Nanomolar LL-37 induces permeability of a biomimetic mitochondrial membrane†
Xin
Jiang‡
a,
Chenguang
Yang‡
bc,
Jie
Qiu
a,
Dongfei
Ma
a,
Cheng
Xu
ad,
Shuxin
Hu
 bc,
Weijing
Han
*a,
Bing
Yuan
bc,
Weijing
Han
*a,
Bing
Yuan
 *a and
Ying
Lu
*bc
*a and
Ying
Lu
*bc
aSongshan Lake Materials Laboratory, Dongguan, Guangdong 523808, China. E-mail: yuanbing@sslab.org.cn
bBeijing National Laboratory for Condensed Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
dCenter for Soft Condensed Matter Physics and Interdisciplinary Research & School of Physical Science and Technology, Soochow University, Suzhou, Jiangsu 215006, China
First published on 16th November 2022
Abstract
LL-37, the only human host cathelicidin peptide, is proposed to be able to induce host cell apoptosis through mitochondrial membrane permeabilization (MMP). Detailed pathways of the LL-37-triggered MMP are however still disputed. It is generally believed that cationic peptides permeate a membrane mostly in conditions of micromolar peptide concentrations and negatively charged membranes, which are not usually satisfied in the mitochondrial circumstance. Herein, using a variety of single-molecule techniques, we show that nanomolar LL-37 specifically induces permeability of a phosphoethanolamine (PE)-rich biomimetic mitochondrial membrane in a protein-independent manner. The insertion dynamics of single LL-37 molecules exhibit different metastable states in bilayers composed of different lipids. Moreover, the PE lipids significantly facilitate adsorption and accumulation of LL-37 on the PE-rich bilayer, and produce deeper insertion of peptide oligomers, especially tetramers, into the bilayer. This work offers an alternative pathway of the LL-37-triggered MMP and apoptosis.
Introduction
LL-37, the only human member of the cathelicidin family of host defense peptides, is one of the typical natural amphipathic cationic peptides which have potent membrane permeabilizing ability over a variety of different host cell and bacterial membranes.1–3 Besides the well-known antimicrobial ability, LL-37 is a multifunctional peptide that links host defense, inflammation, tissue repair and cancer.4–6 Studies have demonstrated the accumulation of exogenous LL-37 at mitochondria.5 Moreover, it is well-documented that LL-37-induced host cell cytotoxicity involves caspase-independent apoptosis via induction of mitochondrial membrane permeabilization (MMP) which is a critical event in the process of physiologic or chemotherapy-induced apoptosis.7–9 Three possible pathways of the LL-37-triggered MMP have been proposed but there is still a lack of definite evidence to support or negate any of the speculations.One theory is that LL-37 activates a GPCR p53-Bax/Bak/Bcl-2 signaling cascade at mitochondria to mediate the apoptotic pathway.8,10,11 There are also some reports suggest that LL-37 disturbs a lipid raft structure in the mitochondrial outer membrane, leading to activation of specific pathways.8,12 It is more likely, though, that LL-37 directly permeates the lipid membrane in a protein-independent manner.8,13–16 However, this last theory is challenged since it has been normally accepted that an amphipathic cationic peptide induces membrane permeability only when it is in high concentrations above a threshold value (e.g., a few μM) and over negatively charged membranes.17–19 As such, most present studies on the action mechanism of amphipathic cationic peptides (e.g., LL-37) are performed using micromolar agents based on a combination of many biochemical techniques such as the fluorescence localization imaging and downstream cytokine detection.5,20–23 The effect of nanomolar peptides, which might also exist in the physiological circumstances, is significantly neglected. On the one hand, LL-37 is produced by specific cells at all times and the physiological concentration of it covers a wide range (from nM to μM).24 Moreover, a globular C1q receptor (p33) protein, which is detected in mitochondria, shows high affinity for LL-37, and, interestingly, shows negative correlation with the cytotoxic effect of LL-37 over host cells.25 This suggests that the p33 protein may recruit LL-37 to the mitochondrial membrane while decrease its local concentration to a quite low level. On the other hand, high concentrations (>4 μM) of LL-37 are shown to be cytotoxic to many different human cell types through permeabilizing the cellular plasma membranes.5,25 This makes the potential mitochondrial disturbance of peptides be covered by the potent cytotoxicity due to plasma membrane permeabilization in related investigations. Therefore, it is of vital importance to understand the membrane permeabilizing effect of LL-37 at low (e.g., nanomolar) concentrations.
The membrane action process of amphipathic cationic peptides normally consists of two steps, including the surface binding and transformation of peptides followed by reorientation of peptides and permeabilization of the membrane.19 To overcome the high energy barrier of peptide reorientation and membrane insertion, oligomerization and fibrillation of peptides might occur.26–28 A variety of structural techniques have been used in previous studies to provide insight into the ensemble average structure of micromolar LL-37.29,30 However, the molecular dynamics of LL-37 interaction with membranes remains unclear. In this work, using a combination of several single-molecule methods, we show that LL-37 at low concentrations (e.g., 1 or 10 nM) is able to induce membrane permeability over a biomimetic mitochondrial membrane. Moreover, the physical molecular mechanism underlying the membrane penetration ability of LL-37 is revealed for the first time. A phosphoethanolamine (PE)-rich bilayer membrane was fabricated to mimic the mitochondrial outer membrane.31,32 A phosphocholine (PC)-rich bilayer mimicking the eukaryotic cell plasma membrane, and a heavily charged phosphatidylic acid (PA)-rich bilayer, were also fabricated for comparison.31 The results obtained using the newly developed surface-induced fluorescence attenuation (SIFA) technique33 in combination with single-molecule imaging and traditional vesicle leakage assay demonstrate that, in comparison of PA which enhances the initial binding of LL-37 on the bilayer surface alone, PE significantly facilitates the accumulation and insertion of LL-37 on/into the bilayer, leading to an enhanced membrane permeabilization effect. Moreover, the PE-triggered oligomerization of LL-37 is found responsible for the insertion of peptides into the bilayer. This work proves an alternative pathway of the LL-37-triggered MMP and apoptosis, which is helpful for the design of advanced chemotherapy drugs.
Results and discussion
Lipid-specific membrane permeabilization by nanomolar LL-37
LL-37 possesses 37 amino acid residues that are distributed among a long amphipathic helix (residues 2–31) and a disordered tail (residues 32–37)34 (Fig. 1a). To understand the membrane permeabilization activity of LL-37 at the molecular level, experiments were performed using nanomolar concentrations of the peptide (e.g., 1 or 10 nM) approaching the minimum physiological concentration.24 In order to characteristically distinguish the mitochondrial outer, plasma, and strongly charged membranes, PE-, PC-, and PA-rich model membrane systems were fabricated, respectively.31,32 The permeabilization of LL-37 through these membranes was studied by monitoring the real-time fluorescence of the Fluo-4 fluorophores encapsulated in Cy5-labeled unilamellar liposomes that were dispersed in a calcium-containing buffer solution and immobilized on the substrate coverslip. Upon permeation, Ca2+ flows into the liposome, thereby enhancing the Fluo-4 fluorescence signal observed using a total internal reflection fluorescence microscope (TIRFM; Fig. 1b; see more details in Materials and methods, ESI†).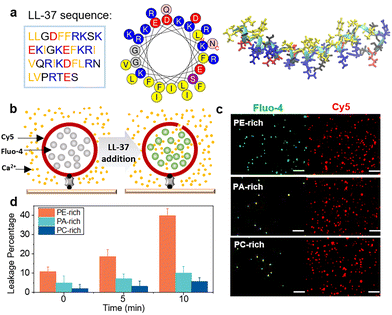 | ||
| Fig. 1 Membrane permeabilization by nanomolar LL-37. (a) Amino acid sequence, helical wheel, and molecular structure of LL-37 (PDB ID: 2K6O). The basic, acidic, and hydrophobic amino acids are shown in blue, red, and yellow, respectively. (b) Schematic illustration of the liposome leakage assay. (c) Typical total internal reflection fluorescence microscopy (TIRFM) images of liposomes labeled with Cy5-PE (red colour in the right channel) and the liposome-encapsulated Fluo-4 (green colour in the left channel) in a solution containing 10 mM Ca2+ and 10 nM label-free LL-37. The images were captured after 10 min of incubation with the peptide. Scale bar in the image is 2 μm. (d) Percentage of leaked vesicles in PE-, PA-, and PC-rich membranes. The statistics in (d) correspond to at least three independent experiments, and the error bars represent the statistical error. | ||
The results illustrated in Fig. S1† demonstrate that after liposome immobilization on the substrate, the three liposome systems with varying lipid composition exhibit similar liposome numbers. In control experiments, the liposomes remain unchanged with no Ca2+ entry for more than 10 min, indicating stability of the system (Fig. S1 and S2†). The fluorescence of individual liposomes in the Fluo-4 channel detected upon the addition of 10 nM wild-type LL-37 in situ confirms that the peptide induces a transmembrane influx of Ca2+. Based on the fluorescent images captured after 10 min incubation (Fig. 1c), PE-rich liposomes exhibit more Fluo-4 fluorescent spots than the other two systems. This suggests that LL-37 permeabilizes PE-rich membranes more effectively than PC- and PA-rich membranes. Indeed, in the PE-rich system, immediate liposome leakage after LL-37 addition is ∼10%, which is significantly higher than the leakage detected in PA- or PC-rich systems (<5%; Fig. 1d and S3†). After 5 min, the leakage percentage of PE-rich liposomes increases to ∼19%, which further reaches 40% after 10 min of incubation with LL-37 (Fig. 1c and d). To confirm the membrane permeabilizing ability of LL-37, Escherichia coli protoplasts, which are also characteristically rich in PE component,35 were prepared and exposed to LL-37 at 10 nM. The protoplasts are effectively permeabilized as well (Fig. S4†). These observations demonstrate that the nanomolar LL-37 has a much stronger permeabilizing ability against the biomimetic mitochondrial (as well as the bacterial) membranes that are rich in PE, than the negatively charged PA-rich membranes or PC-rich eukaryotic plasma membranes.
Membrane-insertion dynamics of monomeric LL-37 peptides
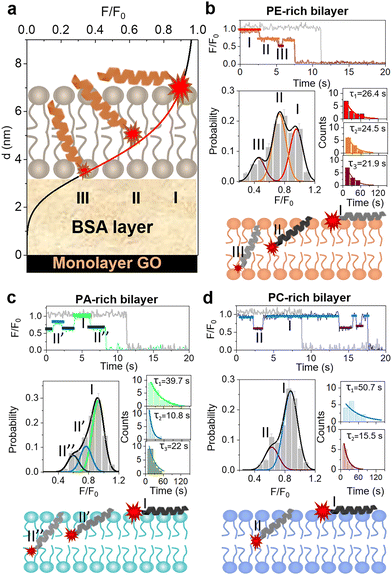
The results observed above suggest that LL-37 exhibits lipid-specific, PE-facilitated membrane permeabilization activity. To elucidate the mechanism underlying this activity, the dynamics of peptide insertion into a bilayer was studied using the recently developed single-molecule surface-induced fluorescence attenuation (SIFA) method. SIFA is able to monitor in real time the Z-position of a fluorophore in a supported lipid bilayer (SLB) with sub-nanometer resolution.33,36,37 A SLB membrane was deposited on a graphene oxide (GO) monolayer, and a BSA layer between them was used as spacer. Considering that the energy transfer between fluorophores and GO depends on the distance between them, changes in the Z-position of an individual fluorophore (i.e., the distance of the probe from GO; d) were deduced from the temporal variation of its normalized fluorescence intensity (normalization against the pristine fluorescence intensity of the dye; F/F0; Fig. 2a; refer to the ESI†). To accurately reflect the effect of membrane composition, different F0 (pristine fluorescence intensity of the dye) values were measured for different membrane systems (Fig. S5†).The N- and C-termini of LL-37 were separately labeled with TAMRA (N-TAMRA-LL-37 and C-TAMRA-LL-37, respectively) in order to study the bilayer insertion states of both termini. First, 1 nM C-TAMRA-LL-37 peptides were added to PE-, PA-, and PC-rich SLBs. It is found that the fluorescence intensity of the peptides does not change significantly with time, which indicates that the C-termini of these peptides remain on the membrane surface (Fig. S6†). The same experiment was performed with labeled N-termini of the peptides, and the obtained results show that N-TAMRA-LL-37 exhibits significant fluctuation in fluorescence intensity. Interestingly, in the case of the PE-rich system, most peptides present obvious changes in fluorescence intensity before photobleaching (as representatively shown in Fig. 2). In addition, three preferred F/F0 values corresponding to the three peaks observed in the fluorescence intensity distribution histograms (states I, II and III) are measured (Fig. 2b). These values refer to the locations of the upper surface (molecular proportion of 36.6%), the central layer between leaflets (46.2%), and the lower surface of the bilayer (15.8%; see Tables S1–S3 in ESI†), which are determined upon conversion to Z-position. The obtained results suggest that the N-terminus of LL-37 undergoes transmembrane insertion and that a favourable state (state II) exists at the bilayer center during the transmembrane translocation of peptides. Notably, the dwell times of states I, II, and III are 26.4, 24.5, and 21.9 s, respectively (refer to Fig. S7† for the definition of dwell time). Considering that these values are much larger than those determined for other AMPs such as melittin (a few seconds only),38 the translocation dynamics of LL-37 across the bilayer is slower than that of melittin.
The PA- and PC-rich membranes were also exposed to N-TAMRA-LL-37, and the results (Fig. 2c and d) show that peptides are not fully inserted into the PA-rich bilayer, as evidenced by the absence of state III in the F/F0 histogram. However, two barely distinguished metastable states constituting 38.0% of all states are detected near the central layer (termed as states II′ and II′′). This indicates that LL-37 cannot be efficiently inserted into a PA-rich bilayer. Moreover, the dwell time of state I in the PA-rich bilayer is much longer (39.7 s) than that in the PE-rich counterpart, which suggests that the peptides are strongly bound to the lipids on the bilayer surface. Similar results are observed in the case of the PC-rich bilayer. Indeed, the peptides interacting with this membrane are mostly located on the membrane surface (88.5%, with a dwell time of 50.7 s), and the inter-leaflet state (state II) can be obviously distinguished. Altogether, monomeric LL-37 peptides can penetrate through a PE-rich bilayer via their N-termini, with a favourable state at the interface between the two leaflets. In contrast, these peptides are inclined to adsorb on the surface of PA- and PC-rich membranes, and only a small proportion of them are half inserted into the bilayers. This strengthened insertion ability of LL-37 into a PE-rich membrane probably attributes to the nanomolar-peptide-induced permeability of the biomimetic mitochondrial membrane.
To further reveal the insights, investigations on the helical structure and oligomerization state, which play key roles in determining the membrane actions of peptides,39,40 were carried out. CD spectra of LL-37 (5 μM) were acquired after incubation with liposomes of different lipid compositions. The peptides exhibit a disordered structure in buffer (Fig. S8a†), but in the presence of liposomes, particularly PE-rich liposomes, their helical architecture is strengthened. Indeed, the helical content in PE-, PA-, and PC-rich membrane systems is 70.0 ± 1.0, 43.2 ± 3.7, and 33.1 ± 1.1%, respectively (Fig. S8b†). The positive correlation between helical content of peptides and leakage possibility of liposomes suggests that the lipid-specific membrane permeabilizing ability of LL-37 may be related to the unique disorder-to-order conformational change of peptides in different membrane environments.
Time-dependent accumulation and assembly of LL-37 in different membrane systems
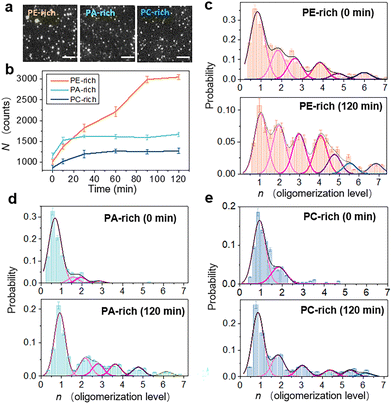
The accumulation and oligomerization degree of peptide molecules on/in the membrane are the principal factors affecting the membrane insertion efficiency of the peptide.26–28 Herein, the accumulation state of LL-37 on/in SLB membranes of varying lipid composition was estimated using the single-molecule fluorescence method. 1 nM TAMRA-labeled LL-37 was added to SLB membranes containing different phospholipid components, and fluorescent images were recorded over time under TIRFM. These images were used to calculate the mean amount of peptides in the field of vision (40 × 40 μm), N, as well as the number of molecules accumulated in individual oligomers, n. Representative fluorescence images of the SLB membranes captured at 90 min after LL-37 addition and the temporal evolution profiles of N for different membrane systems are shown in Fig. 3a and b, respectively (refer to Materials and methods, ESI†). Obviously, LL-37 molecules accumulate on the lipid bilayer over time. Immediately after LL-37 addition (t = 0 min), more peptides are detected on the PA-rich membrane than on the PE- or PC-rich membranes. After 30 min of incubation (t = 30 min), the PA- and PC-rich surfaces are saturated with peptides, but the number of peptides on the PE-rich membrane continues to increase until it reaches approximately twice the number detected on the PA-rich surface (t = 90 min).The value of n was determined according to the single-molecule stepwise photobleaching method,37,41 wherein the number of peptides per single immobile oligomer is obtained by evaluating the steplike photobleaching drops in the fluorescent-intensity trace. Fig. S9† shows the typical fluorescent-intensity traces of photobleaching used to determine the n of individual oligomers, whereas Fig. 3c–e shows the probability distribution histograms of n for different membrane systems, at 0 and 120 min after LL-37 addition. Interestingly, in the presence of PA- or PC-rich membranes, LL-37 exists mostly as monomers or dimers, irrespective of the incubation duration. However, in the presence of the PE-rich membrane, oligomerization tends to occur immediately after peptide addition. Indeed, at t = 0 min, the oligomers (up to hexamers) account for ∼53.5% of the spots, which means that ∼79.3% of all the peptide molecules exist in the form of oligomers. The oligomerization level is significantly enhanced after incubation for 120 min, and the proportion of oligomers increases to ∼73.9%, meaning that ∼91.1% of the peptide molecules are oligomers. The time-related oligomerization process of peptides on/in the PE-rich membrane was also observed in individual oligomers (Fig. S10†). Moreover, formation of oligomers might induce formation of membrane defects with larger sizes (Fig. S11†). To determine the aggregation state of the peptide, E. coli protoplasts were exposed to LL-37 (1 nM). The observations confirmed the PE-facilitated accumulation and oligomerization of LL-37 (Fig. S12†). Comparison between the initial and final oligomerization states of LL-37 on a PE-rich membrane (i.e., Fig. 3c) indicates that the interaction between LL-37 and PE lipids is a time dependent process, which is consistent with the time-dependent increase of peptide number on the PE-rich membrane (orange profile in Fig. 3b). Moreover, comparison between the oligomerization process of peptides on different membranes (Fig. 3c–e) suggests that PE lipids specifically play active roles in facilitating the membrane actions and oligomerization of LL-37 peptides.
Correlation between oligomerization and the membrane insertion ability of LL-37
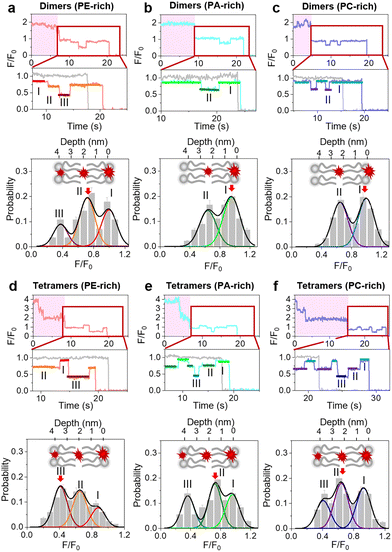
To better understand the positive correlation between oligomerization and the membrane insertion of LL-37, SIFA analysis was also performed on the oligomerized peptides. Briefly, 1 nM N-TAMRA-labeled LL-37 was added to SLBs, and after 120 min of incubation, traces of oligomerized peptides (i.e., with photobleaching steps) were collected for analysis (Fig. S13†). The single-molecule-fluorescence stage (F/F0 < 1; as magnified in Fig. 4) in these traces was used to construct the fluorescence intensity distribution histograms corresponding to different membranes. The results show that in the case of a PE-rich bilayer, LL-37 dimers are preferably located on the upper surface of the membrane, at the center, or on the bottom surface (states I, II, and III, respectively; Fig. 4a and Tables S1–S3†). The highest proportion of dimers (47.4%) is detected in the central layer. Comparatively, the peptides on/in PA- and PC-rich bilayers have two preferred locations only (states I and II), and they tend to be located on the bilayer surface (Fig. 4b and c).The location tendencies of LL-37 trimers are similar to those of the peptide dimers (Fig. S14†). However, the LL-37 tetramers tend to insert more deeply into the bilayer. Indeed, most tetramers (42.3%) are located on the bottom surface of the PE-rich membrane (Fig. 4d). However, the proportions of tetramers on the bottom surfaces of PA- and PC-rich membranes are lower compared to the PE condition (Fig. 4e and f). Overall, these results suggest that oligomerization is correlated with transmembrane insertion of peptides and that the formation of tetramers significantly facilitates partial or complete transmembrane penetration and stable localization of peptides, especially in a PE-rich membrane.
Molecular mechanism of the PE-facilitated insertion and oligomerization of LL-37
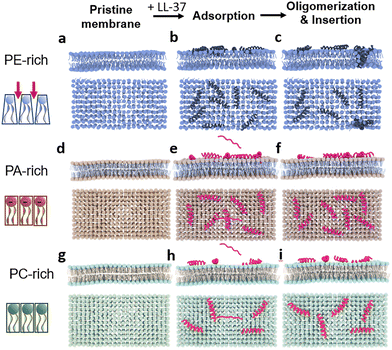
Due to the characteristic pyramidal shape of PE lipids, an abundance of defects might exist on the PE-membrane surface, particularly compared to the continuous and dense surface of a PC or PA film (left column in Fig. 5).42 These defects facilitate the embedding of peptide residues, and triggers the subsequent contact between the hydrophobic part of the surface-bound peptides and the hydrophobic tail chains, thereby stimulating the formation of the amphiphilic helical structure of LL-37. Peptides with this structure aggregate with other peptide molecules, resulting in the formation of aggregates or peptide–lipid complexes. In turn, aggregation increases the disturbance of the membrane structure and enhances the insertion efficiency of LL-37. Interestingly, most LL-37 molecules are inserted into the PE-rich bilayer as oligomers (e.g., fully inserted tetramers or half-inserted dimers), leading to the permeabilization of the membrane (Fig. 5a–c). In comparison, the peptides are easily adsorbed on the surface of PA-rich membranes due to electrostatic interaction with negatively charged lipid heads. However, strong adsorption impedes the contact of peptides with the phospholipid tail chain, resulting in impaired aggregation and membrane insertion. Therefore, a smaller number of LL-37 peptides are observed in the PA-membrane than in the PE-membrane. Moreover, the LL-37 peptides adsorbed on the PA bilayer are mostly monomers, which can hardly be inserted into the membrane (Fig. 5d–f). The electroneutral and dense surface of the PC membrane also hampers the adsorption, transformation, and insertion of LL-37 (Fig. 5g–i).The membrane action of monomeric peptides might be the basis of the oligomerization and membrane insertion (which probably occur simultaneously and cooperatively) of peptides. The membrane insertion of the first monomeric peptide, as a precursor, might recruit more peptides for the formation of oligomers, and decrease the energy barrier for membrane insertion of peptides, leading to the ultimate formation of transmembrane defects (e.g., pores).43 However, the exact relationship between monomer and oligomer peptides during their membrane actions remains an open question.
Conclusions
In this study, the permeabilizing effect of nanomolar LL-37 over membranes of different lipid composition was thoroughly investigated using a combination of several single-molecule methods. Different from traditional models in which membrane-insertion of amphiphilic cationic peptides is principally triggered by the electrostatic affinity between peptides and lipids and requires high local number density of peptides, the PE-specific and oligomerization-facilitated insertion dynamics of LL-37 at nanomolar concentrations is demonstrated herein. CD test and liposome leakage assays quantitatively demonstrate an enhanced helical content and membrane permeabilization efficiency of LL-37 in a PE-rich membrane compared with a PA- or PC-rich membrane. Single-molecule photobleaching and SIFA tests show that approximately twice as many LL-37 peptides accumulate on/in a PE-rich membrane than on/in PA- or PC-rich membranes, most of which exist in the form of oligomers (monomers in the case of PA- or PC-rich systems). Moreover, the LL-37 tetramers tend to be inserted into the bottom of the PE-rich bilayer, while the monomers (as well as dimers and trimers) are inclined to stay in the central layer between leaflets. This work proves that nanomolar LL-37 can directly permeate the lipid membrane in a protein-independent manner, which gains new insights to the membrane activity of the highly concerned amphiphilic cationic peptides. The result might benefit the design of targeting peptides or peptidomimetics to defined cellular populations, such as the oncological therapeutic agents that overcome the Bcl-2-mediated stabilization of mitochondrial membranes. In addition, recent studies demonstrated that LL-37 plays active and complicated roles in modulating different physiological processes such as autophagy.44 This work may be helpful for the understanding of these processes.Author contributions
Y. L. and W. H. conceived, designed, and supervised the project. X. J. and C. Y. performed the experiments. X. J., C. Y. and J. Q. carried out the data analysis. D. M., C. X. and S. H. provided assistance for data curation. X. J. wrote the original draft and collaborated with C. X. on drawing the diagrams. B. Y. modified and finalized the manuscript. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by National Natural Science Foundation of China (no. 12004271, U1932121, 12274308 and 12274307) and Guangdong Basic and Applied Basic Research Foundation (2019A1515110186).Notes and references
- B. P. Lazzaro, M. Zasloff and J. Rolff, Science, 2020, 368, 487 CrossRef PubMed.
- L. J. Zhang and R. L. Gallo, Curr. Biol., 2016, 26, R14–R19 CrossRef PubMed.
- K. Kuroda, K. Okumura, H. Isogai and E. Isogai, Front. Oncol., 2015, 5, 144 Search PubMed.
- E. Piktel, K. Niemirowicz, U. Wnorowska, M. Watek, T. Wollny, K. Gluszek, S. Gozdz, I. Levental and R. Bucki, Arch. Immunol. Ther. Exp., 2016, 64, 33–46 CrossRef CAS PubMed.
- E. Bankell, X. Liu, M. Lundqvist, D. Svensson, K. Sward, E. Sparr and B. O. Nilsson, Biochem. Biophys. Rep., 2022, 29, 101192 CAS.
- V. Armiento, K. Hille, D. Naltsas, J. S. Lin, A. E. Barron and A. Kapurniotu, Angew. Chem., Int. Ed., 2020, 59, 12837–12841 CrossRef CAS PubMed.
- D. Spierings, G. McStay, M. Saleh, C. Bender, J. Chipuk, U. Maurer and D. R. Green, Science, 2005, 310, 66–67 CrossRef CAS PubMed.
- P. Costantini, E. Jacotot, D. Decaudin and G. Kroemer, J. Natl. Cancer Inst., 2000, 92, 1042–1053 CrossRef CAS PubMed.
- K.-M. Debatin, D. Poncet and G. Kroemer, Oncogene, 2002, 21, 8786–8803 CrossRef CAS PubMed.
- S. Farsinejad, Z. Gheisary, S. Ebrahimi Samani and A. M. Alizadeh, Tumor Biol., 2015, 36, 5715–5725 CrossRef CAS PubMed.
- K. Ye, W. X. Meng, H. Sun, B. Wu, M. Chen, Y. P. Pang, J. Gao, H. Wang, J. Wang, S. H. Kaufmann and H. Dai, Nat. Commun., 2020, 11, 3301 CrossRef CAS PubMed.
- E. T. Verjans, S. Zels, W. Luyten, B. Landuyt and L. Schoofs, Peptides, 2016, 85, 16–26 CrossRef CAS PubMed.
- G. Gabernet, A. T. Müller, J. A. Hiss and G. Schneider, MedChemComm, 2016, 7, 2232–2245 RSC.
- A. Marquette and B. Bechinger, Biomolecules, 2018, 8, 18 CrossRef PubMed.
- M. Maduke and D. Roise, Science, 1993, 260, 364–367 CrossRef CAS PubMed.
- T. Kuwana, G. Perkins, M. R. Mackey, M. H. Ellisman, M. Latterich, D. R. Green, R. Schneiter and D. D. Newmeyer, Cell, 2002, 111, 331–342 CrossRef CAS PubMed.
- H. Schroder-Borm, R. Bakalova and J. Andra, FEBS Lett., 2005, 579, 6128–6134 CrossRef PubMed.
- B. Yuan, J. Liu, Z. Deng, L. Wei, W. Li, Y. Dou, Z. Chen, C. Zhang, Y. Xia, J. Wang, M. Zhang, K. Yang, Y. Ma and Z. Kang, NPG Asia Mater., 2021, 13, 18 CrossRef CAS.
- H. W. Huang, Biochemistry, 2000, 39, 8347–8352 CrossRef CAS PubMed.
- Y. E. Lau, A. Rozek, M. G. Scott, D. L. Goosney, D. J. Davidson and R. E. Hancock, Infect. Immun., 2005, 73, 583–591 CrossRef CAS PubMed.
- L. Yang, R. Peltier, M. Zhang, D. Song, H. Huang, G. Chen, Y. Chen, F. Zhou, Q. Hao, L. Bian, M. L. He, Z. Wang, Y. Hu and H. Sun, J. Am. Chem. Soc., 2020, 142, 18150–18159 CrossRef CAS PubMed.
- M. T. Jeena, L. Palanikumar, E. M. Go, I. Kim, M. G. Kang, S. Lee, S. Park, H. Choi, C. Kim, S. M. Jin, S. C. Bae, H. W. Rhee, E. Lee, S. K. Kwak and J. H. Ryu, Nat. Commun., 2017, 8, 26 CrossRef CAS PubMed.
- Y. Feng, F. Chen, J. M. Rosenholm, L. Liu and H. Zhang, Mat. Futures, 2022, 1, 023502 CrossRef.
- A. J. Duplantier and M. L. van Hoek, Front. Immunol., 2013, 4, 143 Search PubMed.
- D. Svensson, L. Wilk, M. Morgelin, H. Herwald and B. O. Nilsson, Biochem. J., 2016, 473, 87–98 CrossRef CAS PubMed.
- Y. Engelberg and M. Landau, Nat. Commun., 2020, 11, 3894 CrossRef PubMed.
- E. Sancho-Vaello, P. Francois, E. J. Bonetti, H. Lilie, S. Finger, F. Gil-Ortiz, D. Gil-Carton and K. Zeth, Sci. Rep., 2017, 7, 15371 CrossRef PubMed.
- E. Sancho-Vaello, D. Gil-Carton, P. Francois, E. J. Bonetti, M. Kreir, K. R. Pothula, U. Kleinekathofer and K. Zeth, Sci. Rep., 2020, 10, 17356 CrossRef CAS PubMed.
- F. Porcelli, R. Verardi, L. Shi, K. A. Henzler-Wildman, A. Ramamoorthy and G. Veglia, Biochemistry, 2008, 47, 5565–5572 CrossRef CAS PubMed.
- X. Li, Y. Li, H. Han, D. W. Miller and G. Wang, J. Am. Chem. Soc., 2006, 128, 5776–5785 CrossRef CAS PubMed.
- E. Zinser, C. D. M. Sperka-Gottlieb, E.-V. Fasch, S. D. Kohlwein, F. Paltauf and G. Daum, J. Bacteriol., 1991, 173, 2026–2034 CrossRef CAS PubMed.
- D. Patel and S. N. Witt, Oxid. Med. Cell. Longevity, 2017, 2017, 4829180 Search PubMed.
- Y. Li, Z. Qian, L. Ma, S. Hu, D. Nong, C. Xu, F. Ye, Y. Lu, G. Wei and M. Li, Nat. Commun., 2016, 7, 12906 CrossRef CAS PubMed.
- G. Wang, J. Biol. Chem., 2008, 283, 32637–32643 CrossRef CAS PubMed.
- E. J. Lugtenberg and R. Peters, Biochim. Biophys. Acta, 1976, 441, 38–47 CrossRef CAS.
- R. J. Stöhr, R. Kolesov, K. Xia, R. Reuter, J. Meijer, G. Logvenov and J. Wrachtrup, ACS Nano, 2012, 6, 9175–9181 CrossRef PubMed.
- J. Xu, G. Qin, F. Luo, L. Wang, R. Zhao, N. Li, J. Yuan and X. Fang, J. Am. Chem. Soc., 2019, 141, 6976–6985 CrossRef CAS PubMed.
- C. Xu, Z. Lin, K. Yang and B. Yuan, Acta Phys. Sin., 2020, 69, 108701 CrossRef.
- S. Q. Zhang, D. W. Kulp, C. A. Schramm, M. Mravic, I. Samish and W. F. DeGrado, Structure, 2015, 23, 527–541 CrossRef CAS PubMed.
- E. Lauwers, Y. C. Wang, R. Gallardo, R. Van der Kant, E. Michiels, J. Swerts, P. Baatsen, S. S. Zaiter, S. R. McAlpine, N. V. Gounko, F. Rousseau, J. Schymkowitz and P. Verstreken, Mol. Cell, 2018, 71, 689–702 CrossRef CAS PubMed.
- L. Dresser, P. Hunter, F. Yendybayeva, A. L. Hargreaves, J. A. L. Howard, G. J. O. Evans, M. C. Leake and S. D. Quinn, Methods, 2021, 193, 80–95 CrossRef CAS PubMed.
- H. Hauser, I. Pascher, R. H. Pearson and S. Sundell, Biochim. Biophys. Acta, 1981, 650, 21–51 CrossRef CAS.
- Y. Lyu, N. Xiang, X. Zhu and G. Narsimhan, J. Chem. Phys., 2017, 146, 155101 CrossRef PubMed.
- R. S. Rekha, S. S. Rao Muvva, M. Wan, R. Raqib, P. Bergman, S. Brighenti, G. H. Gudmundsson and B. Agerberth, Autophagy, 2015, 11, 1688–1699 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods, supplementary images and tables. See DOI: https://doi.org/10.1039/d2nr05409d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |