Open Access Article
Open Access ArticleA green-PAD array combined with chemometrics for pH measurements†
Lisa R.
Magnaghi
 *ab,
Giancarla
Alberti
*ab,
Giancarla
Alberti
 *a,
Bianca M.
Pazzi
a,
Camilla
Zanoni
a and
Raffaela
Biesuz
ab
*a,
Bianca M.
Pazzi
a,
Camilla
Zanoni
a and
Raffaela
Biesuz
ab
aDepartment of Chemistry, University of Pavia, Via Taramelli 12, 27100 Pavia, Italy. E-mail: galberti@unipv.it
bUnità di Ricerca di Pavia, INSTM, Via G. Giusti 9, 50121 Firenze, Italy
First published on 4th October 2022
Abstract
This work presents the development of a green paper-based analytical device (Green-PAD) array for pH detection. The array was obtained with natural dyes extracted from red cabbage (Brassica oleracea) and butterfly pea flower (Clitoria ternatea); a filter paper was used as a substrate. The RGB indexes of the PADs' colors were extracted from the pictures taken using a smartphone or using a specifically developed RGB detector (Arduino-based) to obtain RGB indexes not affected by the light and the photocamera sensitivity. Multi-technique chemometric models were developed for calculating the pH value, starting from the RGB triplet of each sensing PAD. A preliminary and explorative chemometric analysis with PCA (partial component analysis) and TWPCA (3-way PCA) was carried out. Partial least square regression, PLS, was then applied to correlate the color of the PAD's picture with the solutions' pH. Different solutions at various pHs, ranging from 1 to 13, were obtained by titrating orthophosphoric acid with standardized NaOH, and they were used to create PLS models. Some real samples were examined as a test set, and the results were validated with pH-meter measurements. The ability of the PLS to model the experimental data was satisfactory since a good agreement between the experimental and fitted pH values was obtained. The proposed PADs were prepared with natural dyes and filter papers, so they are completely biodegradable and eco-friendly. Their fabrication does not require toxic or expensive reagents and sophisticated equipment. Also, the developed RGB detector, built-up with low-cost components and recycled batteries, adds value making the measurement cheap, easy and feasible also by non-expert people.
Introduction
The first pH definition goes back to 1909 by Sørensen1 as the negative logarithm of the hydrogen ion concentration expressed in molarity. After that, a more rigorous definition of pH in terms of the activity of hydrogen ions in solution was proposed2 (the activity is the effective concentration of the hydrogen ion in solution, which can be considered equal to the ion concentration moving toward high diluted solutions in which less strong interactions between ions take place).3The measurement of pH in various media in which chemical, physical, and biological processes occur is a crucial task. The standard pH measurement techniques are based on quite-expensive instruments or devices for qualitative analyses.4 Traditionally, pH is measured electrochemically using glass electrodes5 or, in the most recent 50 years, using solid-state ISFET sensors.6 Although potentiometric pH measurements using glass electrodes are accurate, have immediate read-out and have a wide range of measurable pHs, they even have drawbacks such as the use of delicate and expensive electrodes and large sample volumes and they require frequent calibrations and trained personnel.7 pH test strips (litmus papers) are another possibility for qualitative and rapid pH determination, but they suffer from poor resolution and visual subjective analysis, unsuitable for quantitative measurements.8 Other attractive optical methods have been proposed for pH measurements,9–12 and among them, colorimetric pH sensors have recently been developed as a new trend inspired by traditional litmus papers.13
Numerous colorimetric pH sensors have been developed recently, both generalized or specific for a pH range or applications.14 They were based on the colorimetric analysis of indicators, i.e., the color variations at different acidity values. Although one advantage of using pH sensors over the pH electrodes is operating at the pH scale's extremes, only a few colorimetric pH sensors have been reported for use in these ranges.15,16 Moreover, for the detection, UV-vis spectroscopy or fluorescence spectroscopy is generally employed, while elaborate data treatment procedures, complex algorithms or smartphone applications are always required for colorimetric analysis with these sensors.13
An emerging approach involves the employment of sensor arrays based on cross-responsive sensor elements. Among them, colorimetric sensor arrays were appropriate for pH determination with a satisfying resolution.7,17,18
Typically, colorimetric pH sensors consist of a solid substrate and a dye sensitive to pH variations. Some studies suggested using paper as the substrate and pigments extracted from fruits and vegetables as dyes for developing this kind of sensor.19–22
Paper-based analytical devices (PADs) were first proposed in 2007 by Martinez et al.23 and then widely applied for environmental analyses, clinical trials and food controls thanks to their simple fabrication and rapid response advantages.24,25
The network of cellulosic fibers gives the paper capillary properties, eliminating the need for a pumping device used in a conventional microfluidic apparatus. Moreover, paper's characteristics such as abundant availability, low cost, biodegradability, the ease of production and surface functionalization make it an appealing candidate as a sensor's substrate.26,27
Compared to synthetic dyes, pigments extracted from fruits and vegetables are eco-friendly, sustainable and accessible.28 In particular, anthocyanin-based dyes can be helpful for the development of pH sensors since their color changes depend on the pH.29 For example, natural dyes’ extracts have been applied as pH sensors in food quality tests.22,30
The present work fits this scenario. A PAD array entirely eco-friendly for pH measurements was developed. The array was obtained using aqueous extracts of pigments from red cabbage (Brassica oleracea) and butterfly pea flower (Clitoria ternatea), and a filter paper was used as a substrate. Among different anthocyanin-based dyes, those extracted from red cabbage (RC) and butterfly pea flower (BPF) were selected because they showed more color changes by varying the pH. Moreover, similar to the synthetic indicators made of combinations of dyes, mixtures of different percentages of the two pure extracts were prepared, aiming to obtain a different color evolution for each PAD of the array.
The RGB space model was applied to correlate the color variations of PADs with the pH of aqueous solutions.
Colorimetric arrays produce data from several sensors, so chemometric analysis is essential since the multidimensional nature of the results. Here, multi-technique chemometric models were developed for calculating the pH value, starting from the RGB triplet of each sensing PAD. The validation of the models was performed by analyzing actual samples.
Different from poorly accurate commercial pH test strips31 or single dye-based colorimetric pH sensors,14 an array with several sensors with overlapping sensitivity patterns was employed here to increase the analytical performance. Moreover, the use of eco-friendly and low-cost materials and reagents is an added advantage compared to other more complex and expensive devices.14
Experimental
Materials and instruments
Cellulose filter paper Whatman grade 1, 11 μm (particle retention), 0.25 psi wet burst, 150 s per 100 mL speed (Herzberg) and 180 μm thickness, was obtained from Laboindustria S.p.A. (Arzergrande, Italy). Red cabbage (Brassica oleracea) and butterfly pea flower tea (Clitoria ternatea) were purchased from a local supermarket (Pavia, Italy).Hydrochloric acid, orthophosphoric acid, and sodium hydroxide were obtained from Merk Life Science S.r.l. (Milan, Italy).
Solutions at different pH values (ranging from 1 to 13) were prepared by titrating orthophosphoric acid with standardized NaOH. The exact pH value was measured using a pH meter.
The tap water sample was obtained from the drinking water supply of Pavia (Italy). The sample was collected after flushing cold water for 20 min from the sink of the laboratory (Chemistry Department, University of Pavia, Italy).
Ammonia cleaner (S.a.i.soc.alcoli Industriali Sas, Italy), Tropical Aloe Vera drink (Eurofood S.p.A., Italy), Schweppes tonic water (Schweppes International Limited, Italy), sprite (Coca-Cola S.r.L., Italy), and white wine vinegar “Gaia” (Formec Biffi S.p.A., Italy) were purchased from a local supermarket (Pavia, Italy).
Instruments
pH measurements were performed using a pH meter (Mettler Toledo mod. SevenMulti) equipped with a combined glass electrode (InLab Pro, Mettler Toledo S.p.A.- Milan, Italy).A pH-indicator paper, pH 1–14 Universal Indicator (Merk Life Science S.r.l. – Milan, Italy), was also used for comparing the data obtained with the PAD array.
Photographs of the PADs' array were taken using an iPad Pro 10,5 (Apple inc., Italy). A portable, led-based lightbox (PULUZ, Shenzhen Puluz Technology Ltd, China) was employed to ensure the reproducibility of the photographs. The GIMP software32 was used for sampling the RGB indexes of each picture.
A specifically developed, Arduino-based RGB detector (see the ESI†) was also employed to collect each PAD's RGB triplet.
The open-source R-based software CAT (Chemometric Agile Tool)33 was used for chemometric data treatment.
Procedures
The pH of each solution measured using a pH meter was used as a reference value for the following chemometric data treatment.
Chemometric data treatment.
When dealing with arrays, the differential sensing approach is used with cross-reactive receptors and pattern recognition algorithms to process a large amount of data acquired which are not usually interpretable by visual inspection or classical dose–response calibrations. Chemometric methods are so routinely used to reduce the data's dimensionality and are shown in a graphical form for interpretation.34Here, chemometric tools were applied to analyze the RGB data set, only centering the data because these indexes are intrinsically scaled from 0 to 255. A multi-technique approach was adopted, combining unsupervised techniques such as principal component analysis (PCA) and three-way principal component analysis (3WPCA) with the supervised partial least square regression (PLS) since it has proven to be the ultimate data elaboration treatment in terms of model robustness and predictive performances.35,36
All the tools exploited are widely reviewed in the literature, so their theoretical aspects will not be discussed.37,38 3WPCA, based on the Tucker3 model,39 was selected since it takes into account the tri-dimensional nature of the data set, which can be considered as a parallelepiped of sizes I × J × K (conventionally termed objects, variables and conditions). Thus, the information related to each of the three modes: RGB indexes (variables), the five types of Green-PADs (objects) and the solution pHs (conditions), is fully separated, allowing a much more straightforward interpretation of the information present in the data set. Indeed, the final result is given by three loading sets and a core array describing the relationship among them. Each of the three loading sets can be interpreted and displayed similarly to a loading plot of the standard PCA.40
PCA and 3WPCA were first run on the entire data set (15 columns (3 RGB indexes per 5 PADs) and 39 lines (13 solutions per 3 replicates). From the resulting score plot of the PCA, the following three pH subintervals are highlighted: the interval from pHs 1 to 4 (acid solutions), the second from pHs 5 to 8 (neutral solutions) and the third from pHs 9 to 13 (alkaline solutions). Then, the PLS tool was applied separately for each pH's subinterval, developing a tailored model correlating the RGB indexes of the PAD sensors with the pH values of the solutions in the range under investigation.
The training set required to build up the PLS models comprised 3 replicates of each solution; so the training input matrix had 15 columns (3 RGB indexes per 5 PADs) and, respectively, 12 lines (4 solutions per 3 replicates) for the interval from pH 1 to 4 and for that from pH 5 to 8, while 15 lines (5 solutions per 3 replicates) for the interval from pH 9 to 13.
The test set used to validate the PCA and PLS models comprised three replicates of real samples commercially available and characterized by different pH values: in particular, we selected Schweppes tonic water, sprite, white wine vinegar and aloe vera drink as acidic samples, tap water as neutral and ammonia cleaner as alkaline. Moving back now to matrix dimensions, the test input matrix had 15 columns (3 RGB indexes per 5 PADs) and, respectively, 12 lines (4 solutions per 3 replicates) for samples with pHs from 1 to 4, while 3 lines (1 solution per 3 replicates) for samples with pHs from 5 to 8 and 3 lines (1 solution per 3 replicates) for samples with pH from 9 to 13.
Results and discussion
Preparation and analysis of green-PADs
The devices described here were prepared by employing a filter paper as a substrate. The currently proposed μPADs were fabricated by paper patterning using different procedures such as wax printing or screen-printing, photolithography, inkjet etching, plasma treatment, and laser printing.41–47 Most of these techniques involve complex procedures and are expensive, making them inaccessible in restricted resources laboratories. Besides, patterning of μPADs by wax requires a strict flow control of the hydrophobic barrier into the porous paper's matrix, determining inhomogeneity and irreproducibility.47 A simple and alternative approach is the cutting of the paper in the desired dimensions and shapes, avoiding the formation of the containment barriers of the liquid.26 This last patterning method was adopted here by creating square PADs by cutting a sheet of filter paper. Indeed, our objective was to prepare devices with low-cost and sustainable materials and techniques affordable for resource-limited settings. For obtaining PADs, totally green, natural pigments extracted from fruit and vegetables were employed. Among different anthocyanin-based dyes, those extracted from red cabbage (RC) and butterfly pea flower (BPF) were selected because they showed more color changes by varying the pH. Similar to the synthetic indicators made of combinations of dyes, three mixtures of different percentages of the two pure extracts were prepared, so different color evolution for each PAD of the array occurred.Fig. 1 shows the UV-vis spectra of the extracts (single or in mixtures) at neutral pH, i.e., without adding acids or alkaline solutions, in the wavelength range of 400–800 nm. As can be seen from the graph, in the spectrum of the 100% RC extract, the characteristic broad peak at 550 nm is evident, and it is due to the presence of acylated anthocyanins, mainly cyanidin 3-diglucoside-5-glucoside derivatives with various acylated groups linked to the diglucoside.48,49 By increasing the percentage of BPF, a bathochromic shift and split into two peaks at 570 and 620 nm arises. These peaks are those characteristics of ternatins and polyacylated anthocyanins (i.e., malonylated delphinidin 3,3′,5′-triglucosides, with 3′,5′-side chains with alternating D-glucose and p-coumaric acid units) responsible for the blue color of the butterfly pea flower extract.50
The extracts of red cabbage and butterfly pea flower were previously employed as pH indicators since the solution acidity influences their color, and this is due to the reversible structural transformation of the anthocyanins that lead to color changes.48–50 A comparison with the color palette at various pHs of the PADs of 100% RC and 100% BPF with those previously reported48 for aqueous extracts of red cabbage and butterfly pea flower50 was performed, confirming the correct preparation of the solutions and the preservation of the anthocyanins.
A volume of 0.2 mL of the extract was selected to load the PADs since it was verified to be enough to cover the whole surface of the PAD without overflow from the borders. Also, immersion of the paper in 5 mL of the extract was considered, but inhomogeneity of the color of the PAD and low sorption kinetics occurred.
Moreover, it was necessary to define the loading ratio quantity of the extract/volume of sample solution since it affects the color intensity. Different experiments were performed, considering drop-coating 0.2 mL of the sample, or immersing the PAD in 5 mL of the sample solution for 10 seconds, i.e., the minimum time required to impregnate the paper. The second strategy was adopted as the better uniformity of the color.
Finally, the waiting time before taking the photographs (or placing the PAD in the RGB detector) at room temperature was selected. Five minutes is the time required because it is enough to obtain homogeneous color and avoid the drying out of the paper.
Some trials with other filter papers were performed, aiming to reduce the waiting time; however, since the similar porosity, their use has not proved advantageous.
Regarding the stability, it would be underlined that the PADs prepared are disposable sensors, and the natural dye extracts were not stable. So the PADs have to be immediately used after their preparation.
Color analysis and chemometrics
The Green-PADs of the array showed very distinct colors when immersed in aqueous solutions at different pHs ranging from 1 to 13, as shown in Fig. 2. | ||
| Fig. 2 Green-PAD array. A5 = 100%RC; A4 = 75%RC-25%BPF; A3 = 50%RC-50%BPF; A2 = 25%RC-75%BPF; A1 = 100% BPF. Three replicates for each pH. | ||
As is well known, the PADs' color can be described by models with different bases, pros and cons51 that will not be detailed here. The RGB model was selected to quantify the color change with the solution's pH. The open-source GIMP software32 was used to acquire the RGB indexes of the PADs' images.
The RGB value matrixes, adequately organized, were thus subjected to multivariate analysis, applying only the centering as the data pre-treatment since the RGB indexes are intrinsically scaled from 0 to 255.
A multi-technique approach was adopted; unsupervised techniques were first applied to visualize and rationalize the overall data set, i.e., 3WPCA and PCA.
The 3WPCA was applied to compare the different responses of each kind of PADs to the solution pHs. The entire data set was employed, so the five types of Green-PADs as objects, the pH of 13 solutions as conditions, and the RGB indexes of the entire array as variables.
Table 1 reports the explained variance percentage after unfolding; comparing the lowest value obtained after unfolding, which was in the case of conditions mode, and the % variance explained by the Tucker3 model (68.09%), we could observe that no significant loss in the information was detected when the overall color evolution is taken into account. Furthermore, all the percentages of the explained variance are pretty good considering the intrinsically high variability of the data employed.
| Mode | Axis 1 | Axis 1 & 2 |
|---|---|---|
| Objects | 47.84 | 89.10 |
| Variables | 56.71 | 81.96 |
| Conditions | 50.41 | 72.47 |
Fig. 3 shows the triplot, in which the loading values of the three modes (objects, conditions and variables) are reported altogether. The objects, i.e., the five types of Green-PADs obtained with five different extracts, show loading values arranged along the horizontal axis (axis 1) due to the different brightness, which increases with the decrease in the percentage of BPF and the corresponding increase in the % of RC. This assumption is confirmed by the loadings of the variables, i.e., the R, G, and B indexes: they all have a positive value on the x-axis, which indicates that they all increase, moving from the left to the right of the plot, leading to brighter Green-PAD colors when % RC is higher. Conversely, on the y-axis, R has a positive loading value. In contrast, G and B have negative values, suggesting that by the change of the solution pH, the R index increases while the G and B indexes decrease, which correspond to the numerical effect of the significant clear color variations for the PADs immersed in solutions at the extremes of the pH range. The conditions are the pHs: it can be observed that there is a clear distinction for pH values lower than 4 and pH higher than 9, which is easily recognizable, and given both for the differences in the color shade (separation along axis 2) and the brightness (separation along axis 1); conversely, for pH values from 5 to 8, the difference is smaller as expected since the similar color of the PADs in this pH range.
 | ||
| Fig. 3 3WPCA applied to the Green-PAD array: triplot of loadings values. A1 = 100%RC; A2 = 75%RC-25%BPF; A3 = 50%RC-50%BPF; A4 = 25%RC-75%BPF; A5 = 100% BPF. Three replicates for each pH. | ||
PCA was also applied to the entire data set to visualize the color transition and identify the main clusters.
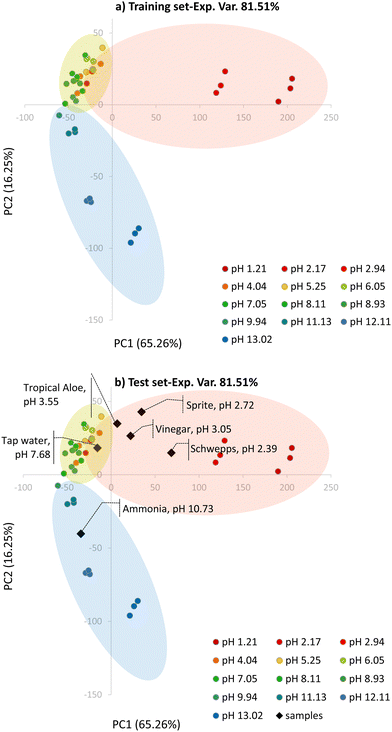
The model was obtained considering only the first two components, which explain 81.51% of the experimental variance; Fig. 4a shows the resultant score plot.
Three main clusters, partially overlapped, can be qualitatively distinguished in the score plot: a first cluster (red ellipsoid) for samples at pH lower than 5 that are separated along the PC1 axis and their score values on this component are inversely proportional to the pH. A second cluster (little green ellipsoid) is highlighted for samples at pH between 5 and 8 that are mainly separated along the PC2 axis, and their score values decrease with the increasing pH. The third cluster (blue ellipsoid) is for samples at pH from 9 to 13; in this case, samples are separated alongside both PC1 and PC2, with PC1 score values increasing and PC2 score values decreasing with increasing pH.
The PCA model was validated by a projection of the test set, as shown in Fig. 4b: all samples are correctly located in the corresponding clusters of the score plot.
After validation, the PCA model was investigated to divide the entire pH range into subintervals for the following PLS analysis. The subintervals identified correspond to the PCA clusters previously described.
After defining the three pH subintervals, the PLS tool was applied. Three PLS models (named A for the acid pH range, N for the neutral pH range and B for the basic pH range) were developed, using the corresponding three training sets as detailed in the previous paragraph, “Chemometric data treatment.”
The models were then validated by predicting the test samples and comparing the experimental values, measured using a pH meter, with those obtained by the Green-PAD array–PLS models.
Table 2 reports the number of components used to build the PLS models, the % explained variance in cross-validation (CV), and the root mean square error in CV (RMSECV).
| Model | n. comp. | %Exp.Var. CV | RMSECV |
|---|---|---|---|
| A | 5 | 90.30 | 0.3287 |
| N | 5 | 90.81 | 0.3326 |
| B | 4 | 93.68 | 0.3674 |
For models A and N, the minimum of the RMSECV is obtained with 5 components, whereas for model B, it is obtained with 4 components. The explained variance % is high in all cases, about 90%. The RMSECV values, around 0.3, are higher than those achievable using a pH meter, but it was expected since the lower robustness and precision of the RGB acquired from the PAD photographs compared to standard glass electrodes.
Fig. 5 shows the plots of experimental vs. fitted values for each model; a pretty good agreement between the experimental and fitted data for all the models can be observed.
The models were validated by applying them to the analysis of actual samples of different pHs. In the case of unknown samples, RGB indexes describing the colors of the PAD were projected in the PCA model (see Fig. 4b) to identify the most suitable PLS model to be exploited to calculate the pH value.
Table 3 shows the relative error % between the samples' pH values measured using a pH meter and those calculated by the Green-PAD array–PLS models. For comparison, the estimated pH values obtained by the commercial litmus paper were also reported (the pH color chart and image of the litmus paper after immersion in each sample are reported in the ESI,† Fig. S3).
| Sample | pHGE | pHLP | pHPADs | RE % |
|---|---|---|---|---|
| pHGE = value obtained by glass electrodes.pHLP = value obtained by the Litmus paper.pHPADs= value obtained by Green-PADs.RE % = relative error. | ||||
| Schweppes | 2.39 | 2–3 | 2.4(1) | 1.7 |
| Sprite | 2.72 | 3 | 2.9(3) | 4.2 |
| White wine vinegar | 3.05 | 3–4 | 2.8(2) | 0.6 |
| Tropical aloe vera | 3.55 | 4 | 3.5(4) | 1.1 |
| Tap water | 7.68 | 8 | 7.4(4) | 1.2 |
| Ammonia cleaner | 10.73 | 10–11 | 10.7(1) | 0.7 |
As can be seen, despite the scarce precision of the measurements with PADs, a good agreement between the pH values was obtained.
As is well known, digital images can be different depending on the type of camera, focus, and environment brightness. Therefore, it was thought to couple the developed PADs to a color detector that was not subject to this environmental variability. A device called the RGB detector was thus developed (thanks to a collaboration with Eng. Dario Pistoia). The device comprises a 4-led color sensor, and it is based on an Arduino hardware platform. It is simple and economical and can also be used for pH measurements out-of-lab. The description of the RGB detector is reported in the ESI†.
The same experiments previously described were carried out by registering the RGB indexes using the detector to create the PLS models for the array of PADs at the three pH subintervals and, subsequently, their application for determining the pH of real samples.
Table 4 reports the number of components used to build the PLS models, the % explained variance in cross-validation (CV), and the root mean square error in CV (RMSECV).
| Model | n. comp. | %Exp.Var. CV | RMSECV |
|---|---|---|---|
| A | 3 | 99.82 | 0.0537 |
| N | 5 | 92.08 | 0.2892 |
| B | 5 | 98.65 | 0.1708 |
For models B and N, the minimum of the RMSECV is obtained with 5 components, whereas for model A, it is obtained with 3 components. The explained variance % is high in all cases, higher than 92%. The RMSECV values are lower than those obtained for the previous model based on the RGB acquired by the PAD photographs, indicating higher precision and better agreement with the values measured using a pH meter. In particular, the RMSECV for model A is similar to that achievable with glass electrodes at pHs lower than 2.
Fig. 6 shows the plots of experimental vs. fitted values for each model. The results of the validation are summarized in Table 5.
 | ||
| Fig. 6 Experimental vs. fitted plot for (a) model A, (b) model N, and (c) model B. RGB indexes acquired by the RGB detector. | ||
| Sample | pHGE | pHLP | pHPADs | RE % |
|---|---|---|---|---|
| pHGE = value obtained by glass electrodes.pHLP = value obtained by the Litmus paper.pHPADs= value obtained by Green-PADs.RE % = relative error. | ||||
| Schweppes | 2.39 | 2–3 | 2.35(3) | 1.7 |
| Sprite | 2.72 | 3 | 2.84(1) | 4.2 |
| White wine vinegar | 3.05 | 3–4 | 3.03(2) | 0.6 |
| Tropical aloe vera | 3.55 | 4 | 3.51(1) | 1.1 |
| Tap water | 7.68 | 8 | 7.77(6) | 1.2 |
| Ammonia cleaner | 10.73 | 10–11 | 10.80(3) | 0.7 |
As expected, the data obtained with the detector (see Table 5) are certainly more precise than those obtained from the RGB data of digital images. The differences between the pH values determined with the PADs and those measured with the detector are of the same order of magnitude as those obtained from the photographic data. This aspect was also predictable. Indeed, all photographs were taken simultaneously for both arrays of Green-PADs (for PLS models and for the samples); therefore, photographs were not affected by brightness, focus, or exposure variations.
Since the RGB detector guarantees objectivity to the measurements under any operating conditions, it is undoubtedly the best choice when it is necessary to perform determinations at different times with different photographic devices.
Conclusions
A simple, easy-to-use, low-cost, and completely green paper-based array for pH measurement was proposed. The array was prepared using filter paper scraps and aqueous extracts of vegetables, allowing rapid and economical preparation.Natural dyes extracted from red cabbage (Brassica oleracea) and butterfly pea flower (Clitoria ternatea) were selected because of a wide range of color variations with pH changes.
The RGB space model was applied to correlate the PADs' color variation with the pH of aqueous solutions.
Multi-technique chemometric models were developed for calculating the pH value, starting from the RGB triplet of each sensing PAD. The RGB indexes were acquired by the photographs of the PADs or using a homemade RGB detector.
Unsupervised techniques were first applied to visualize and rationalize the overall data set, i.e., 3WPCA and PCA. This approach allowed in identifying pH subintervals and thus developing tailored PLS models. Three PLS models (named A for the acid pH range from 1 to 4, N for the neutral pH range from 5 to 8, and B for the basic pH range from 9 to 13) were developed and then validated by predicting the test samples. The experimental values, measured using a pHmeter, were compared with those obtained by the Green-PAD array–PLS models showing a good agreement. As expected, better data reproducibility was obtained by employing the RGB detector since it ensures objectivity to measurements under any operating conditions.
Similarly, in the case of unknown samples, RGB indexes describing PAD's colors, anyhow they were acquired, can be projected in the PCA model to identify the most suitable PLS model to be used to calculate the pH values, thus increasing the accuracy of the results.
Author contributions
Giancarla Alberti: conceptualization, methodology, and writing – original draft. Lisa Rita Magnaghi: formal analysis, data curation, and writing – review and editing. Bianca Maria Pazzi: investigation and writing – review and editing. Camilla Zanoni: writing – review and editing. Raffaela Biesuz: formal analysis and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Eng. Dario Pistoia for the realization of the RGB detector and MIUR for funding the Camilla Zanoni's PhD grant.References
- S. P. L. Sørensen, C. R. Trav. Lab. Carlsberg, 1909, 8, 1 Search PubMed.
- S. P. L. Sørensen and K. L. Linderstrøm-Lang, C. R. Trav. Lab. Carlsberg, 1924, 15, 6 Search PubMed.
- R. P. Buck, S. Rondinini and A. K. Covington, et al. , Pure Appl. Chem., 2002, 74(11), 2169–2200 CrossRef CAS.
- O. R. Chanu, A. Kapoor and V. Karthik, Mater. Today: Proc., 2021, 40, S64–S68 CAS.
- G. Palleschi, G. Volpe and D. Compagnone, et al. , Talanta, 1994, 41(6), 917–923 CrossRef CAS.
- P. Bergveld, Sens. Actuators, B, 2003, 88, 1–20 CrossRef CAS.
- Y. Li, Y. Wang, S. Chen, Z. Wang and L. Feng, Anal. Chim. Acta, 2021, 1154, 338275 CrossRef CAS PubMed.
- H. D. Duong, et al. , Mater. Sci. Eng., C, 2020, 107, 110323 CrossRef CAS PubMed.
- C. G. Frankær, K. J. Hussain, T. C. Dorge and T. J. Sørensen, ACS Sens., 2018, 4, 26–31 CrossRef PubMed.
- C. G. Frankær, M. Rosenberg, M. Santella, K. J. Hussain, B. W. Laursen and T. J. Sørensen, ACS Sens., 2019, 4, 764–773 CrossRef.
- B. Y. Chu, T. T. Qi and J. F. Liao, et al. , Sens. Actuators, B, 2012, 166, 56–60 CrossRef.
- J. D. Moore, R. L. Lord, G. A. Cisneros and M. J. Allen, J. Am. Chem. Soc., 2012, 134, 17372–17375 CrossRef CAS.
- L. R. Magnaghi, S. Compagnin, C. Zanoni, G. Alberti, P. Quadrelli and R. Biesuz, ISMEC Acta, 2021, 10, OC20 Search PubMed.
- A. Steinegger, O. S. Wolfbeis and S. M. Borisov, Chem. Rev., 2020, 120(22), 12357–12489 CrossRef CAS PubMed.
- A. Safavi and H. Abdollahi, Anal. Chim. Acta, 1998, 367, 167–173 CrossRef CAS.
- A. Safavi and M. Bagheri, Sens. Actuators, B, 2003, 90, 143–150 CrossRef CAS.
- R. Gotor, P. Ashokkumar, M. Hecht, K. Keil and K. Rurack, Anal. Chem., 2017, 89, 8437–8444 CrossRef CAS PubMed.
- S. Capel-Cuevas, M. P. Cuellar, I. de Orbe-Paya, M. C. Pegalajar and L. F. Capitan-Vallvey, Microchem. J., 2011, 97, 225–233 CrossRef CAS.
- D. B. L. Terci and A. V. Rossi, Quim. Nova, 2002, 25, 684–688 CrossRef.
- L. B. Golasz, J. Silva and S. B. Silva, Food Sci. Tech., 2013, 33, 155–162 CrossRef.
- P. M. A. Khan and M. Farooqui, J. Adv. Sci. Res., 2011, 2(4), 20–27 CAS.
- S. Pourjavaher, H. Almasi and S. Meshkini, et al. , Carbohydr. Polym., 2017, 156, 193–201 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., Int. Ed., 2007, 46, 1318–1320 CrossRef CAS PubMed.
- Y. Xu, M. Liu, N. Kong and J. Liu, Microchim. Acta, 2016, 183, 1521–1542 CrossRef CAS.
- T. Ozer, C. McMahon and C. S. Henry, Annu. Rev. Anal. Chem., 2020, 13, 85–109 CrossRef.
- Y. He, Y. Wu, J.-Z. Fu and W.-B. Wu, RSC Adv., 2015, 5, 78109–78127 RSC.
- P. S. Sruthi, S. Balasubramanian and P. S. Kumar, et al. , Anal. Chim. Acta, 2021, 1182, 338953 CrossRef.
- X. Zhang, S. Lu and X. Chen, Sens. Actuators, B, 2014, 198, 268–273 CrossRef CAS.
- N. Kapilraj, S. Keerthanan and M. Sithambaresan, J. Chem., 2019, 2019, 1–6 CrossRef.
- M. Pavai, J. Mihaly and A. Paszternak, Food Anal. Methods., 2015, 8, 2243–2249 CrossRef.
- N. A. Metheny, et al. , Crit. Care Nurse, 2017, 37(3), 50–58 CrossRef PubMed.
- The GIMP Development Team, GNU Image Manipulation Program (GIMP), free software. Ver. 2.10.22 Available at: https://www.gimp.org.
- R. Leardi, C. Melzi and G. Polotti, CAT (Chemometric Agile Tool), R-based software freely downloadable from https://gruppochemiometria.it/index.php/software.
- K. L. Diehl and Eric V. Anslyn, Chem. Soc. Rev., 2013, 42, 8596–8611 RSC.
- L. R. Magnaghi, C. Zanoni, G. Alberti, P. Quadrelli and R. Biesuz, Talanta, 2022, 241, 123230 CrossRef CAS PubMed.
- L. R. Magnaghi, F. Capone, G. Alberti, C. Zanoni, B. Mannucci, P. Quadrelli and R. Biesuz, ACS Food Sci. Technol., 2021, 1(5), 819–828 CrossRef CAS.
- R. G. Brerenton, Chemometrics, Wiley, 2003, ISBN 0-471-48977-8 Search PubMed.
- G. E. P. Box, J. S. Hunter and W. G. Hunter, Statistics for Experimenters: Design, Innovation, and Discovery, Wiley, 2005, ISBN 978-0-471-71813-0 Search PubMed.
- P. M. Kroonenberg, Three Mode Principal Component Analysis, DSWO Press, Leiden, 1983 Search PubMed.
- R. Leardi, C. Armanino, S. Lanteri and L. Alberotanza, J. Chemometrics, 2000, 14, 187–195 CrossRef CAS.
- M. Younas, A. Maryam and M. Khan, et al. , Microfluid. Nanofluid., 2019, 23, 38 CrossRef.
- Y. Lu, W. Shi, J. Qin and B. Lin, Anal. Chem., 2010, 82, 329–335 CrossRef CAS PubMed.
- W. Dungchai, O. Chailapakul and C. S. Henry, Analyst, 2011, 136(1), 77–82 RSC.
- S. A. Klasner, A. K. Price and K. W. Hoeman, et al. , Anal. Bioanal. Chem., 2010, 397(5), 1821–1829 CrossRef CAS PubMed.
- K. Abe, K. Kotera, K. Suzuki and D. Citterio, Anal. Bioanal. Chem., 2010, 398(2), 885–893 CrossRef CAS PubMed.
- X. Li, J. Tian, T. Nguyen and W. Shen, Anal. Chem., 2008, 80(23), 9131–9134 CrossRef CAS PubMed.
- R. Ghosh, S. Gopalakrishnan and R. Savitha, et al. , Sci. Rep., 2019, 9, 7896 CrossRef.
- X. Wu and R. L. Prior, J. Agric. Food Chem., 2005, 53, 3101–3113 CrossRef CAS PubMed.
- W. Wiczkowski, D. Szawara-Nowak and J. Topolska, Food Res. Int., 2013, 51(1), 303–309 CrossRef CAS.
- X. Fu, Q. Wu, J. Wang, Y. Chen, G. Zhu and Z. Zhu, Molecules, 2021, 26, 7000 CrossRef CAS.
- S. Y. Kahu, R. B. Raut and K. M. Bhurchandi, Color Res. Appl., 2019, 44, 8–33 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nj03675d |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2022 |