 Open Access Article
Open Access ArticleCharacterization of lipase from Candida rugosa entrapped in alginate beads to enhance its thermal stability and recyclability†
Alice
Vetrano
 a,
Francesco
Gabriele
a,
Raimondo
Germani
b and
Nicoletta
Spreti
a,
Francesco
Gabriele
a,
Raimondo
Germani
b and
Nicoletta
Spreti
 *a
*a
aDepartment of Physical and Chemical Sciences, University of L’Aquila, Via Vetoio – Coppito, I-67100 L’Aquila, Italy. E-mail: nicoletta.spreti@univaq.it
bCEMIN, Centre of Excellence on Nanostructured Innovative Materials, Department of Chemistry, Biology and Biotechnology, University of Perugia, Via Elce di Sotto 8, I-06123 Perugia, Italy
First published on 4th May 2022
Abstract
Lipase from Candida rugosa has been immobilized in different formulations of calcium alginate beads, prepared by ionotropic gelation, which differ from each other in CaCl2 concentration and hardening time, to investigate the effects of immobilization conditions on enzyme properties. Morphological studies on all hydrated beads, performed by SEM equipped with a Peltier plate, revealed a different internal compactness. Despite this, all types of beads had an immobilization yield of 100% measured with the Bradford method and about 94% evaluated from the residual activity of the preparation solutions; moreover, all entrapped biocatalysts catalyzed the complete hydrolysis of p-nitrophenyl acetate, even after one month of storage in distilled water at 4 °C. When the internal microstructure of the beads was more compact, the rate of hydrolysis of the most hydrophobic p-nitrophenyl dodecanoate was halved, probably due to mass transfer limiting effects. The immobilized lipase had better resistance to temperature inactivation than the free form: enzyme residual activity at 50 °C after a week were approximately 70% and 20% for the immobilized and free forms respectively. An excellent recyclability in water at 25 °C of entrapped enzyme was also found, having residual activity greater than 80% at the tenth reaction cycle. The best bead formulation was then used for the resolution of (R)-1-phenylethanol in aqueous solution starting from racemic (R,S)-1-phenylethyl acetate. The enantioselectivity found (E = 10) was slightly higher but did not differ significantly from that of free lipase towards the same substrate (E = 4).
1. Introduction
The employment of enzymes as biocatalysts has emerged as an important tool in many industrial processes, such as the synthesis of active pharmaceuticals, detergents, food ingredients and fine chemicals.1 However, although bioconversions are more environmentally friendly and more sustainable, some disadvantages limit the application of enzymes both in academic research and mainly in industry. These can be ascribed to the lack of operational stability and to the difficult recovery and reuse of biocatalysts, especially for their application in industrial processes, where multiple high yield cycles are required. Many technological implementations offer the possibility to overcome these drawbacks, such as reaction medium or protein and substrate engineering.1 Immobilization techniques have also proven to be a very powerful tool for implementing enzyme properties.2 In fact, immobilization, in addition to allowing easier recovery and reuse of the enzyme reducing operating costs, can improve other enzyme features, such as its stability in many reaction media, activity, selectivity, specificity, and resistance to temperature, chemicals and inhibitors.3–5 The modification of enzyme properties due to its immobilization can sometimes be associated with changes in its structure in a more active form. Many other causes must also be considered, such as an increase in rigidity of the enzyme structure that avoids distortions, a protective effect of the support under harsh conditions, a decrease in substrate and/or product inhibition or the creation of their partition towards or away from the enzymatic environment.3 Furthermore, in addition to the enzyme, the performance of an immobilized biocatalyst strongly depends both on the support materials (classic inorganic or organic supports and new materials with desired properties) and on the method of immobilization.6–9 Several approaches have been investigated to improve enzyme performances, such as binding (physical, ionic or covalent) to a support, cross-linking, which involves the formation of covalently linked protein-protein aggregates, or entrapment, in which the enzyme is physically confined within a three-dimensional polymeric network.10–12 Each of them can affect enzyme properties and a universal method for enzyme immobilization still does not exist. Therefore, the appropriate choice of the best immobilization process must be made to obtain good catalytic activity, stability and reusability of the enzymes.13Lipases (triacylglycerol ester hydrolase, E.C. 3.1.1.3) are ubiquitous enzymes widely employed to catalyze a wide range of enantio- and regioselective reactions such as hydrolysis, esterification, transesterification, aminolysis and ammoniolysis.14–18 Unlike the usual esterases, lipases are able to hydrolyze long-chain acyl glycerols and contain an amphiphilic peptide lid domain covering the active site of the enzyme, which, in the presence of a hydrophobic interface, undergoes a conformational rearrangement allowing the enzyme to switch to the active state, a phenomenon known as interfacial activation.19
Lipase from Candida rugosa (CRL), whose structural features, mechanism of action and catalytic versatility are well known, exists as several isoenzymes, with a high structural homology, but different carbohydrate content, isoelectric point and substrate specificity.20–23 CRL is one of the most used enzymes for biotransformations, but the use of the free form is not convenient, as it is deactivated when exposed to temperatures higher than 50 °C for a long time24 and, above all, the enzyme is not recyclable and its separation from the final products requires many time-consuming steps. For these reasons, several methods have been reported to improve enzyme stability and recyclability: physical adsorption on solid supports,25–29 cross-linking,30–32 covalent binding33–35 and encapsulation on a solid matrix.36–39 Adsorption is the simplest and cheapest method, but the leaching of the enzyme from the support is a significant disadvantage for practical uses, compromising its reusability. Cross-linked enzyme aggregates, employing bifunctional reagents, can be either used as carrier-free macroparticles or immobilized on different supports30,31 or they can be trapped in alginate beads.32 In these cases, the operational stability and reusability of the biocatalyst have been improved and the immobilization protocol can be decisive due to the variety of enzyme structural conformations. Covalent binding to a support generally prevents enzyme leaching from the surface, but this method could have the disadvantage of a possible irreversible deactivation. Reusability, storage stability and tolerance to organic solvents were improved when CRL, chemically modified with a monomer of the metal–organic frameworks was used to the “in situ” synthesis of immobilized CRL composite,33 as well as better lipase activity was achieved as a result of irreversible enzyme immobilization onto a ternary alginate/nanocellulose/montmorillonite composite.34 The covalent immobilization of CRL onto magnetic beads offered important advantages in terms of enzyme reusability, thanks to the magnetically easy recovery of the biocatalyst from the reaction media, and can be applied for synthetic purpose,35 for food applications,40,41 and for biodiesel production.42,43 Moreover, CRL, encapsulated in silica sol gels in the presence of magnetic sporopollenin/Fe3O4 nanoparticles,36,37 β-cyclodextrin-grafted38 and in the presence of N-methylglucamine based calix[4]arene magnetic nanoparticles39 exhibited high thermal stability, reusability and excellent enantioselective capability.
Naturally-derived polymers turn out to be more advantageous in the entrapment of biomolecules than synthetic ones thanks to their intrinsic properties such as biocompatibility, non-toxicity, biodegradability and renewability that make them very attractive supports in many applications in biomedical, pharmaceutical and food sectors.44 Among them, alginate, an anionic polysaccharide derived from brown algae consisting of β-D-mannuronate (M) and α-L-guluronate (G) as monomeric units, is by far the most widely used polymer for immobilization and microencapsulation technologies, thanks to its ability to easily form the desired three-dimensional structures in an aqueous environment by coordinating divalent cations.45 Many alginate-based supports have been developed for enzyme immobilization and for the enhancement of enzyme properties, in terms of operational stability and reusability, together with their biotechnological applications, have recently been reviewed.44 CRL was also entrapped in Ca-alginate beads and, thanks to the strong affinity of the polysaccharide for the enzyme, its thermal stability was improved as the energy barrier of the first deactivation step was higher than that of the free enzyme.24 Nevertheless, the main problem was the CRL leaching from the beads,46 that can be partially overcome by covering their surface with chitosan or silicate.47 Esterification reactions were successfully performed in aqueous media using biphasic alginate beads, consisting of a solid matrix of calcium alginate and hexadecane,48 or in organic solvents when CRL was immobilized in polyvinyl alcohol (PVA), alginate and boric acid beads.49 In the latter case, the biocatalyst was more compatible with both water and organic media, since a certain amount of water molecules was also allowed in dehydrating solvents. The immobilization of CRL in magnetic alginate beads made possible to easily collect and reuse the biocatalyst for 6 cycles, but its activity was lower with respect to the enzyme entrapped in alginate beads.50
In this paper, CRL was immobilized within six different formulations of calcium-alginate beads. The operating conditions, such as the concentration of CaCl2 and the residence time in the hardening solution, have been taken into account to evaluate their effect on: (i) loading efficiency; (ii) biocatalytic hydrolysis toward two substrates with different hydrophobicity p-nitrophenyl acetate (p-NPA) and p-nitrophenyl dodecanoate (p-NPD); (iii) recyclability and thermostability of the immobilized biocatalyst, decisive properties for industrial applications, and (iv) internal structure. Finally, the best formulation was chosen to perform the kinetic resolution of the racemic ester (R,S)-1-phenylethyl acetate.
2. Experimental section
2.1 Chemicals
Alginic acid sodium salt from brown algae (low viscosity), lipase from Candida rugosa (CRL, type VII, >1000 U mg−1 solid), p-nitrophenyl acetate (p-NPA), p-nitrophenyl dodecanoate (p-NPD), racemic (R,S)-1-phenylethanol and (R)-(+)-1-phenylethanol were purchased from Sigma Aldrich. Racemic (R,S)-1-phenylethyl acetate was obtained from Merck. Coomassie Brilliant Blue G-250 dye was supplied by Bio-Rad. Enzyme and substrate were used with no further purification. All other chemicals used were of analytical grade.2.2 Entrapment of CRL in calcium alginate beads
After an optimization of the shape and resistance to magnetic stirring, the best six bead formulations were reported in Table 1; they differed from each other in the concentration of the calcium chloride solution and in the residence time in the solution itself. CRL (2 mg ml−1) was added in a 5% (w/v) alginate aqueous solution and the mixture was stirred thoroughly to ensure complete mixing. Two ml of the resulting solution were withdrawn with a syringe with a 23G needle (inner diameter 600 μm) and dropped at a distance of about 2 cm into a 2% or 5% calcium chloride solution, maintained under mechanical stirring at 50 rpm, obtaining about 120 beads with a total weight of 1.2 ÷ 1.4 g. After the formation of the beads, they were left in the calcium chloride solution for a time ranging from 10 min to 1 h, to increase their mechanical strength. Finally, the beads were filtered under vacuum and rinsed with distilled water to remove the excess of calcium chloride solution.| CaCl2, % (w/v) | Residence time, min | |
|---|---|---|
| a Sodium alginate solution 5% (w/v) and CRL 2 mg ml−1. | ||
| Beads 1 | 2 | 10 |
| Beads 2 | 5 | 10 |
| Beads 3 | 2 | 30 |
| Beads 4 | 5 | 30 |
| Beads 5 | 2 | 60 |
| Beads 6 | 5 | 60 |
2.3 Immobilization efficiency
Loading efficiency was determined by checking the amount of CRL in both the bead preparation and washing solutions using the Bradford method.51 A calibration curve was obtained by measuring the absorbance of solutions containing from 0 to 0.3 mg ml−1, prepared from a stock solution with a CRL concentration of 2 mg ml−1, at λ = 595 nm. The resulting calibration curve had a R2 correlation coefficient of 0.99. The loss of enzyme from beads over time was assessed similarly. In addition, the activity of the solution used to prepare the beads was also measured and subtracted from that of the free enzyme in order to determine the immobilized activity and therefore the immobilization yield (%):52 | (1) |
 | (2) |
2.4 CRL activity assay
Hydrolytic activity of lipase was determined by spectrophotometric measurements, by using Shimadzu UV-160A instrument, following the hydrolysis reaction of p-nitrophenyl acetate (p-NPA); the procedure was adapted from literature to this case of study.53 Beads (1.2 ÷ 1.4 g – 4 mg CRL) were placed in a reaction vessel containing 9 ml of distilled water; then, 1 ml of substrate stock solution (100 mM p-NPA in CH3CN) was added to the vessel in order to start the reaction. The reaction was carried out at room temperature under mild stirring and monitored at different times, taking 20 μl from the reaction solution, placing them in the 1 ml cuvette, where 0.98 ml of distilled water were already present. Spectrophotometric measurements were performed at λ = 348 nm, which corresponds to the isosbestic point of the equilibrium between p-nitrophenol/p-nitrophenoxide (p-NP), with a molar extinction coefficient as 5400 M−1 cm−1. Once the reaction was completed, the beads were filtered under vacuum, washed, placed in a container with distilled water and stored at 4 °C.Activity of encapsulated lipase was also determined with a more hydrophobic substrate, p-nitrophenyl dodecanoate (p-NPD), dissolved in CH3CN; 1 ml of a 100 mM stock solution was placed in the reaction vessel containing 9 ml of tert-butyl alcohol as a solvent and the beads containing the enzyme (1.2 ÷ 1.4 g – 4 mg CRL). The reaction was monitored at different times following the appearance of p-NP at 348 nm.
2.5 CRL reusability
The stability of the encapsulated CRL and its reuse were tested under the same conditions described in the previous section. After each cycle, the biocatalyst was filtered, washed with water several times to remove any product adsorbed on the beads and reintroduced into a fresh reaction medium. The substrate hydrolysis reaction was assayed at appropriate time intervals up until its complete conversion to product.2.6 Thermostability
Thermal stability of free and immobilized CRL was studied. Both forms of the enzyme were incubated in distilled water at 25 and 50 °C for various periods, from 8 h up to one week. The p-NPA hydrolysis reaction was conducted at the incubation temperatures for 30 min in pure water to determine both the initial and the remaining activity.2.7 Kinetic resolution of racemic 1-phenylethyl acetate
Hydrolysis reaction of (R,S)-1-phenylethyl acetate was performed by placing 1 ml from a stock solution (100 mM in CH3CN) into the reaction vessel containing 9 ml of water and 1.2 ÷ 1.4 g of beads. At different times, the beads were separated by filtration under vacuum, the aqueous phase was extracted with ethyl ether and, after the solvent was removed, isopropanol was added to the flask. The conversion, as well as the enantiomeric excess value (ee%) of the reagents and products were determined by HPLC analysis with an Agilent – 1220 Infinity II instruments equipped with a chiral column (Lux Cellulose-1) using 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hexane/isopropanol as eluent at a flow rate of 0.6 ml min−1. The retention times were 3.6, 3.9, 14.7, 18.5 min for (R)-1-phenylethylacetate, (S)-1-phenylethylacetate, (R)-1-phenylethanol, and (S)-1-phenylethanol, respectively. The enantiomeric excess of the substrate (ees) and product (eep), conversion (c) and enantioselectivity (E), the latter determined with the actual values of c and ee, were calculated by applying the following equations:54
1 hexane/isopropanol as eluent at a flow rate of 0.6 ml min−1. The retention times were 3.6, 3.9, 14.7, 18.5 min for (R)-1-phenylethylacetate, (S)-1-phenylethylacetate, (R)-1-phenylethanol, and (S)-1-phenylethanol, respectively. The enantiomeric excess of the substrate (ees) and product (eep), conversion (c) and enantioselectivity (E), the latter determined with the actual values of c and ee, were calculated by applying the following equations:54 | (3) |
 | (4) |
 | (5) |
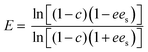 | (6) |
2.8 Morphological characterization
The shape and size of the beads were analyzed using Leica S8APO stereoscopic microscope with EC3 camera connected to a computer. The diameters distribution of the beads was evaluated by using the variance coefficient (CV), which indicates the deviation of each diameter (Dn) from the average value (Dm), and was determined as follows: | (7) |
Sphericity factor (SF), indicating the roundness of the beads, was determined by using the following equation:55,56
 | (8) |
The aspect ratio (AR) gives a good description of large bead deformations but is less accurate on smaller ones. AR varies from unity for a sphere to infinity for an elongated particle and was determined using the following equation:
 | (9) |
The surface morphology and the internal structure of the hydrate systems was investigated using a scanning electron microscope (SEM) equipped with a Peltier cooling-device MK3 Cool stage Carl Zeiss SUPRA with a working distance of about 8 mm and high voltage of 10 KV. Analyses were done around 0 °C in variable pressure mode (20 Pa) using a BSE detector (Signal A BSD4).
3. Results and discussion
3.1 Immobilization of CRL onto Ca-alginate beads
Different experimental conditions were tested to optimize the best composition to obtain stable Ca-alginate beads. Both their size and shape have a noticeable effect on their chemical and mechanical stability, and the production of monodisperse and spherical beads is preferable. The main factors affecting the size and shape of Ca-alginate beads have been previously reported and reviewed.55,56 Being alginate a family of linear binary copolymers of mannuronic (M) and guluronic (G) acids, the chemical properties of the commercial alginate used to prepare the beads, namely the molecular weight (MW = 37.6 ± 0.2 kDa) and the composition (M/G = 1.77), influenced their cross-link ability; the data of alginate characterization are reported in ESI† (Tables S1, S2 and Fig. S1). Increasing sodium alginate concentrations were tested to obtain spheres with good mechanical strength, but below 5% (w/v) beads were fragile and broke rapidly under mild magnetic stirring. Among the other parameters that affect the size and shape of the beads, the concentration of calcium chloride in gelation solution and the hardening time in the reticulation bath play an important role. The beads were prepared by extrusion dripping, one of the most popular methods, in which an alginate solution containing the enzyme is extruded through a capillary and dropped into a calcium chloride solution at room temperature. Briefly, CRL was dispersed in the aqueous alginate solution, added dropwise in two different calcium chloride solutions, i.e., 2 and 5% (w/v) and maintained under magnetic stirring at 50 rpm for 10, 30 or 60 min, to obtain six different formulations indicated as Beads 1–6, as reported in Table 1.Fig. 1 shows a schematic representation of the synthetic process for preparing the immobilized CRL on Ca-alginate beads.
3.2 Immobilization efficiency
The amount of the enzyme in the alginate beads, its loss in the preparation procedure and over time were evaluated using the Bradford method for all six different types of beads. In the preparation and washing solutions, the amount of CRL was found to be below the detection limit of the method (22 μg ml−1). Furthermore, to evaluate the effective amount of immobilized enzyme, the catalytic activity of the preparation solution was measured, and the immobilization yield, obtained from eqn (1), resulted of about 94.4%. The immobilization efficiency was also determined as previously described in eqn (2) and turns out to be 76%. These parameters indicated that 18% of the immobilized activity was lost due to entrapment procedure because the enzyme was inactivated or inaccessible to the substrate. As regards the enzyme loss over time, withdrawals of the storage water were taken at different times for one month and subjected to the Bradford assay. The only preparations that had a detectable enzyme loss after 48 h of storage in the aqueous solution, which remained fairly constant over time, were Beads 1 (1%) and Beads 2 (0.6%), i.e., those with a shorter residence time in the calcium chloride solution (10 min) (Table S3, ESI†). In the other cases, there was an oscillation of the absorbance values that were below the sensitivity of the detection method.3.3 Hydrolytic activity of enzymatic Ca-alginate beads
Before evaluating the hydrolytic activity of the lipase from Candida rugosa in alginate beads, blank tests were performed to rule out that enzyme-free beads catalyze the p-NPA hydrolysis reaction.Once this was established, activity tests were performed with the CRL-containing beads in pure water, rather than in buffer solution. Indeed, notwithstanding the pH of the medium is a critical parameter in enzymatic reactions, since it affects ionization state of enzyme leading to changes in the active site,57 the presence of salts in the reactor vessel could lead to corrosion issues in view of industrial applications, as biofuel production.58
The first catalytic tests were performed with all the bead formulations with p-NPA at a concentration of 10 mM. Fig. 2 shows the conversion percentage of the substrate over time for the six types of beads.
 | ||
Fig. 2 Substrate conversion percentages for all types of beads at 30 min ( ), 1 h ( ), 1 h ( ), 2 h ( ), 2 h ( ) and 3 h ( ) and 3 h ( ); [p-NPA] = 10 mM, T = 25 °C. ); [p-NPA] = 10 mM, T = 25 °C. | ||
The figure clearly shows that, regardless of the bead formulation, all reactions were complete within three hours with an initial reaction rate of 130 ± 10 μM min−1, and only slight differences were observed during the reaction between the different types of beads. Therefore, it was not possible to discriminate them in terms of conversion efficiency if used immediately after their preparation.
In Section 3.2, we proved that there was no leakage of enzyme from the beads during long-term storage, except for Beads 1 and Beads 2, where the loss was still less than or equal to 1%, but we had no information regarding its activity after a long period of storage in water. Therefore, a reaction cycle was performed one month after preparation to evaluate whether the enzyme in the beads was still active. For all types of beads, complete substrate conversion was achieved within four hours indicating that there were no significant changes in the activity of CRL after a month of storage (Fig. S2, ESI†).
3.4 CRL recyclability
One of the most useful advantages of enzyme immobilization is its reusability, which is of great importance in the production of biocatalysts. For this reason, many papers in the literature deal with the stability and recyclability of CRL immobilized on solid supports by adsorption,25,27,29 cross-linking,30 covalent binding,33–35 or entrapment.36,38,47,50 Although in most of them the enzyme showed significant stabilization and recyclability, the deactivation of proteins can frequently occur. This effect is often due to the leakage of enzyme from the support or to its conformational limitation; furthermore, low substrate diffusion can also contribute to the loss of catalytic activity.To assess the reusability of CRL within Ca-alginate beads, the two extreme formulations, Beads 1 and Beads 6, were chosen to perform ten catalytic cycles, in order to evaluate the influence of both CaCl2 concentration and residence time in the solution itself. These tests were carried out with 10 mM substrate and reactions were followed until to completion. Fig. 3 shows the results of repeated uses obtained after 3 h of reaction, which is the time required to the biocatalyst to complete the first cycle.
 | ||
Fig. 3 Residual activity of immobilized CRL in Ca-alginate beads in water after 3 h of reaction: Beads 1 ( ) and Beads 6 ( ) and Beads 6 ( ); [pNPA] = 10 mM, T = 25 °C. ); [pNPA] = 10 mM, T = 25 °C. | ||
The data obtained clearly indicated that Beads 1 and Beads 6 were able to convert all the substrate into product for the first four and three cycles respectively and, at the tenth cycle, for both formulations the loss of activity was less than 20%. Despite this slightly decrease of relative activity, the reaction time from 3 h to the first cycle increases up to 7 h to the tenth one to reach the complete hydrolysis of the substrate, due to a halving of initial reaction rate for both formulations (from about 120 μM min−1 to 60 μM min−1). The results obtained so far showed that there were no differences in terms of catalytic efficiency and recyclability between the different formulations.
The use of alginate beads in the immobilization of CRL according to our formulations, compared to other systems, shows numerous advantages, as can be seen from Table 2. The preparation of the support is simple and requires only two steps, and high immobilization efficiency, recyclability and storage stability are achieved. Furthermore, our biocatalyst is active in distilled water, avoiding the buffer which is probably the cause of the observed enzyme leakage46,47 and induces swelling of the beads increasing their fragility (data not shown).
| Immobilization method | Preparation step | Biocatalyst | Immobilization yield | Substrate | Recycle times | Residual activity | Operational condition | Storage stability (days) | Residual activity | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Covalent immobilization onto a solid carrier (MOF) | 2 | Candida rugosa lipase (CRL) type VII | CRL-ZIF-8 8.18%, mCRL-ZIF-8 9.45% | p-Nitrophenyl palmitate | 7 | CRL-ZIF-8 45.8%, mCRL-ZIF-8 52.3% | Isopropanol- PBS pH 7,5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) 35 °C 9) 35 °C |
7 | CRL-ZIF-8 34.42%, mCRL-ZIF-8 42.18% | 32 |
| Covalent immobilization onto a ternary support (CRL-ALG/NC/MMT) | 5 | Candida rugosa lipase (CRL) type VII | 2.9% | Levulinic acid | 9 | 47.5% | Ethanol 50 °C | — | — | 34 |
| Covalent immobilization on magnetic beads | 5 | Candida rugosa lipase (CRL) | — | Racemic ibuprofen | 5 | 100% | Cyclohexane 37 °C | — | — | 35 |
| Sol–gel encapsulation in presence of magnetic sporopollenin/Fe3O4 nanoparticles | 4 | Candida rugosa lipase (CRL) | Fe-A-Spo-E 36%, Fe-EP-Spo-E 89% | p-Nitrophenyl palmitate | 7 | Fe-A-Spo-E 21%, Fe-EP-Spo-E 63% | Isopropanol – PBS pH 7 37 °C | 50 | Fe-A-Spo-E 45%, Fe-EP-Spo-E 40% | 36 |
| Sol–gel encapsulation e with Fe3O4 or Fe3O4-Spo | 5 | Candida rugosa lipase (CRL) type VII | Fe3O4 78.6%, Fe3O4-Spo 76.2% | Racemic naproxen methyl ester | 5 | Fe3O4-E 36%, Fe3O4-Spo 64% | Aqueous phase–organic solvent (PBS pH 7 – isooctane) | 42 | Fe3O4 >70%, Fe3O4-Spo >90% | 37 |
| Sol–gel encapsulation with b-cyclodextrin grafted magnetic nanoparticles (CD-APS-NP) | 5 | Candida rugosa lipase (CRL) type VII | 2.2% | p-Nitrophenyl palmitate | 6 | ∼50% | PBS buffer (pH 7.0) 35 °C | — | — | 38 |
| Sol–gel Encapsulation in the presence of magnetic calix[4]arene nanoparticles | 4 | Candida rugosa lipase (CRL) | 3.2% | Racemic naproxen methyl ester | 5 | 28% | Aqueous phase–organic solvent (PBS – isooctane) | — | — | 39 |
| Covalent immobilization on magnetite coated with nanosilica | 5 | Candida rugosa lipase (CRL) | ∼80% |
n-Butyric acid and 1-butanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
17 | 50% | n-Heptane at 45 °C | — | — | 59 |
| Encapsulation in chitosan nanoparticles | 3 | Candida rugosa lipase (CRL) | — | Olive oil | 7 | 53% | Water–oil (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 37 °C 1) 37 °C |
— | — | 60 |
| Entrapment in Ca-alginate beads | 2 | Candida rugosa lipase (CRL) | 35% | p-Nitrophenyl butyrate | 3 | 72% | Tris–HCl buffer (pH 7.2); 30 °C | — | — | 47 |
| Covalent immobilization of lipase onto the silica nanoflowers-NH2 | 3 | Candida antarctica lipase | ∼57% | Levulinic acid and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) 10) |
8 | 68% | Tert-butyl methyl ether 40 °C | — | — | 61 |
| Freeze dried calcium alginate beads | 3 | Candida antarctica lipase B (CALB) | — | p-Nitrophenyl butyrate | 6 | ∼90% | Distilled water 35 °C | 16 | ∼100% | 62 |
| Immobilization on magnetic sol–gel hybrid organic-inorganic (Fe3O4 MNPs@TEOS-TSD@ CALB) | 4 | Candida antarctica lipase B (CALB) | 90% | Waste cooking oil | 10 | 16.5% | Methanol; 40 °C | — | — | 63 |
| Covalent immobilization on polydopamine functionalized magnetic mesoporous biochar (MPCB-DA) | 6 | Bacillus licheniformis lipase | 46.5% | p-Nitrophenyl palmitate | 10 | 56% | Tris–HCl (50 mM, pH 8.5); 40 °C | 70 | 88% at 25 °C | 64 |
| Covalent immobilization on hydroxyapatite/glycyrrhizin/lithium-based metal–organic framework (HA/GL/Li-MOF) nanocomposites | 5 | Thermomyces lanuginosus lipase (TLL) | TLL@HA/GL/Li-MOF 71%, TLL@Li-MOF 68% | p-Nitrophenyl palmitate | 10 | TLL@HA/GL/Li-MOF ∼55%, TLL@Li-MOF ∼45% | Carbonate bufer (100 mM, pH 9.0); 60 °C | 30 | TLL@HA/GL/Li-MOF ∼30%, TLL@Li-MOF ∼20% | 65 |
| Entrapment inside Ca-Alginate beads | 2 | Candida rugosa lipase (CRL) type VII | 94.4% | p-Nitrophenyl acetate | 10 | >80% | Distilled water 25 °C | 30 | ∼100% | Current work |
3.5 Morphological studies
The morphological characterization was performed to determine the macro- and microstructure of beads prepared with different concentrations of calcium chloride and residence times in the hardening solution; also in this case, Beads 1 and Beads 6 were chosen.Analyses were initially performed using the stereomicroscope and the results are shown in Fig. 4.
Both Beads 1 and Beads 6 showed fairly uniform dimensions and the size distribution of 25 beads was measured, since the variance coefficient (CV), determined using eqn (7), was 0.9 and 3.7% respectively. Table 3 reports some of the dimensionless shape indicators quantified using eqn (8) and (9) (Section 2.8).
| Beads 1 | Beads 6 | |
|---|---|---|
| Average diameter (Dm) | 3.2 mm | 3.4 mm |
| Sphericity factor (SF) | 0.031 | 0.069 |
| Aspect ratio (AR) | 1.06 | 1.15 |
Both bead formulations have similar dimensions of about 3 mm, but better spherical shape was obtained with Beads 1, which are those prepared with lower calcium chloride concentration and shorter hardening time, since SF < 0.05 and AR was slightly higher than unity.56 On the other hand, Beads 6, as already visible from the stereomicroscope image, are less spherical; their SF value was very similar to that of alginate particles reported in the literature and prepared with our same experimental conditions, i.e. concentration of alginate and CaCl2, and hardening time.66 Then, the analyses by the scanning electron microscope were performed on both the external surface and the internal structure (by cutting them with a scalpel). Generally, in literature, SEM morphological studies are conducted on dehydrated beads, providing information that often does not correspond to the real systems employed in catalytic tests, which are hydrated. To avoid dehydration of the samples, the application of variable-pressure equipment (VP-SEM) and Peltier cooling-device allows the investigation of wet samples and hydrated systems in SEM.67 Thanks to the aid of this methodology, in this study we were able to investigate the samples in their operating conditions. The SEM images were acquired at different magnifications and the most significant at 70x and 300x are shown in Fig. 5.
 | ||
| Fig. 5 SEM images at 70x magnification of the external structure and at 70x and 300x magnifications of the internal structure of Beads 1 and Beads 6. | ||
Although no significant differences were detected from their external surface, the analysis of the cross section highlighted less internal compactness of Beads 1 than that of Beads 6 which seemed denser and more homogeneous. Further SEM analyses were then performed to understand if the cause of this different internal morphology was due to the CaCl2 concentration and/or to the hardening time. For these analyses, Beads 2 (CaCl2 5%, 10 min) and Beads 5 (CaCl2 2%, 60 min) were selected and SEM images, reported in Fig. S3 (ESI†) of both the whole beads and the internal structure highlighted that the internal compactness depends on the gelation time and not on CaCl2 concentration.
3.6 Reaction with p-nitrophenyl dodecanoate
The p-nitrophenyl dodecanoate (p-NPD) was then chosen to evaluate the efficiency of CRL immobilized on Beads 1 and 6 on a more hydrophobic substrate. Being poorly soluble in water, p-NPD hydrolysis reaction was carried out in a sterically hindered alcohol, i.e., tert-butyl alcohol, in order to avoid or at least reduce the competitive transesterification reaction rate. In this case, the conversion efficiency of the two formulations was very different. In fact, Beads 1 were able to completely hydrolyze the substrate in 6 h, while the rate of the reaction catalyzed by Beads 6 was very low: after 6 h, little more than 40% of product was formed and increased slowly over time (Fig. S4, ESI†). The reaction did not complete even after 24 h, being the conversion equal to 80%. This result clearly indicated that the internal structure of Beads 6 compared to Beads 1 limited the mass transfer of reagents and/or products in the reaction medium and this drawback was evident with large, hydrophobic substrates and therefore much more similar to natural ones. On the other hand, Beads 1, thanks to their lower compactness, were able to ensure the free diffusion of reagents within their porous structure.Furthermore, the time taken by Beads 1 to completely hydrolyze p-NPD was twice than that required for the reaction with p-NPA. Therefore, to determine if the increase in reaction time was due to the substrate or to the solvent, the reaction of p-NPA in tert-butyl alcohol was performed for comparison purpose (Fig. S5, ESI†). After 48 h, only 58% of conversion was achieved, and this result can be explained by the dehydration of the beads that became smaller and smaller over time. The loss of water from the confined environment in which the enzyme is located led to the observed slowdown of the reaction. This hypothesis was confirmed by experiments performed by varying the amount of water inside the reaction medium; the time required for complete hydrolysis of the p-NPA was decreased from 24 to 4 h with a water percentage of 5% and 75% respectively (Fig. S6, ESI†).
Therefore, given all the considerations made so far, the Beads 1 formulation, being one of the fastest preparations and ensuring better mass transfer, thus allowing their possible use with a wide range of substrates, has been chosen to perform thermostability tests and kinetic resolution of (R,S)-1-phenylethyl acetate.
3.7 Thermostability
One of the advantages of enzyme immobilization is the improvement of thermal stability. Therefore, the stability of both free and immobilized lipase in Beads 1 were determined by incubating them for different times and the residual activity was measured at 25.0 ± 0.1 and 50.0 ± 0.1 °C (Fig. 6). | ||
Fig. 6 Thermal stability of free ( / / ) and immobilized lipase ( ) and immobilized lipase ( / / ) at 25 °C (empty symbols) and 50 °C (closed symbols). ) at 25 °C (empty symbols) and 50 °C (closed symbols). | ||
The effect of immobilization on Ca-alginate beads on CRL stability is clearly highlighted by the figure at both investigated temperatures. Immobilized CRL activity at 25 °C did not decrease after 7 days of incubation while the free form lost 30% of its initial activity after 5 days and 60% after one week. Even more evident was the stabilization effect at 50 °C. In fact, the residual activity of free lipase, after only 8 h, was lower than 40% and continued to decrease over time until reaching a value of about 20% after one week of incubation. On the other hand, the remaining activity of immobilized CRL was about 70% after 1 day of heat treatment and remained unchanged throughout the week.
Moreover, stability tests were carried out with Beads 6 at 50 °C to assess the effect of their higher compactness on the enzyme thermal stability. They showed that the different operational conditions used in the preparation of the two formulations did not affect the stability of the enzyme, being the loss of activity after 24 h equal to 66% (data not shown).
The data reported in Fig. 6 show the trends of residual activity over time, but do not consider the differences in the reaction rate before incubation. In particular, hydrolysis rate with free CRL was 30% higher than that of immobilized one both at 25 °C (183 vs. 132 μM min−1) and 50 °C (520 vs. 370 μM min−1). This effect could be due to a slower diffusion of the substrate inside the support. However, after a week of incubation the reaction rate of Beads 1 at 25 °C and 50 °C was 2 times higher than that of the free enzyme, thanks to the improvement in enzymatic stabilization following the entrapment.
3.8 Kinetic resolution of (R,S)-1-phenylethyl acetate
Many studies have been devoted to the kinetic resolution of racemic substrates with pharmacological activity, such as naproxen30,37,68 and ibuprofen35 by using immobilized CRL, since the enantiomers of these non-steroidal anti-inflammatory drugs demonstrate different therapeutic activities.Given the excellent recyclability and thermal stability of Beads 1, and therefore their possible application in industrial processes, they were used in preliminary tests for the resolution of (R)-1-phenylethanol in aqueous solution starting from racemic 1-phenylethyl acetate as model substrate. In the literature, several papers report about the stereoselective kinetic resolution of rac-1-phenylethyl acetate catalyzed by other lipases,69–73 in which very high enantiomeric excess towards the (R)-enantiomer were obtained, but with not too satisfying yields. More recently, marine microbial GDSL lipase MT6 showed opposite stereoselectivity, as it hydrolyzed racemic 1-phenylethyl acetate to generate (S)-1-phenylethanol instead of (R)-1-phenylethanol.74 During reaction course, as the conversion increased, the ee of the product (eep) decreased and, at the optimal reaction time (12 h), a conversion of 28.5% with an ee value of the substrate (ees) higher than 97% was obtained. On the other hand, CRL solubilized in phosphate buffer at pH 7.2 showed no enantiopreference for the (R)-acetate with an ee value of only 44% and an enantioselectivity factor of 4.75
Here, the course and selectivity of the kinetic resolution of (R,S)-1-phenylethyl acetate were checked, as reported in materials and methods, by chiral HPLC and all the chromatograms are shown in Fig. S7 (ESI†). The percentage of enantiomeric excess (ees and eep), conversion (c) as well as enantioselectivity (E), determined using eqn (3)–(6), are reported in Table 4.
| Reaction time (h) | ee s, % | ee p, % | c, % | E |
|---|---|---|---|---|
| 8 | 3.5 | 61.5 | 5.3 | 4.3 |
| 24 | 17.6 | 62.8 | 21.9 | 5.2 |
| 32 | 18.1 | 61.9 | 22.6 | 5.1 |
| 48 | 23.1 | 64.5 | 26.4 | 5.8 |
| 72 | 49.5 | 61.7 | 44.5 | 6.8 |
| 96 | 83.3 | 60.9 | 57.8 | 10.3 |
As observed from these results, while ees increased with reaction time and conversion, eep remained almost constant. After 96 h of reaction, the highest values of enantioselectivity (E = 10.3), conversion (c = 57.8%) and enantiomeric excess of substrate (ees = 83.3%) were observed. These values are slightly higher but do not differ so much from those reported in the literature regarding free lipase in phosphate buffer, which showed an ees value of 44% and an enantioselectivity factor of 4.75 Given the intrinsic chirality of the matrix, we performed these tests to assess whether the immobilization improved the enantioselectivity of the enzyme towards the chosen substrate. This has not happened, but the entrapment of CRL improved its stability under non-physiological conditions and temperatures and made it recyclable for multiple cycles, which is essential for industrial applications.
4. Conclusion
In this study, lipase from Candida rugosa was efficiently trapped in Ca-alginate beads prepared using different operating conditions. Morphological studies highlighted that, despite differing in their internal microstructure, all formulations exhibited high immobilization efficiency, good recyclability and stability, both operational and thermal. The effectiveness of the immobilized CRL was instead dependent on the hydrophobicity of the substrate. The higher internal compactness of Beads 6, compared to Beads 1, did not affect the hydrolysis rate of p-NPA, but limited the mass transfer of the more hydrophobic substrate p-NPD causing a notable decrease in its reaction rate. Further studies will be performed to improve the stereoselectivity of the immobilized biocatalyst by modifying either the operational conditions or the bead composition.Author contributions
A. Vetrano: investigation, validation, writing-original draft; F. Gabriele: investigation, visualization, writing-review & editing; R. Germani: conceptualization, writing-review & editing; N. Spreti: conceptualization, writing-review & editing, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge financial support from the Ministero dell’Università e della Ricerca, MIUR (Rome, Italy). The Authors thank Dr Lorenzo Arrizza (Microscopy Centre, University of L’Aquila) for his kindly assistance in SEM analysis.References
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS PubMed.
- R. A. Sheldon and S. van Pelt, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC.
- R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- R. C. Rodrigues, J. J. Virgen-Ortíz, J. C.-S. dos Santos, Á. Berenguer-Murcia, A. R. Alcantara, O. Barbosa, C. Ortiz and R. Fernandez-Lafuente, Biotechnol. Adv., 2019, 37, 746–770 CrossRef CAS PubMed.
- R. C. Rodrigues, Á. Berenguer-Murcia, D. Carballares, R. Morellon-Sterling and R. Fernandez-Lafuente, Biotechnol. Adv., 2021, 52, 107821 CrossRef CAS PubMed.
- J. Zdarta, A. S. Meyer, T. Jesionowski and M. Pinelo, Catalysts, 2018, 8, 92 CrossRef.
- J. C.-S. D. Santos, O. Barbosa, C. Ortiz, A. Berenguer-Murcia, R. C. Rodrigues and R. Fernandez-Lafuente, ChemCatChem, 2015, 7, 2413–2432 CrossRef.
- S. Cantone, V. Ferrario, L. Corici, C. Ebert, D. Fattor, P. Spizzo and L. Gardossi, Chem. Soc. Rev., 2013, 42, 6262–6276 RSC.
- R. A. Sheldon, Adv. Synth. Catal., 2007, 349, 1289–1307 CrossRef CAS.
- S. García-Embid, F. Di Renzo, L. De Matteis, N. Spreti and J. M. de la Fuente, Appl. Catal., A, 2018, 560, 94–102 CrossRef.
- L. De Matteis, R. Germani, M. V. Mancini, F. Di Renzo and N. Spreti, Appl. Catal. A Gen., 2015, 492, 23–30 CrossRef CAS.
- L. De Matteis, R. Germani, M. V. Mancini, G. Savelli, N. Spreti, L. Brinchi and G. Pastori, J. Mol. Catal. B: Enzym., 2013, 97, 23–30 CrossRef CAS.
- C. Garcia-Galan, Á. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Adv. Synth. Catal., 2011, 353, 2885–2904 CrossRef CAS.
- B. P. Dwivedee, S. Soni, M. Sharma, J. Bhaumik, J. K. Laha and U. C. Banerjee, ChemistrySelect, 2018, 3, 2441–2466 CrossRef CAS.
- P. Choudhury and B. Bhunia, Biopharm J., 2015, 1, 41–47 Search PubMed.
- B. Andualema and A. Gessesse, Biotechnology, 2012, 11, 100–118 CrossRef CAS.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS.
- A. Houde, A. Kademi and D. Leblanc, Appl. Biochem. Biotechnol., 2004, 118, 155–170 CrossRef CAS PubMed.
- R. D. Schmid and R. Verger, Angew. Chem., Int. Ed., 1998, 37, 1608–1633 CrossRef PubMed.
- J. Barriuso, M. E. Vaquero, A. Prieto and M. J. Martínez, Biotechnol. Adv., 2016, 34, 874–885 CrossRef CAS PubMed.
- P. Domínguez De María, J. M. Sánchez-Montero, J. V. Sinisterra and A. R. Alcántara, Biotechnol. Adv., 2006, 24, 180–196 CrossRef PubMed.
- S. Benjamin and A. Pandey, Yeast, 1998, 14, 1069–1087 CrossRef CAS PubMed.
- E. Subroto, R. Indiarto, A. D. Pangawikan, S. Huda and V. P. Yarlina, Food Res., 2020, 4, 1391–1401 Search PubMed.
- M. Matsumoto and K. Ohashi, Biochem. Eng. J., 2003, 14, 75–77 CrossRef CAS.
- H. Aghaei, M. Ghavi, G. Hashemkhani and M. Keshavarz, Int. J. Biol. Macromol., 2020, 162, 74–83 CrossRef CAS PubMed.
- R. D.-M. Ferreira, R. Brackmann, E. B. Pereira and R. D.-C. da Rocha, Appl. Biochem. Biotechnol., 2020, 190, 839–850 CrossRef CAS PubMed.
- L. T. Izrael Živković, L. S. Živković, V. P. Beškoski, K. R. Gopčević, B. M. Jokić, D. S. Radosavljević and I. M. Karadžić, J. Mol. Catal. B: Enzym., 2016, 133, S533–S542 CrossRef.
- L. Izrael-Zivkovic, L. Zivkovic, B. Jokic, A. Savic and I. Karadzic, J. Serb. Chem. Soc., 2015, 80, 1113–1125 CrossRef.
- L. T. Izrael Živković, L. S. Živković, B. M. Babić, M. J. Kokunešoski, B. M. Jokić and I. M. Karadžić, Biochem. Eng. J., 2015, 93, 73–83 CrossRef.
- S. Salgın, M. Çakal and U. Salgın, Prep. Biochem. Biotechnol., 2020, 50, 148–155 CrossRef PubMed.
- S. Velasco-Lozano, F. López-Gallego, J. Rocha-Martin, J. M. Guisán and E. Favela-Torres, J. Mol. Catal. B: Enzym., 2016, 130, 32–39 CrossRef CAS.
- J. C. Wu, V. Selvam, H. H. Teo, Y. Chow, M. M.-R. Talukder and W. J. Choi, Biocatal. Biotransform., 2006, 24, 352–357 CrossRef CAS.
- B. Zou, L. Zhang, J. Xia, P. Wang, Y. Yan, X. Wang and I. O. Adesanya, Appl. Biochem. Biotechnol., 2020, 192, 132–145 CrossRef CAS PubMed.
- F. N.-N. Mohd Hussin, N. Attan and R. A. Wahab, Enzyme Microb. Technol., 2020, 136, 109506 CrossRef CAS PubMed.
- M. P. Marszałł and T. Siódmiak, Catal. Commun., 2012, 24, 80–84 CrossRef.
- E. Ozyilmaz, K. Etci and M. Sezgin, Prep. Biochem. Biotechnol., 2018, 48, 887–897 CrossRef CAS PubMed.
- E. Yilmaz, M. Sezgin and M. Yilmaz, J. Mol. Catal. B: Enzym., 2011, 69, 35–41 CrossRef CAS.
- E. Ozyilmaz, S. Sayin, M. Arslan and M. Yilmaz, Colloids Surf., B, 2014, 113, 182–189 CrossRef CAS PubMed.
- S. Sayin, E. Yilmaz and M. Yilmaz, Org. Biomol. Chem., 2011, 9, 4021–4024 RSC.
- W. Xie and X. Zang, Food Chem., 2018, 257, 15–22 CrossRef CAS PubMed.
- W. Xie and X. Zang, Food Chem., 2017, 227, 397–403 CrossRef CAS PubMed.
- W. Xie and J. Wang, Energy Fuels, 2014, 28, 2624–2631 CrossRef CAS.
- W. Xie and M. Huang, Energy Convers. Manag., 2018, 159, 42–53 CrossRef CAS.
- M. Bilal and H. M.-N. Iqbal, Int. J. Biol. Macromol., 2019, 130, 462–482 CrossRef CAS PubMed.
- K. Yong and D. J. Mooney, Prog. Polym. Sci., 2012, 37, 106–126 CrossRef PubMed.
- S. S. Betigeri and S. H. Neau, Biomaterials, 2002, 23, 3627–3636 CrossRef CAS PubMed.
- K. Won, S. Kim, K. J. Kim, H. W. Park and S. J. Moon, Process Biochem., 2005, 40, 2149–2154 CrossRef CAS.
- C. H. Ng and K. L. Yang, Enzyme Microb. Technol., 2016, 82, 173–179 CrossRef CAS PubMed.
- R. Dave and D. Madamwar, Process Biochem., 2006, 41, 951–955 CrossRef CAS.
- C. H. Yang, C. C. Yen, J. J. Jheng, C. Y. Wang, S. S. Chen, P. Y. Huang, K. S. Huang and J. F. Shaw, Molecules, 2014, 19, 11800–11815 CrossRef PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- J. Boudrant, J. M. Woodley and R. Fernandez-Lafuente, Process Biochem., 2020, 90, 66–80 CrossRef CAS.
- Y. Ji, Z. Wu, P. Zhang, M. Qiao, Y. Hu, B. Shen, B. Li and X. Zhang, Biochem. Eng. J., 2021, 169, 107962 CrossRef CAS.
- C. S. Chen, Y. Fujimoto, G. Girdaukas and C. J. Sih, J. Am. Chem. Soc., 1982, 104, 7294–7299 CrossRef CAS.
- B. B. Lee, P. Ravindra and E. S. Chan, Chem. Eng. Technol., 2013, 36, 1627–1642 CAS.
- E. S. Chan, B. B. Lee, P. Ravindra and D. Poncelet, J. Colloid Interface Sci., 2009, 338, 63–72 CrossRef CAS PubMed.
- Y. Liu, Q. Jin, L. Shan, Y. Liu, W. Shen and X. Wang, Ultrason. Sonochem., 2008, 15, 402–407 CrossRef CAS PubMed.
- E. Torsner, Corros. Eng., Sci. Technol., 2010, 45, 42–48 CrossRef CAS.
- E. Onoja, S. Chandren, F. I.-A. Razak and R. A. Wahab, J. Biotechnol., 2018, 283, 81–96 CrossRef CAS PubMed.
- M. Weng, C. Xia, S. Xu, Q. Liu, Y. Liu, H. Liu, C. Huo, R. Zhang, C. Zhang and Z. Miao, Colloid Polym. Sci., 2022, 300, 41–50 CrossRef CAS.
- B. Jia, C. Liu and X. Qi, Fuel Process. Technol., 2020, 210, 106578 CrossRef CAS.
- S. Zhang, W. Shang, X. Yang, S. Zhang, X. Zhang and J. Chen, Bull. Korean Chem. Soc., 2013, 34, 2741–2746 CrossRef CAS.
- E. Parandi, M. Safaripour, M. H. Abdellattif, M. Saidi, A. Bozorgian, H. Rashidi Nodeh and S. Rezania, Fuel, 2022, 313, 123057 CrossRef CAS.
- J. Zhao, M. Ma, X. Yan, G. Zhang, J. Xia, Z. Zeng, P. Yu, Q. Deng and D. Gong, Food Chem., 2022, 379, 132–148 Search PubMed.
- A. Ameri, F. Asadi, M. Shakibaie, A. Ameri, H. Forootanfar and M. Ranjbar, Appl. Biochem. Biotechnol., 2022, 194, 2108–2134 CrossRef CAS PubMed.
- A. K. Tamo, I. Doench, A. M. Helguera, D. Hoenders, A. Walther and A. O. Madrazo, Polymers, 2020, 12, 1–24 Search PubMed.
- A. Wassilkowska and T. Woźniakiewicz, Solid State Phenom., 2015, 231, 139–144 Search PubMed.
- E. Yilmaz, K. Can, M. Sezgin and M. Yilmaz, Bioresour. Technol., 2011, 102, 499–506 CrossRef CAS PubMed.
- I. Bustos-Jaimes, Y. García-Torres, H. C. Santillán-Uribe and C. Montiel, J. Mol. Catal. B: Enzym., 2013, 89, 137–141 CrossRef CAS.
- F. Kartal and A. Kilinc, Biotechnol. Prog., 2012, 28, 937–945 CrossRef CAS PubMed.
- I. Bustos-Jaimes, W. Hummel, T. Eggert, E. Bogo, M. Puls, A. Weckbecker and K. E. Jaeger, ChemCatChem, 2009, 1, 445–448 CrossRef CAS.
- Y. Gao, R. Zhong, J. Qin and B. Lin, Chem. Lett., 2009, 38, 262–263 CrossRef CAS.
- E. M. Hill, J. M. Broering, J. P. Hallett, A. S. Bommarius, C. L. Liotta and C. A. Eckert, Green Chem., 2007, 9, 888–893 RSC.
- D. Deng, Y. Zhang, A. Sun and Y. Hu, Chin. J. Catal., 2016, 37, 1966–1974 CrossRef CAS.
- F. Bellezza, A. Cipiciani, G. Cruciani and F. Fringuelli, J. Chem. Soc., Perkin Trans. 1, 2000, 4439–4444 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nj01160c |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2022 |


