cancer receptor with a posing score of −14.2074 and RMSD value of 1.56 Å. Compounds (1–3) possessed a good bioavailability score with a 0.55 value.
1. Introduction
Pawpaw or Papaya (Carica papaya Linn.) is an evergreen tree that belongs to the family Caricaceae, it is indigenous to Central America and the South of Mexico. It is commonly grown in the subtropical and tropical regions and cultivated in many countries worldwide.1,2 C. papaya is considered the most valuable species in the family due to its nutritional and therapeutic benefits.3 Its leaves are traditionally used as a cardiotonic, vermifuge, febrifuge and as a treatment for colic, dengue fever, beriberi, asthma, cancer and stomach troubles in India and Australia.4,5 In addition, the different parts of the Pawpaw plant have been used for their therapeutic applications and strong activities as an antibacterial, antiviral, antitumor, antidiabetic, anti-inflammatory and management of neurodegeneration.6 Numerous scientific studies have confirmed that C. papaya contains alkaloids, glycosides, tannins, saponins, flavonoids and glycosides which may be responsible for its therapeutic activity.5Cancer is a major cause of mortality with a higher incidence in developed and developing countries. Worldwide, about 19.3 million new cancer cases and an estimated 10.0 million deaths due to cancer were reported in 2020. Female breast cancer was the most commonly diagnosed cancer, with an estimated 11.7% of new cases.7 In Egypt, breast cancer incidence accounts for about 38.85% of total diagnosed female cancer cases.8
Despite the advances in cancer research and clinical trials of promising new therapies, there is still a great demand for the discovery of new safe and effective drugs with low adverse effects on human health.9 Natural products have a strong role in the development of anti-cancer agents, thus various drug discovery programs continue to invest in this outstanding source.10
Many scientific studies have reported the effect of C. papaya leaves extract on the treatment of breast cancer, cervical carcinoma, hepatocellular carcinoma, osteosarcoma, lung adenocarcinoma and many other types of cancer.6,11
The EGFR tyrosine kinase is critical for hormone receptor positive breast cancer and upregulation of EGFR leads to aberrant signalling.12 It has been reported that 57% of breast carcinomas express EGFRwt.13 In addition, triple negative breast cancer (TNBC) characterized by low expression of estrogen, progesterone and Her2 receptors, is associated with overexpression of EGFR.14 In addition to the role of EGFR in breast cancer progression, aromatase enzyme is critical for breast cancer development and progression. Aromatase catalyzes the final rate-limiting step in estrogen biosynthesis. Aromatase catalyzes a three-step reaction on androgen substrates. The third step of the reaction leads to the aromatization of the A-ring. Aromatase is also highly expressed in breast cancer producing higher levels of estrogen.15 However, breast cancer cells constantly develop resistance to aromatase inhibitors by acquiring estrogen receptor mutations, truncation and upregulation of ER-related transcription factors activator protein 1 (AP1) and NF-κB, aromatase inhibitors are effective in breast cancer treatment.16–18
Even though previous studies on the effect of C. papaya and its leaves on breast cancer, the effect of its bioactive compounds and possible mechanism of action on specific cancer targets still needs more exploration. This provoked us to carry out an extensive phytochemical study of C. papaya Linn. leaves to isolate the active metabolites and test their effect on the MCF-7 breast cancer cell line as well as evaluation of their epithelial growth factor receptor (EGFRwt) kinase and aromatase (CYP19A) enzyme inhibition activity. In addition, the explanation of the potent compounds possible binding mode to their targets by in silico molecular docking studies.
2. Result and discussion
2.1. Structure elucidation of the isolated compounds
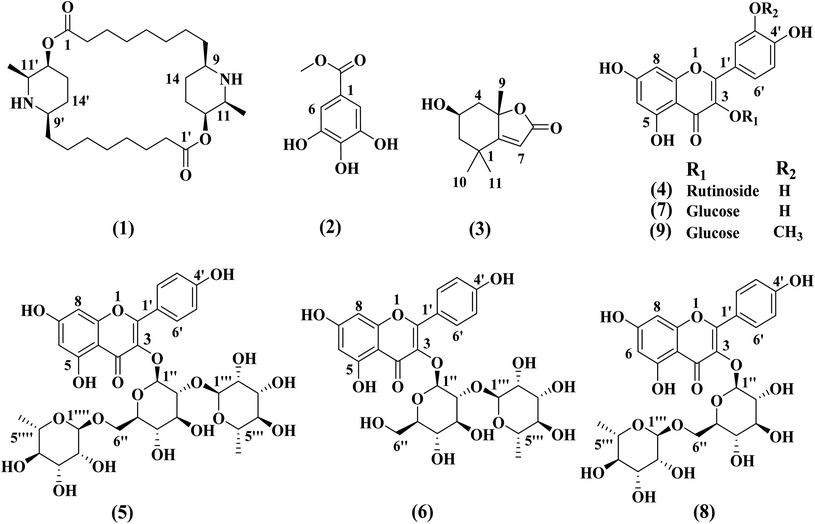
The chemical structures of compounds (1–9) was determined based on different spectroscopic data including 1D and 2D NMR, MS data, as well as comparison of the data with the previously reported in the literature. The structures of all the isolated compounds are declared in Fig. 1. The compounds were elucidated as carpaine (1),19 methyl gallate (2),20 loliolide (3),21 rutin (4),22 clitorin (5),23 kaempferol-3-O-neohesperidoside (6),24 isoquercetin (7),25 nicotiflorin (8)26 and isorhamnetin-3-O-β-D-glucopyranoside (9).27 The compounds 3, 4, 5–7 and 9 were isolated for the first time from the genus Carica.2.2. MTT cytotoxicity assay in breast cancer cells line (MCF-7)
Breast cancer is a common malignancy among females linked with several risk factors as age, familial factors, reproductive factors, lifestyle and hormonal factors. Breast cancer usually starts from ductal hyperproliferation and then develops into either benign or metastatic tumors influenced by exposure to carcinogens.28 Extensive studies investigated the role of dietary and natural products in the reduction of development and progression of breast tumors. Natural products exhibit several anticancer activities by different mechanism such as direct inhibition of tumor cell proliferation, metastasis and angiogenesis of breast tumor cells.29The cytotoxic effect of the isolated compounds from C. papaya on MCF-7 cells line was evaluated using MTT assay which measures metabolic activity as an indicator of cellular viability and proliferation. The results (Table 1) showed that methyl gallate (2) and clitorin (5) exhibited the most potent cytotoxic effects against MCF-7 cell lines with the IC50 value 1.11 ± 0.06 and 2.47 ± 0.14 μM, respectively. Moreover, kaempferol-3-O-neohesperidoside (6), nicotiflorin (8) and isorhamnetin-3-O-β-D-glucopyranoside (9) showed strong effect with IC50 values higher than the standard drug staurosporine (IC50 = 10.2 ± 0.58) (Table 1). The possible mechanism of their potential effect should be investigated for the development of targeted drug therapy for breast tumors.
| No. | Compound | IC50 (μM) |
|---|---|---|
| 1 | Carpaine | 13.7 ± 0.78 |
| 2 | Methyl gallate | 1.11 ± 0.06 |
| 3 | Loliolide | 28.2 ± 1.61 |
| 4 | Rutin | 25.6 ± 1.46 |
| 5 | Clitorin | 2.47 ± 0.14 |
| 6 | Kaempferol-3-O-neohesperidoside | 3.58 ± 0.2 |
| 7 | Isoquercetin | 13.1 ± 0.75 |
| 8 | Nicotiflorin | 4.94 ± 0.28 |
| 9 | Isorhamnetin-3-O-β-D-glucopyranoside | 9.51 ± 0.54 |
| Staurosporine (positive control) | 10.2 ± 0.58 |
2.3. EGFRwt kinase activity
Herein, we studied the inhibitory activity of the isolated compounds on EGFRwt.30,31 The results (Table 2) revealed that all the tested compounds possess EGFRwt potent inhibition activity at nM concentration with IC50 values ranging from 37.3 ± 1.9 to 100.20 ± 5.1 nM. Methyl gallate (2) and nicotiflorin (8) exhibited the highest inhibitory activities comparable with erlotinib (IC50 = 35.94 nM) with IC50 values of 37.3 ± 1.9 and 41.08 ± 2.1 nM, respectively. On the other hand, compounds 1, 3–7 and 9 had an interference effect with EGFRwt but to lesser extent than methyl gallate (2) and nicotiflorin (8).| No. | Compound | IC50 (nM) | |
|---|---|---|---|
| EGFRwt | Aromatase | ||
| 1 | Carpaine | 47.59 ± 2.4 | 107.90 ± 6.32 |
| 2 | Methyl gallate | 37.30 ± 1.9 | 94.13 ± 5.51 |
| 3 | Loliolide | 68.82 ± 3.5 | 207.60 ± 12.2 |
| 4 | Rutin | 44.51 ± 2.3 | 147.60 ± 8.64 |
| 5 | Clitorin | 89.58 ± 4.6 | 77.41 ± 4.53 |
| 6 | Kaempferol-3-O-neohesperidoside | 64.46 ± 3.3 | 334.60 ± 19.6 |
| 7 | Isoquercetin | 83.40 ± 4.2 | 354.20 ± 20.7 |
| 8 | Nicotiflorin | 41.08 ± 2.1 | 92.84 ± 5.44 |
| 9 | Isorhamnetin-3-O-β-D-glucopyranoside | 100.20 ± 5.1 | 436.40 ± 25.6 |
| Erlotinib (positive control) | 35.94 ± 1.8 | ||
| Letrozole (positive control) | 77.72 ± 4.55 | ||
2.4. Aromatase (CYP19A) enzyme activity
Breast cancer is mostly reliant on estrogen or progesterone, especially in postmenopausal females. Generally, there are two common treatment approaches for breast cancer, modulation of estrogen receptor by selective estrogen receptor modulators or inhibition of aromatase enzyme by aromatase inhibitors. Aromatase is the key enzyme that acts on androgen precursors for the synthesis of estrogen. Aromatase inhibitors are used to either block the production of estrogen or block the action of estrogen on its receptors and for treatment of estrogen dependant breast cancer.32,33 In the current study, the investigation of the effect of isolated compounds on aromatase (CYP19A) revealed that all the tested compounds potently inhibited the effect of aromatase enzyme with IC50 range from 77.41 ± 4.53 to 436.40 ± 25.6 nM (Table 2).The IC50 values of compounds clitorin (5) and nicotiflorin (8) were 77.41 ± 4.53 and 92.84 ± 5.44 nM, respectively. The results are demonstrated in Fig. 2 and Table 2. It is noteworthy that clitorin (5) was more effective than the standard drug letrozole (IC50 = 77.72 ± 4.55). In addition, the nicotiflorin (8) showed a dual potent effect on both EGFR and aromatase comparable to the standard drugs which could be scaffold to the development of safe effective therapy for breast cancer.
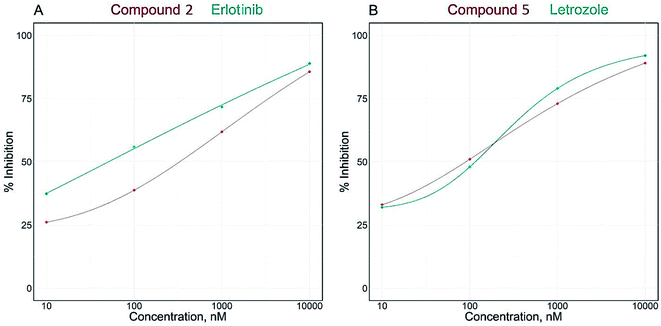 | ||
| Fig. 2 IC50 dose response curve of enzymatic inhibitory activities of isolated compounds 2 and 5 against EGFRwt (A) and aromatase (CYP19A) (B). | ||
2.5. Molecular docking study
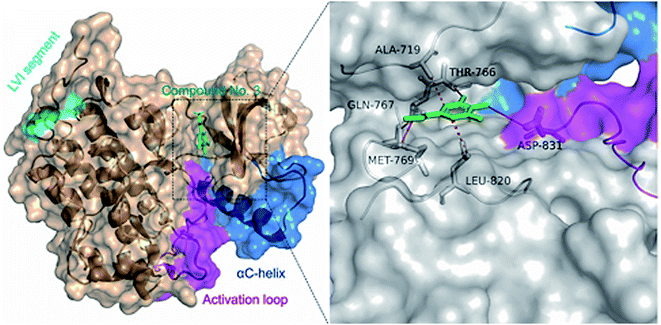
In order to understand the binding mode of the most potent compounds with the tested enzymes, a molecular docking analysis was conducted. EGFR kinase domain includes essential regulatory elements including αC helix (amino acids 729–744) and activation loop (amino acids 831–852).34 These elements are important for allosteric regulation and conformational changes and controlling EGFR activation. Our molecular docking analysis showed that methyl gallate (2) bound with the EGFR (PDB ID: 1M17) through hydrogen bonding with MET-769, THR-766, GLN-767 amino acid residues. In addition, methyl gallate (2) bound with ASP-831 in the activation loop of EGFR kinase domain. The binding pose score was −4.5287 kcal mol−1 with a root mean square deviation (RMSD) value of 1.69 Å in comparison to standard inhibitor erlotinib (−6.7615 kcal mol−1, RMSD = 1.24). Other hydrophobic interactions are shown in (Fig. 3).The catalytic cleft of aromatase comprises amino acids Ile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 305, Ala
305, Ala![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 306, Asp
306, Asp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 309 and Thr
309 and Thr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310 from the I-helix, Phe
310 from the I-helix, Phe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 221 and Trp
221 and Trp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 224 from the F-helix, Ile
224 from the F-helix, Ile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 133 and Phe
133 and Phe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134 from the B–C loop, Val
134 from the B–C loop, Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370, Leu
370, Leu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372 and Val
372 and Val![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 373 from the K-helix-β3 loop, Met
373 from the K-helix-β3 loop, Met![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 374 from β3, Leu
374 from β3, Leu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 477 and Ser
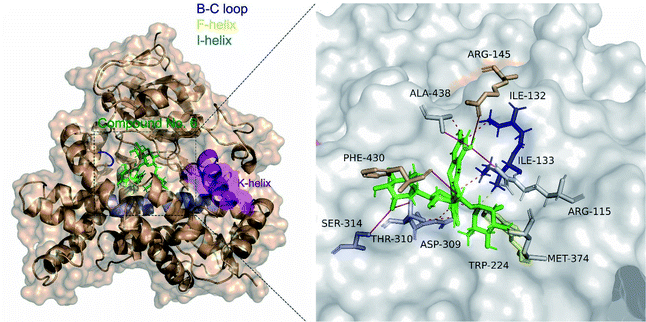
477 and Ser![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478 from the β8–β9 loop.35 Molecular docking analysis showed that clitorin (5) interacted with human aromatase cytochrome P450 (CYP19A1) (PDB ID: 3S79) with a posing score −14.2074 kcal mol−1 with RMSD value of 1.56 in comparison to standard inhibitors erlotinib (−11.2837 kcal mol−1, RMSD = 1.24) and letrozol (−7.2807 kcal mol−1, RMSD = 1.28). Clitorin (5) interacted through H-bonds with several amino acids in the catalytic cleft of aromatase. It formed H-bonds with ARG-145, ALA-438, PHE-430, ASP-309, SER-314 in the catalytic cleft I-helix, MET 374 and MET 311 (2 H-bonds) as hydrogen donors, while interacted as hydrogen acceptor with CYS 437 and MET 374 amino acid residues. It also interacted through hydrophobic interaction with ILE-133 from the BC-loop in the catalytic cleft (Fig. 4).
478 from the β8–β9 loop.35 Molecular docking analysis showed that clitorin (5) interacted with human aromatase cytochrome P450 (CYP19A1) (PDB ID: 3S79) with a posing score −14.2074 kcal mol−1 with RMSD value of 1.56 in comparison to standard inhibitors erlotinib (−11.2837 kcal mol−1, RMSD = 1.24) and letrozol (−7.2807 kcal mol−1, RMSD = 1.28). Clitorin (5) interacted through H-bonds with several amino acids in the catalytic cleft of aromatase. It formed H-bonds with ARG-145, ALA-438, PHE-430, ASP-309, SER-314 in the catalytic cleft I-helix, MET 374 and MET 311 (2 H-bonds) as hydrogen donors, while interacted as hydrogen acceptor with CYS 437 and MET 374 amino acid residues. It also interacted through hydrophobic interaction with ILE-133 from the BC-loop in the catalytic cleft (Fig. 4).
2.6. ADME pharmacokinetics and drug-likeness properties
The nine identified compounds were screened for their ADME pharmacokinetics and drug-likeness using the websites servers36,37 as previously described38 (Table 3). All tested compounds had good membrane permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p values ≤5). The compounds (1–3) possessed good bioavailability scores with 0.55 value. Furthermore, compounds (4–9), showed promising drug-likeness scores. Detailed molecular properties, absorption, distribution, metabolism and excretion in silico assessment are shown in Table 3.
p values ≤5). The compounds (1–3) possessed good bioavailability scores with 0.55 value. Furthermore, compounds (4–9), showed promising drug-likeness scores. Detailed molecular properties, absorption, distribution, metabolism and excretion in silico assessment are shown in Table 3.
| Molecule | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| MW | 478.72 | 184.15 | 196.25 | 610.52 | 740.66 | 594.52 | 464.38 | 594.52 | 478.4 |
| TPSA | 76.66 | 86.99 | 46.53 | 269.43 | 308.12 | 249.2 | 210.51 | 249.2 | 199.51 |
| MLOGP | 3.75 | 0.18 | 1.49 | −3.89 | −4.77 | −3.43 | −2.59 | −3.43 | −2.37 |
| No. atoms | 34 | 13 | 14 | 43 | 52 | 42 | 33 | 42 | 34 |
| nON | 6 | 5 | 3 | 16 | 19 | 15 | 12 | 15 | 12 |
| nOHNH | 2 | 3 | 1 | 10 | 11 | 9 | 8 | 9 | 7 |
| No. rotb | 0 | 2 | 0 | 6 | 8 | 6 | 4 | 6 | 5 |
| Fraction Csp3 | 0.93 | 0.12 | 0.73 | 0.44 | 0.55 | 0.44 | 0.29 | 0.44 | 0.32 |
| Rotatable bonds | 0 | 2 | 0 | 6 | 8 | 6 | 4 | 6 | 5 |
| H-Bond acceptors | 6 | 5 | 3 | 16 | 19 | 15 | 12 | 15 | 12 |
| H-Bond donors | 2 | 3 | 1 | 10 | 11 | 9 | 8 | 9 | 7 |
| Molvolume | 497.37 | 152.63 | 187.48 | 496.07 | 611.91 | 488.05 | 372.21 | 488.05 | 389.73 |
| Lipinski violations | 0 | 0 | 0 | 3 | 3 | 3 | 2 | 3 | 2 |
| Ghose violations | 2 | 0 | 0 | 4 | 4 | 4 | 1 | 4 | 0 |
| Veber violations | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Egan violations | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Muegge violations | 1 | 1 | 1 | 4 | 5 | 3 | 3 | 3 | 3 |
| ESOL class | Poorly soluble | Very soluble | Very soluble | Soluble | Soluble | Soluble | Soluble | Soluble | Soluble |
| GI absorption | High | High | High | Low | Low | Low | Low | Low | Low |
| BBB permeant | No | No | Yes | No | No | No | No | No | No |
| Pgp substrate | Yes | No | No | Yes | No | Yes | No | Yes | Yes |
| CYP1A2 inhibitor | No | No | No | No | No | No | No | No | No |
| CYP2C19 inhibitor | No | No | No | No | No | No | No | No | No |
| CYP2C9 inhibitor | No | No | No | No | No | No | No | No | No |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No | No | No | No | No |
| BBB score | 2.48 | 2.68 | 3.68 | 1.21 | 1 | 1.24 | 1.61 | 1.24 | 1.57 |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 |
| Drug-likeness model score | −1.49 | −0.65 | −1.02 | 0.91 | 0.9 | 0.88 | 0.68 | 0.9 | 0.59 |
3. Experimental section
3.1. General experimental procedures
1H- and 13C-NMR spectra were acquired at 25 °C using a Varian Inova 400 MHz NMR spectrometer. High-resolution mass spectra were measured using a Thermo scientific LTQ/XL Orbitrap, with FTMS analyzer, a mass range of 100–2000 and a resolution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. For LC-ESI-MS, gradient separation was obtained using a Sun Fire C-18 analytical HPLC column (5 mm, 4.6 × 150 mm, Waters) with a mobile phase of 0–100% MeOH over 30 min at a flow rate of 1 mL min−1. HPLC separation was performed on Agilent 1260 Infinity semi-preparative HPLC system with an Agilent Eclipse XDB-C18 column (5 μm, 10 × 250 mm, Agilent technologies, USA) monitored using an Agilent photodiode array detector. Detection was carried out at 220, 254, 280, 350 and 400 nm. All chemical reagents were purchased from Sigma-Aldrich (USA) and used without further purification. Medium pressure liquid chromatography (MPLC) separations were carried out using Biotage system with normal silica and reversed-phase pre-packed columns. UV-Detection was carried out at 220 and 254 nm. TLC was performed on pre-coated TLC plates with silica gel 60 F254 (layer thickness 0.2 mm, Merck, Darmstadt, Germany).
000. For LC-ESI-MS, gradient separation was obtained using a Sun Fire C-18 analytical HPLC column (5 mm, 4.6 × 150 mm, Waters) with a mobile phase of 0–100% MeOH over 30 min at a flow rate of 1 mL min−1. HPLC separation was performed on Agilent 1260 Infinity semi-preparative HPLC system with an Agilent Eclipse XDB-C18 column (5 μm, 10 × 250 mm, Agilent technologies, USA) monitored using an Agilent photodiode array detector. Detection was carried out at 220, 254, 280, 350 and 400 nm. All chemical reagents were purchased from Sigma-Aldrich (USA) and used without further purification. Medium pressure liquid chromatography (MPLC) separations were carried out using Biotage system with normal silica and reversed-phase pre-packed columns. UV-Detection was carried out at 220 and 254 nm. TLC was performed on pre-coated TLC plates with silica gel 60 F254 (layer thickness 0.2 mm, Merck, Darmstadt, Germany).
3.2. Plant material
C. papaya leaves were collected in June 2019 from the Research Farm, Faculty of Agriculture, Minia University, Minia, Egypt. The plant was identified by Prof. Mahmoud A. H. Abdou, Department of Horticulture, Faculty of Agriculture, Minia University. A voucher sample (Mn-Ph-Cog-044) was kept in the Herbarium of the Pharmacognosy Department, Faculty of Pharmacy, Minia University, Minia, Egypt. The plant materials were dried in shade, finely powdered and the powder was used for further analysis.3.3. Extraction and isolation
The air-dried powdered leaves of C. papaya (1.8 kg) were extracted with MeOH–H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v, 4 × 8 L each) in a closed glass container at room temperature for three times at one week interveals. The total hydromethanolic extract (THME) was concentrated under reduced pressure at 40 °C to afford a dark yellowish-green residue (160 g). THME was suspended in distilled water (500 mL) and successively partitioned between n-hexane (500 mL, 4×), methylene chloride (MC) (500 mL, 4×) and ethyl acetate (EtOAc) (500 mL, 4×); each fraction was concentrated under reduced pressure to give n-hexane (80 g), MC (16 g), EtOAc (25 g) and aqueous (30 g), respectively. A part of MC fraction (10 g) was chromatographed on Biotage flash chromatography system using prepacked silica column using n-hexane–EtOAc gradients (10
2, v/v, 4 × 8 L each) in a closed glass container at room temperature for three times at one week interveals. The total hydromethanolic extract (THME) was concentrated under reduced pressure at 40 °C to afford a dark yellowish-green residue (160 g). THME was suspended in distilled water (500 mL) and successively partitioned between n-hexane (500 mL, 4×), methylene chloride (MC) (500 mL, 4×) and ethyl acetate (EtOAc) (500 mL, 4×); each fraction was concentrated under reduced pressure to give n-hexane (80 g), MC (16 g), EtOAc (25 g) and aqueous (30 g), respectively. A part of MC fraction (10 g) was chromatographed on Biotage flash chromatography system using prepacked silica column using n-hexane–EtOAc gradients (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 9
0, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8.5
1, 8.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, 8
1.5, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7.5
2, 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 and 7
2.5 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) to afford compound 1 (20 mg), compound 2 (40 mg) and compound 3 (50 mg).
3, v/v) to afford compound 1 (20 mg), compound 2 (40 mg) and compound 3 (50 mg).
Furthermore, a part of EtOAc fraction (10 g) was subjected to MPLC Biotage system using prepacked RP-18 column chromatography with a mobile phase H2O–MeOH gradient (9.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 9
0.5, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8.5
1, 8.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, 8
1.5, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7.5
2, 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 and 7
2.5 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), v/v to afford four major fractions; fraction A (1.8 g), fraction B (2.6 g), fraction C (1.5 g) and fraction D (1.4 g). Fraction B was subjected to semi-preparative HPLC to afford compound 4 (8 mg), compound 5 (6 mg), compound 6 (3 mg), compound 7 (4 mg), compound 8 (5 mg) and compound 9 (7 mg).
3), v/v to afford four major fractions; fraction A (1.8 g), fraction B (2.6 g), fraction C (1.5 g) and fraction D (1.4 g). Fraction B was subjected to semi-preparative HPLC to afford compound 4 (8 mg), compound 5 (6 mg), compound 6 (3 mg), compound 7 (4 mg), compound 8 (5 mg) and compound 9 (7 mg).
Compound 1 (carpaine) was separated as a white crystalline. Its positive HR-ESI-MS: m/z 479.3828 [M + H]+ (calcd for C28H51N2O4: 479.3849). The 1H-NMR (400 MHz, CD3OD) δH: 4.98 (2H, br s, H-12, H-12′), 3.48 (2H, q, J = 6.4 Hz, H-11, H-11′), 3.18 (2H, m, H-9, H-9′), 2.36–2.52 (4H, m, H-2, H-2′), 1.81–2.01 (4H, m, H-13, H-13′), 1.68–1.78 (4H, m, H-14, H-14′), 1.68–1.78 (4H, m, H-3, H-3′), 1.68–1.78 (4H, m, H-7, H-7′), 1.34–1.43 (4H, m, H-4, H-4′), 1.34–1.43 (4H, m, H-5, H-5′), 1.34–1.43 (4H, m, H-6, H-6′), 1.34–1.43 (4H, m, H-8, H-8′), and 1.28 (6H, d, J = 6.7 Hz, CH3-11, 11′). 13C-NMR (100 MHz, CD3OD): δC: 174.2 (C-1, C-1′), 69.2 (C-12, C-12′), 58.3 (C-9, C-9′), 55.6 (C-11, C-11′), 34.9 (C-2, C-2′), 34.2 (C-8, C-8′), 29.9 (C-6, C-6′), 29.3 (C-5, C-5′), 29.0 (C-4, C-4′), 27.9 (C-13, C-13′), 26.3 (C-7, C-7′), 25.5 (C-14, C-14′), 24.0 (C-3, C-3′) and 15.7 (CH3 attached to C-11, C-11′). The spectral data of compound 1 are shown in Fig. S1–S6.†
Compound 2 (methyl gallate) was separated as a whitish-grey powder. The 1H-NMR (400 MHz, CD3OD) δH: 3.81 (3H, s, OCH3-1) and 7.04 (2H, s, H-2, H-6). The 1H-NMR spectrum is illustrated in Fig. S7.†
Compound 3 (loliolide) was separated as a white amorphous powder. The positive HR-ESI-MS: m/z 197.1179 [M + H]+ (calcd for C11H16O3: 197.1177). The 1H-NMR (400 MHz, CD3OD) δH: 5.75 (1H, s, H-7), 4.22 (1H, m, H-3), 2.48 (1H, dt, J = 13.5, 2.4 Hz, H-4a), 1.99 (1H, dt, J = 14.4, 3.3 Hz, H-2a), 1.76 (3H, s, CH3), 1.74 (1H, dd, J = 13.6, 3.8 Hz, H-4b), 1.55 (1H, dd, J = 14.4, 3.4 Hz, H-2b), 1.47 (3H, s, CH3) and 1.28 (3H, s, CH3). 13C-NMR (100 MHz, CD3OD) δC: 185.7 (C-6), 174.4 (C-8), 113.3 (C-7), 88.9 (C-5), 67.2 (C-3), 48.0 (C-2), 46.5 (C-4), 37.2 (C-1), 31.0 (C-9, CH3), 27.4 (C-11, CH3) and 27.0 (C-10, CH3). The spectral data of compound 3 are demonstrated in Fig. S8–S13.†
Compound 4 [Rutin (syn.: quercetin-3-O-rutinoside or sophorin or rutoside)] was separated as a yellow powder. The positive HR-ESI-MS: m/z 611.1607 [M + H]+ (calcd for C27H30O16: 611.1612). The full spectral data are shown in Fig. S14–S18.† The 1H-NMR (400 MHz, DMSO-d6) spectral data of the aglycone displayed signals at δH: 12.59 (1H, br s, OH–C5), 7.55 (1H, br s, H-2′), 7.54 (1H, d, J = 8.2 Hz, H-6′), 6.84 (1H, d, J = 8.2 Hz, H-5′), 6.38 (1H, d, J = 1.9 Hz, H-8), 6.19 (1H, d, J = 1.9 Hz, H-6); 3-glucosyl unit; 5.34 (d, J = 7.3 Hz, H-1′′) and rhamnosyl unit attached to C-6′′ of glucosyl unit; 4.38 (1H, br s, H-1′′′) and 0.99 (3H, d, J = 6.3 Hz, H-6′′′). 13C-NMR experiment (100 MHz, DMSO-d6), the aglycone showed signals at δC: 177.3 (C-4), 164.1 (C-7), 161.2 (C-5), 156.6 (C-2), 156.4 (C-9), 148.4 (C-4′), 144.7 (C-3′), 133.3 (C-3), 121.7 (C-5′), 121.2 (C-1′), 116.5 (C-6′), 115.2 (C-2′), 103.9 (C-10), 98.7 (C-6), 93.6 (C-8), carbons of glucosyl unit; δC 101.2 (C-1′′), 76.3 (C-3′′), 75.9 (C-5′′), 74.1 (C-2′′), 70.1 (C-4′′) and 67.6 (C-6′′) and carbons of rahmnosyl unit attached to C-6′′ of glucosyl unit δC: 100.7 (C-1′′′), 71.8 (C-4′′′), 70.6 (C-3′′′), 70.4 (C-2′′′), 68.2 (C-5′′′) and 17.7 (C-6′′′).
Compound 5 [clitorin (syn.: kaempferol 3-O-(2′′,6′′-di-α-O-rhamnopyranosyl)-β-glucopyranoside)] was separated as a yellowish powder. The positive HR-ESI-MS: m/z 741.2243 [M + H]+ (calcd for C33H40O19: 741.2242). 1D and 2D spectral data are illustrated in Fig. S19–S23.† The 1H-NMR spectral data (400 MHz, DMSO-d6) the aglycone displayed signals at δH: 12.64 (1H, br s, OH–C5), 7.95 (2H, d, J = 8.7, H-2′, H-6′), 6.87 (2H, d, J = 8.8, H-3′, H-5′), 6.40 (1H, d, J = 2.0 Hz, H-8), 6.19 (1H, d, J = 2.0 Hz, H-6); 3-glucosyl unit; 5.48 (d, J = 7.0 Hz, H-1′′), 3.66 (1H, m, H-6′′a), 3.22 (1H, m, H-6′′b), rhamnosyl unit attached to C-2′′ of glucosyl unit; 5.05 (1H, br s, H-1′′′), 0.96 (3H, d, J = 6.4 Hz, H-6′′′) and rhamnosyl unit attached to C-6′′ of glucosyl unit; 4.32 (1H, br s, H-1′′′) and 0.81 (3H, d, J = 6.4 Hz, H-6′′′). 13C-NMR experiment (100 MHz, DMSO-d6), the aglycone showed signals at δC: 177.2 (C-4), 164.1 (C-7), 161.2 (C-5), 159.8 (C-4′), 156.9a (C-9), 156.4a (C-2), 132.6 (C-3), 130.7 (C-2′), 130.7 (C-6′), 98.7 (C-6), 93.7 (C-8), 104.0 (C-10), 121.0 (C-1′), 115.1 (C-3′), 115.1 (C-5′), carbons of 3-glucosyl unit; δc 98.7 (C-1′′), 77.3b (C-3′′), 77.1b (C-5′′), 75.6 (C-2′′), 70.5c (C-4′′) and 68.26d (C-6′′), carbons of rhamnosyl unit attached to C-2′′ of glucosyl unit; δc 100.6e (C-1′′′), 71.81f (C-4′′′), 70.5c (C-3′′′), 70.3c (C-2′′′), 68.30d (C-5′′′) and 17.3g (C-6′′′), carbons of rhamnosyl unit attached to C-6′′ of glucosyl unit; δc 100.8e (C-1′′′), 71.78f (C-4′′′), 70.6c (C-3′′′'), 70.3c (C-2′′′), 68.3 (C-5′′′) and 17.7g (C-6′′′), (a, b, c, d, e, f and g signals may be interchanged).
Compound 6 [kaempferol-3-O-neohesperidoside (syn.: kaempferol-3-O-glucorhamnoside or kaempferol 3-O-(2′′-O-α-L-rhamnopyranosyl)-β-D-glucopyranoside)] was separated as a yellow amorphous powder. The positive HR-ESI-MS: m/z 595.1659 [M + H]+ (calcd for C27H31O15: 595.1657). The 1H-NMR and positive HR-ESI-MS spectral data are shown in Fig. S24–S25.† The 1H-NMR spectral data (400 MHz, DMSO-d6) of the aglycone displayed signals at δH: 12.64 (1H, br s, OH–C-5), 8.04 (2H, d, J = 8.8 Hz, H-2′, H-6′), 6.88 (2H, d, J = 8.8 Hz, H-3′, H-5′), 6.43 (1H, d, J = 2.0 Hz, H-8), 6.20 (1H, d, J = 2.0 Hz, H-6); 3-glucosyl unit; 5.65 (d, J = 7.2 Hz, H-1′′), 3.67 (1H, m, H-6′′a), 3.48 (1H, m, H-6′′b) and rhamnosyl unit attached to C-2′′ of glucosyl unit; 4.13 (1H, br s, H-1′′′) and 0.75 (3H, d, J = 6.0, H-6′′′).
Compound 7 [isoquercetin (syn.: quercetin-3-O-β-D-glucopyranoside or isoquercitrin or isotrifoliin)] was separated as a yellow powder. The positive HR-ESI-MS: m/z 465.1028 [M + H]+ (calcd for C21H21O12: 465.1027). The 1H-NMR and positive HR-ESI-MS spectral data are shown in Fig. S26–S27.† The 1H-NMR spectral data (400 MHz, DMSO-d6) of the aglycone displayed signals at δH: 12.63 (1H, br s, OH–C5), 7.58 (2H, m, H-2′, H-6′), 6.84 (1H, d, J = 8.8 Hz, H-5′), 6.40 (1H, d, J = 2.0 Hz, H-8), 6.20 (1H, d, J = 2.0 Hz, H-6); 3-glucosyl unit; 5.46 (d, J = 7.6 Hz, H-1′′) 3.57 (1H, m, H-6′′a), 3.43 (1H, m, H-6′′b), 3.26 (2H, m, H-2′′, H-3′′) and 3.09 (2H, m, H-4′′, H-5′′).
Compound 8 [nicotiflorin (syn.: kaempferol-3-O-rutinoside or nicotifloroside or nictoflorin)] was separated as a yellow amorphous powder. The positive HR-ESI-MS: m/z 595.1661 [M + H]+ (calcd for C27H30O16: 595.1663). The 1H-NMR and positive HR-ESI-MS spectral data are shown in Fig. S28–S29.† The 1H-NMR (400 MHz, DMSO-d6) spectral data of the aglycone displayed signals at δH: 12.55 (1H, br s, OH–C5), 7.98 (2H, d, J = 8.8 Hz, H-2′, H-6′), 6.87 (2H, d, J = 8.8 Hz, H-3′, H-5′), 6.41 (1H, d, J = 1.6 Hz, H-8), 6.20 (1H, d, J = 1.6 Hz, H-6); 3-glucosyl unit; 5.31 (d, J = 7.6 Hz, H-1′′), 3.66–3.33 (2H, m, H-6) and rhamnosyl unit attached to C-6′′ of glucosyl unit; 4.37 (1H, br s, H-1′′′) and 0.96 (3H, d, J = 6.0 Hz, H-6′′′).
Compound 9 (isorhamnetin 3-O-β-D-glucopyranoside) was separated as a yellow amorphous powder. The positive HR-ESI-MS: m/z 479.1190 [M + H]+ (calcd for C22H22O12: 479.1189). 1D and 2D spectral data are illustrated in Fig. S30–S34.† The 1H-NMR spectral data (400 MHz, DMSO-d6) of the aglycone displayed signals at δH: 12.61 (1H, br s, OH–C5), 7.94 (1H, d, J = 2.0 Hz, H-2′), 7.49 (1H, dd, J = 8.4, 2.0 Hz, H-6′), 6.92 (1H, d, J = 8.4 Hz, H-5′), 6.45 (1H, d, J = 2.0 Hz, H-8), 6.21 (1H, d, J = 2.0 Hz, H-6), 3.84 (3H, s, OCH3-3′); 3-glucosyl unit; 5.57 (d, J = 7.2 Hz, H-1′′), 3.57 (1H, m, H-6′′a), 3.39 (1H, m, H-6′′b), 3.23 (2H, m, H-2′′, H-3′′) and 3.11 (2H, m, H-4′′, H-5′′). 13C-NMR experiment (100 MHz, DMSO-d6), the aglycone showed signals at δC: 177.4 (C-4), 164.2 (C-7), 161.2 (C-5), 156.4 (C-9), 156.3 (C-2), 149.4 (C-4′), 146.9 (C-5′), 133.0 (C-3), 122.0 (C-2′), 121.1 (C-1′), 115.2 (C-3′), 113.5 (C-6′), 104.0 (C-10), 98.7 (C-6), 93.7 (C-8), 55.7 (OCH3), carbons of 3-glucosyl unit; δc 100.8 (C-1′′), 77.5 (C-5′′), 76.4 (C-3′′), 74.3 (C-2′′), 69.8 (C-4′′) and 60.6 (C-6′′).
3.4. MTT cytotoxicity assay in breast cancer cells line (MCF-7)
The cell viability assay was performed to evaluate the effect of the isolated compounds (1–9) from C. papaya leaves on breast cancer cell line (MCF-7) using the in vitro MTT assay protocol as previously described by Hamed et al., 2021.39 Briefly, breast cancer cell line (MCF-7) were obtained from American Type Culture Collection and cultured in DMEM medium (Invitrogen/Life Technologies) supplemented with 10% FBS (Hyclone, USA). Insulin (10 μg mL−1, Sigma-Aldrich, USA) and 1% penicillin–streptomycin. the cells were seeded in 96-well plates (VWR, Switzerland) at cell density 1.2–1.8 × 104 cells per well. Then cells were treated with the isolated compounds (1–9) at serial concentrations in triplicates for 24 h. DMSO (0.1%, w/v) was used as a blank control and staurosporine was used as a positive control. After treatments, the medium was removed from all wells of the plate and 20 μL of the MTT reagent (Sigma-Aldrich, USA) (0.5 mg mL−1) in PBS was added and incubated for 3.5 h at 37 °C. The solution was removed and 50 μL of DMSO was added, incubated for 5 min and the absorbance was measured at 570 nm.3.5. EGFRwt kinase inhibition activity
Evaluation of the inhibitory activity of the isolated compounds (1–9) against EGFRwt kinase was carried out using EGFRwt Kinase Assay Kit (BPS biosciences) according to manufacturer's instructions.40,41 In brief, the master mixture was constructed from EGFRwt enzyme, their substrates, ATP and kinase assay enzymatic buffer were incubated with the tested compounds for 40 min at 30 °C to achieve the enzymatic reaction. Then, the reaction was stopped by the addition of a detecting reagent (Kinase-Glo Max reagent), followed by incubation at room temperature for another 15 min. Finally, luminescence was measured using the microplate Robonik P2000 ELISA Reader. All samples and controls were tested in duplicate and the results are presented as percentage enzyme inhibition and compared to erlotinib selected as reference drugs due to their potent inhibitory activity of EGFRwt.3.6. Aromatase (CYP19A) inhibition activity
The in vitro aromatase inhibitory activity of the compounds (1–9) was evaluated using (Bio Vision, Aromatase (CYP19A) Inhibitor Screening Kit (Fluorometric)) in comparison to letrozole as the reference drug. This method was carried out according to Acar Çevik et al., 2020 procedure.42 The compounds were dissolved in DMSO in concentrations ranging from 10 to 104 nM. The recombinant human aromatase stock was prepared according to the protocol and the samples were added and the plate was incubated for 10 min at 37 °C to allow tested compounds to interact with the aromatase. After incubation, 30 μL of the aromatase substrate/NADP+ mixture was added to each well and the fluorescence at Ex/Em = 488/527 nm was measured immediately (within 1 min).3.7. Molecular docking simulation
Molecular docking was performed as briefly described to understand the binding affinity of the potent compounds in comparison with the reference drugs at the molecular level.43 The crystal structure of both the epidermal growth factor receptor tyrosine kinase (EGFRwt) (PDB ID: 1M17) and Human placental aromatase cytochrome P450 (CYP19A1) for the breast cancer receptor (PDB ID: 3S79) were obtained from the Protein data bank. The molecular docking simulation was conducted using the “Molecular Operating Environment (MOE 2014.9) and preparation and optimization of both ligands and receptors were carried out according to induced fit MOE protocol.44 The parameters of scoring were Triangle Matcher, scoring was set at London dG and rescoring at GBVI/WSA dG. The docking score, root mean square deviation (RMSD) and ligand–receptor complexes were tested for interaction analysis. The 3D images were made using the MOE visualizing tool.3.8. Drug like properties and ADME prediction of isolated compounds
The drug likeliness, molecular properties prediction ADME and pharmacokinetic parameters of the isolated compounds were calculated using a set of software including “MolSoft,” “Molinspiration”, “PreADME” and “SwissADME” websites servers.36,373.9. Statistical analysis
Results are expressed as mean ± standard deviation (SD) based on triplicate experiments. The IC50 values of the tested compounds were determined using curve fitting in the R programming language and associated packages including Magrittr,45 drc46 and ggplot2.47 Graphs and figures were generated using R console and PyMOL (PyMOL Molecular Graphics System, Schrödinger).4. Conclusion
Due to the reported various secondary metabolites of C. papaya Linn. leaves, in addition to their promising cytotoxic activity against breast cancer cell line supported by protein kinase inhibition activities and molecular docking study. They could be considered as potential candidates against breast cancer. Therefore, further investigations could have a supportive role in the pharmaceutical field towards the development of new breast anti-cancer drugs.Conflicts of interest
The authors declare no conflict of interest.References
- C. Ramona, B. Ana, C. Mihai and F. Stãnicã, J. Hortic. Sci. Biotechnol., 2017, 21, 130–136 Search PubMed.
- F. A. Carvalho and S. S. Renner, in Genetics and genomics of papaya, Springer, 2014, pp. 81–92 Search PubMed.
- A. Daagema, P. Orafa and F. Igbua, Eur. J. Food Res. Rev., 2020, 13, 52–66 Search PubMed.
- A. Nugroho, H. Heryani, J. S. Choi and H.-J. Park, Asian Pac. J. Trop. Biomed., 2017, 7, 208–213 CrossRef.
- Y. H. Chai, S. Yusup and M. S. Ruslan, Curr. Nutr. Food Sci., 2020, 16, 1287–1298 CrossRef CAS.
- S. P. Singh, S. Kumar, S. V. Mathan, M. S. Tomar, R. K. Singh, P. K. Verma, A. Kumar, S. Kumar, R. P. Singh and A. Acharya, Daru, J. Pharm. Sci., 2020, 28, 735–744 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- S. K. Mohamed, MJN, 2021, 12, 111–119 CrossRef.
- J. C. Bailar and H. L. Gornik, N. Engl. J. Med., 1997, 336, 1569–1574 CrossRef PubMed.
- A. L. Harvey, R. Edrada-Ebel and R. J. Quinn, Nat. Rev. Drug Discovery, 2015, 14, 111–129 CrossRef CAS PubMed.
- C. I. Liyongo, C. M. Ashande, C. F. Mawi, B. M. wa Mbembo, J.-J. A. Domondo, M. I. S. Matondang, J.-P. K. Kayembe and P. T. Mpiana, BIoEx J., 2020, 2, 663–675 CrossRef.
- J. L. Hsu and M.-C. Hung, Cancer Metastasis Rev., 2016, 35, 575–588 CrossRef CAS PubMed.
- H. Ge, X. Gong and C. K. Tang, Int. J. Cancer, 2002, 98, 357–361 CrossRef CAS PubMed.
- C. Choccalingam, L. Rao and S. Rao, Breast Cancer: Basic Clin. Res., 2012, 6, 21–29 Search PubMed.
- N. Harada, J. Steroid Biochem. Mol. Biol., 1997, 61, 175–184 CrossRef CAS PubMed.
- S. Chumsri, T. Howes, T. Bao, G. Sabnis and A. Brodie, J. Steroid Biochem. Mol. Biol., 2011, 125, 13–22 CrossRef CAS PubMed.
- R. Clarke, M. C. Liu, K. B. Bouker, Z. Gu, R. Y. Lee, Y. Zhu, T. C. Skaar, B. Gomez, K. O'Brien and Y. Wang, Oncogene, 2003, 22, 7316–7339 CrossRef CAS PubMed.
- Y. Zhou, C. Yau, J. W. Gray, K. Chew, S. H. Dairkee, D. H. Moore, U. Eppenberger, S. Eppenberger-Castori and C. C. Benz, BMC Cancer, 2007, 7, 1–15 CrossRef PubMed.
- D. Thi Hoa Vien and T. Van Loc, Int. J. Environ. Agric. Biotech., 2017, 2, 2394–2397 Search PubMed.
- S. El-Sayyad, M. Mohamed, M. Kamel, K. Mohamed and A. El-Hifnawy, Bull. Pharm. Sci. Assuit Univ., 1999, 22, 123–130 CAS.
- J. Panyo, K. Matsunami and P. Panichayupakaranant, Pharm. Biol., 2016, 54, 1522–1527 CrossRef CAS PubMed.
- B. Wang and H. Hu, Asian J. Chem., 2013, 25, 2012–2014 Search PubMed.
- K. Yoshitama, T. Oyamada and S. Yahara, J. Plant Res., 1997, 110, 379–381 CrossRef CAS.
- T. Iwashina, M.-a. Yamaguchi, M. Nakayama, T. Onozaki, H. Yoshida, S. Kawanobu, H. Ono and M. Okamura, Nat. Prod. Commun., 2010, 5, 1903–1906 CAS.
- H. D. Smolarz, Pharm. Biol., 2002, 40, 390–394 CrossRef CAS.
- S.-Y. Lee, J.-S. Kim and S.-S. Kang, Korean J. Pharmacogn., 2012, 43, 206–212 Search PubMed.
- D.-M. Wang, W.-J. Pu, Y.-H. Wang, Y.-J. Zhang and S.-S. Wang, Molecules, 2012, 17, 4595–4603 Search PubMed.
- Y.-S. Sun, Z. Zhao, Z.-N. Yang, F. Xu, H.-J. Lu, Z.-Y. Zhu, W. Shi, J. Jiang, P.-P. Yao and H.-P. Zhu, Int. J. Biol. Sci., 2017, 13, 1387–1397 Search PubMed.
- Y. Li, S. Li, X. Meng, R.-Y. Gan, J.-J. Zhang and H.-B. Li, Nutrients, 2017, 9, 728–766 CrossRef PubMed.
- K. Nakai, M.-C. Hung and H. Yamaguchi, Am. J. Cancer Res., 2016, 6, 1609–1632 CAS.
- A. E. Maennling, M. K. Tur, M. Niebert, T. Klockenbring, F. Zeppernick, S. Gattenlöhner, I. Meinhold-Heerlein and A. F. Hussain, Cancers, 2019, 11, 1826–1845 CrossRef CAS PubMed.
- R. Kharb, K. Haider, K. Neha and M. S. Yar, Arch. Pharm., 2020, 353, 2000081–2000101 CrossRef CAS PubMed.
- A. Ali, N. U. Jan, S. Ali, B. Ahmad, A. Ali, S. Samrana, A. Jahan, H. Ali, I. A. Khan and H. Rahim, Chem. Biol. Drug Des., 2020, 95, 233–239 CrossRef CAS PubMed.
- M. Landau, S. J. Fleishman and N. Ben-Tal, Structure, 2004, 12, 2265–2275 Search PubMed.
- D. Ghosh, J. Griswold, M. Erman and W. Pangborn, Nature, 2009, 457, 219–223 Search PubMed.
- A. E. Allam, H. K. Assaf, H. A. Hassan, K. Shimizu and Y. A. Elshaier, RSC Adv., 2020, 10, 29983–29998 Search PubMed.
- R. F. Abdelhameed, E. S. Habib, A. K. Ibrahim, K. Yamada, M. S. Abdel-Kader, S. A. Ahmed, A. K. Ibrahim, J. M. Badr and M. S. Nafie, Molecules, 2021, 26, 940–953 CrossRef CAS PubMed.
- M. S. Nafie, A. M. Amer, A. K. Mohamed and E. S. Tantawy, Bioorg. Med. Chem., 2020, 28, 115828 CrossRef CAS PubMed.
- A. N. Hamed, N. A. Abdelaty, E. Z. Attia, M. N. Amin, T. F. Ali, A. H. Afifi, U. R. Abdelmohsen and S. Y. Desoukey, Nat. Prod. Res., 2021, 1–5 Search PubMed.
- J. L. Nakamura, Expert Opin. Ther. Targets, 2007, 11, 463–472 CrossRef CAS PubMed.
- S. A. Elmetwally, K. F. Saied, I. H. Eissa and E. B. Elkaeed, Bioorg. Chem., 2019, 88, 102944–102960 Search PubMed.
- U. Acar Çevik, B. Kaya Çavuşoğlu, B. N. Sağlık, D. Osmaniye, S. Levent, S. Ilgın, Y. Özkay and Z. A. Kaplancıklı, Molecules, 2020, 25, 1642–1657 CrossRef PubMed.
- M. E. Abouelela, H. K. Assaf, R. A. Abdelhamid, E. S. Elkhyat, A. M. Sayed, T. Oszako, L. Belbahri, A. E. E. Zowalaty and M. S. A. Abdelkader, Molecules, 2021, 26, 1767–1796 CrossRef CAS PubMed.
- E. S. Tantawy, A. M. Amer, E. K. Mohamed, M. M. Abd Alla and M. S. Nafie, J. Mol. Struct., 2020, 1210, 128013–128022 CrossRef CAS.
- P. Lemenkova, Carpathian J. Electr. Comp. Eng., 2020, 13, 5–12 Search PubMed.
- C. Ritz, J. C. Strebig and M. C. Ritz, Creative Commons: Mountain View, CA, USA, 2016 Search PubMed.
- H. Wickham, Wiley Interdiscip. Rev. Comput. Stat., 2011, 3, 180–185 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07000b |
| This journal is © The Royal Society of Chemistry 2022 |