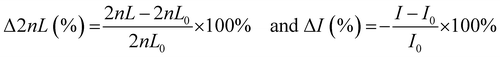Shining light in blind alleys: deciphering bacterial attachment in silicon microstructures†
Heidi
Leonard
 *a,
Xin
Jiang
*a,
Xin
Jiang
 a,
Sofia
Arshavsky-Graham
a,
Sofia
Arshavsky-Graham
 a,
Liran
Holtzman
a,
Liran
Holtzman
 a,
Yuri
Haimov
a,
Daniel
Weizman
a,
Sarel
Halachmi
a,
Yuri
Haimov
a,
Daniel
Weizman
a,
Sarel
Halachmi
 bc and
Ester
Segal
bc and
Ester
Segal
 *a
*a
aDepartment of Biotechnology and Food Engineering, Technion-Israel Institute of Technology, Haifa 3200003, Israel. E-mail: heidi.leonard1@gmail.com; esegal@technion.ac.il
bFaculty of Medicine, Technion-Israel Institute of Technology, Haifa, 3200003, Israel
cDepartment of Urology, Bnai Zion Medical Center, Haifa, 3104800, Israel
First published on 17th May 2022
Abstract
With new advances in infectious disease, antifouling surfaces, and environmental microbiology research comes the need to understand and control the accumulation and attachment of bacterial cells on a surface. Thus, we employ intrinsic phase-shift reflectometric interference spectroscopic measurements of silicon diffraction gratings to non-destructively observe the interactions between bacterial cells and abiotic, microstructured surfaces in a label-free and real-time manner. We conclude that the combination of specific material characteristics (i.e., substrate surface charge and topology) and characteristics of the bacterial cells (i.e., motility, cell charge, biofilm formation, and physiology) drive bacteria to adhere to a particular surface, often leading to a biofilm formation. Such knowledge can be exploited to predict antibiotic efficacy and biofilm formation, and enhance surface-based biosensor development, as well as the design of anti-biofouling strategies.
New conceptsThe quest for control over biofilm formation has revealed the importance of understanding and quantifying the interactions between bacteria and the surfaces they colonize. By using an easily engineered silicon substrate with periodic micro-topographies, we introduce a platform based on silicon diffraction gratings that enables real-time and label-free monitoring of bacterial surface adhesion and colonization. Such a platform does not require sophisticated microscopy tools or any labels as do most of the currently used methods, and it can be conveniently modified with different topographies and surface chemistries, presenting endless study opportunities in a single tool. We demonstrate the application of the platform to study how different bacterial species, ranging from standard laboratory strains to genetically engineered mutants and clinical isolates, preferentially colonize surfaces with varying topography, wettability and charge and monitor over the formation of a biofilm. Using the internal optical properties of the silicon diffraction gratings, we resolve not only bacterial adhesion, but also the process of bacterial infiltration into the studied microstructures. The platform devises a new toolbox to investigate how different material properties guide bacterial attachment, in real-time and in a non-destructive manner, which is imperative for mediating infections and biofouling. |
Introduction
Bacteria often thrive in surface-associated communities, in which the colonized surface provides both protection and a stable anchor for the development of a dense cellular network.1 These populations of bacterial cells that adhere to a surface and to each other by a variety of secreted substances and appendages are often classified as biofilms.2,3 The latter can manifest themselves as medical nuisances and threats, such as antibiotic-resistant urinary tract infections4 or as part of natural ecosystems, such as the geochemical recycling of sediment.5,6 It has been reported that surface-associated bacterial networks are more resistant to environmental stresses, such as antibiotics, than their free-floating planktonic counterpart cells.7 Thus, elucidating and quantifying the attachment behaviour of bacterial cells to their substrates and the development of biofilms is crucial to mediating problems of persistent infections,8 microbiome research,9 microbial ecology,10 biofouling,11 and even the design of synthetic microbial populations.12–15Although traditionally, the behaviour of microbes has been successfully studied in pure liquid cultures, new methods and materials for quantitatively examining bacterial attachment and colony formation on surfaces have recently emerged.16 Copious structurally hard and soft materials, specifically lamellar and nano-/micro-patterned materials, have been examined for their effect on bacterial colonization and adhesion;12,17–28 however, very few of these studies have harnessed the resulting intrinsic optical properties29 of the materials for real-time, quantitative evaluation of bacterial adhesion. Methods such as Raman spectroscopy,30,31 quartz crystal microbalances,32,33 and total internal reflectance fluorescence34,35 have been used to monitor bacterial adhesion and biofilm formation on surfaces, but employ substrates that can be difficult to chemically or structurally manipulate, while Raman spectroscopy only reveals the chemical signature of the bacteria,30 as opposed to a phenotypic quantification of adhered cells. Highly sensitive techniques, such as resonant mass sensors for cell characterization, require the precise positioning of a single cell on a cantilever tip,36 while supercritical-angle fluorescence requires immobilization of the specifically monitored cells.37 Furthermore, using traditional imaging techniques to elucidate bacterial behaviour38 can be difficult to quantify populations of cells, expensive, and destructive to the microbial networks.39,40
While many efforts focus on understanding mammalian cell interfaces with materials,41–44 in this work, we investigate bacterial cell responses to manipulated surfaces of silicon topologies by intrinsic phase-shift reflectometric interference spectroscopy measurements, termed PRISM.45,46 In this method, bacterial cells colonize on diffractive, patterned Si microstructures, while zero-order reflectance spectra are continuously collected during illumination with a broadband white light source.45–51 The resulting reflectance spectra exhibit optical interference fringes due to the reflection of the incident light at the two interfaces of the Si microstructures (top and bottom). As bacteria occupy the empty spaces within the microstructures, the refractive index of the filling medium increases, resulting in increased values of optical path difference. This corresponds to 2nL, in which n represents the refractive index of the filling medium and L refers to the height of the grating.52 Additionally, light scattering effects induce changes in the intensity of the reflected light.46 Thus, bacterial adhesion to chemically and structurally modified substrates can be monitored in real-time without the use of labels or sophisticated microscopes. This allows for the elucidation of long-disputed theories, such as the application of the Derjaguin–Landau–Verwey–Overbeek (DLVO) colloid stability theory to bacteria,53 and revealing tendencies of clinically relevant pathogenic bacteria. This work extensively studies the interactions of various bacterial species and strains with chemically manipulated abiotic surfaces to highlight the paramount role that a material surface has on the initial formation of complex microbial communities.
Results & discussion
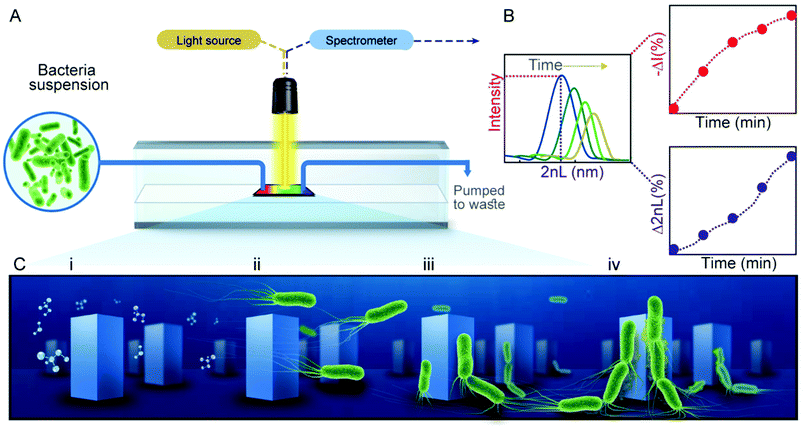
Concept of the assay – PRISM monitors bacteria adhesion
PRISM assays to monitor bacterial adhesion are performed in transparent flow cells, each housing a Si photonic chip comprised of arrayed microstructures (Fig. 1A), in the form of periodic pillars (Checkerboard format, ∼1.3 μm in width, 3 μm in depth and ∼2.9 μm between the pillars) or pores (∼3.2 μm in width, 4 μm in depth), see Fig. S1 (ESI†) for respective electron micrographs of the gratings. For PRISM adhesion assays, the photonic chips are illuminated at a normal incident angle by a collimated broadband light source through the flow cell, while being exposed to various solutions at a constant flow rate (see Fig. 1A). Using frequency analysis of the resulting reflectance spectra, a single peak is obtained, where its position and amplitude correspond to 2nL and the intensity of the reflected light, respectively (Fig. 1B). The percent of the change in both parameters is calculated over time as Δ2nL (%) and −ΔI (%), corresponding to cells infiltration and adhesion to the microstructures, respectively. Specifically, as more bacterial cells collect within the microstructured grating, the 2nL values rises as a function of the increasing refractive index45 and the intensity decreases (corresponding to a positive change in −ΔI (%) value) due to light scattering induced by the cells46 (Fig. 1B). Thus, both parameters contribute complementary information: the 2nL value is indicative of bacteria trapped within the microstructures and the intensity value reveals bacteria residing on top of the microstructures.By manipulating the topology and chemical functionality of the Si photonic chips, as well as examining the responses of a variety of bacteria species and strains, effects of different stages of bacteria attachment to the abiotic surfaces can be observed. Specifically, we focus on the attachment stages outlined in Fig. 1C, which include the preconditioning of the microstructures with an aqueous solution of motility buffer (MB) (Fig. 1C-i), followed by the introduction of cells to the topologies (Fig. 1C-ii), their subsequent attachment to the surface (Fig. 1C-iii), and the initial formation of a community as cells attach to each other (Fig. 1C-iv).
Wetting and fluid flow precondition the substrate for bacteria
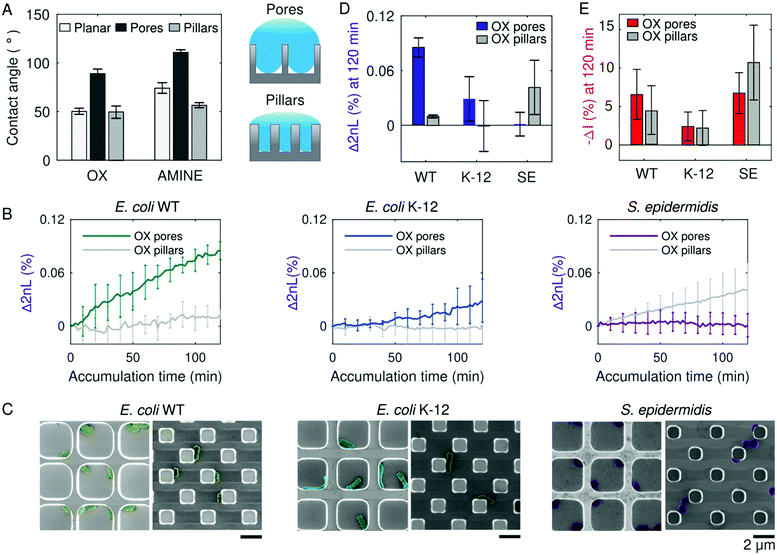
The molecular conditioning of a surface, which is the first step toward the formation of bacterial colonies, occurs before bacteria cells are present (Fig. 1C-i). During this stage, an aqueous solution of MB is introduced to the Si surface, resulting in its coating with ions from the surrounding medium upon sufficient wetting. The introduction of a liquid to a surface brings about two key players involved in bacteria cell attachment – the first being surface wettability,26,54 which is partially dependent on the structural topology of the surface;55 and the second being surface charge, which is dependent on the surface chemistry.56,57Consequently, Si photonic chips containing one of two topologies (i.e., pillars or pores) are utilized to observe the role of substrate wettability on bacteria colonization. The topologies are either kept as oxidized (termed as OX) surfaces, garnering a negative charge in neutral pH MB, or subjected to amino-silanization (termed as AMINE), garnering a more positively charged surface in neutral pH MB.58,59 Contact angle measurements conducted in MB on the OX Si topologies reveal that the pillars exhibit a similar contact angle value of 55° as the planar surface, whereas the pores topology deviates and displays a greater contact angle value of 99° (Fig. 2A). The AMINE-functionalized topologies also exhibit the same trend for the pores vs. planar (123° vs. 82°), while the pillars depict a reduced contact angle value of 62°. This suggests that the planar Si surfaces follow a Young wetting regime of a smooth surface, and the pore topologies follow the Cassie–Baxter model of wetting.60 The latter suggests that air is trapped underneath the buffer within the pores, resulting in an air–liquid interface of micro-pockets of vapor, as schematically illustrated in Fig. 2A.61 The pillar topologies on the other hand follow the Wenzel model, suggesting that the liquid can entirely infiltrate into a rough surface (i.e., micropillars).62,63 Moreover, for the same surface topography, the contact angle values demonstrate that the AMINE surfaces, silanized with APTES, are more hydrophobic in nature as compared to the respective OX substrates.64
During PRISM assays, Si chips are first exposed to a sterile MB without bacteria to acquire a stable baseline signal. Thus, due to their intrinsic wetting properties, pillar and pore topologies have already provided a different liquid microenvironment before bacteria cells were introduced. As preliminary tests, the PRISM responses of three non-pathogenic bacteria strains, namely the laboratory mutant Escherichia coli (E. coli) K-12, clinically isolated E. coli (ATCC 25922, termed as WT), and Staphylococcus epidermidis (S. epidermidis ATCC 14990), were observed over time, with Δ2nL (%) demonstrating a different rate and degree of accumulation of cells in the topologies (Fig. 2B). By utilizing MB, as opposed to a nutrient-rich growth medium, only events related to bacterial attachment are observed during the experiment, though cell motility is preserved.51,65
As a complimentary analysis, scanning electron microscopy (SEM) images of the Si chips after 120 min of bacteria accumulation during PRISM assays (Fig. 2C) reveal how the rod-shaped E. coli cells position themselves alongside the OX pillars in straight lines, while in OX pores they either adhere to the corners (as with E. coli WT) or reside in the middle of the pore (as with E. coli K-12). Contrarily, individual S. epidermidis cells position themselves within the corners of the OX pores, while in the OX pillar topologies, S. epidermidis cells tend to cohere to other cells. The microscopic menisci formed in the pore substrates underneath the MB may also explain why S. epidermidis cells tend to aggregate in the corners of the pores (Fig. 2C), as it has been previously observed that certain types of bacteria prefer to congregate at air–liquid interface66 or perhaps this is where they accumulate the most according to Brownian motion.24,67 It is intriguing that not only are differences in adhesion noticeable between bacterial species, but also amongst bacterial strains, demonstrating the complexity of the process of cell adhesion to a surface.68
Furthermore, Fig. 2D and E summarize changes in 2nL and corresponding intensity values, respectively, for E. coli WT, E. coli K-12, and S. epidermidis accumulated after 120 min on OX pore and pillar substrates. The 2nL changes indicate that both strains of E. coli (WT and K-12) exhibit a higher degree of accumulation within OX microstructured pores than within the OX pillar substrates, while S. epidermidis cells favor adhesion to OX pillar substrates over OX pore substrates. This preferred adhesion to specific microstructure is not apparent in the intensity changes, as similar values are obtained for pores and pillars for each strain. This suggests that the cells reside in a similar manner on the upper plane of the microstructures, while the degree of infiltration is different.69 Thus, the complementary signals indicate a preferred infiltration of the cells into the microstructures versus their total adhesion to the upper or bottom planes of the microstructures and provide superior information compared to microscopic imaging. For instance, while SEM images of S. epidermidis indicate the presence of the cells within the pores (Fig. 2C-right), it does not allow to conclude on the degree of infiltration. On the contrary, PRISM assay indicates that these cells do not infiltrate to the bottom of the pores but reside in their upper plane due to the small 2nL change and high intensity change. It should be also noted that the adhesion observed in the SEM micrographs may be affected by the bacteria fixation procedure,70 while PRISM provides genuine real-time information. These results confirm that even before the presence of bacteria, wetting of the pores and pillars provide different surrounding micro-environments that ultimately affect bacteria adhesion. It is also interesting to note that the E. coli WT exhibits more adhesion to oxidized pores than E. coli K-12, despite being of the same bacterial species. This was also observed on planar Si, imaged by SEM, see Fig. S2 (ESI†). Nevertheless, it should be emphasized that planar Si provides only limited information as it does not allow for real-time monitoring of bacterial adhesion. In the quest to uncover what other factors besides wetting affect bacterial adhesion, we proceed by investigating the role of cell motility and cellular phenotype in bacterial adhesion.71
Bacterial mechanics guide cells to the surface
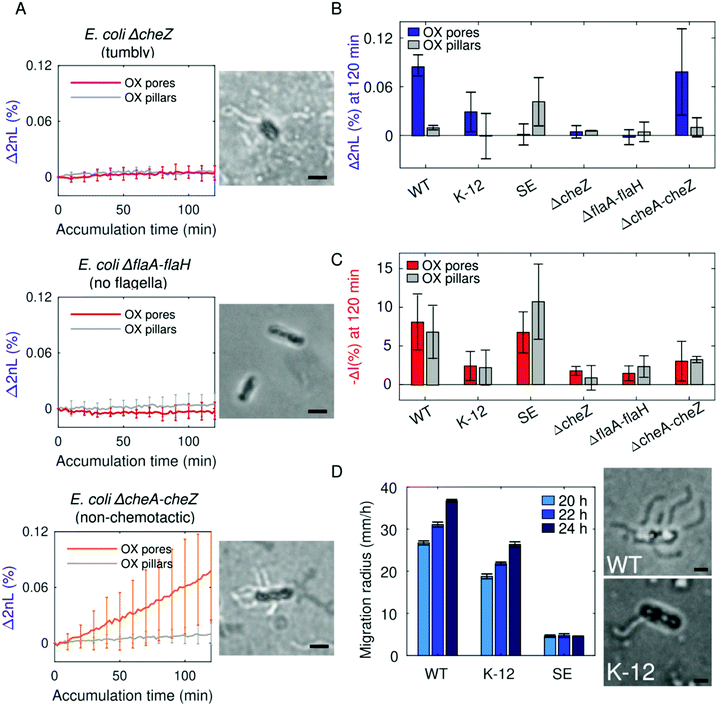
Noting the opposite adhesion preferences to pore and pillar substrates between species, the differences in the magnitude of attachment between species, and knowing that bacterial cells must reach a close enough distance to attach to a surface, we investigated the role of bacteria motility. More specifically, we determine whether cells reach the microtopologies as part of active navigation guided by cell mechanics (e.g., flagellar motors and chemotactic responses) or if the cells passively fall into the interstitial space of the microstructures. By employing PRISM, genetically modified E. coli mutants in the presence of OX substrates are monitored (Fig. 3). Specifically, non-motile E. coli without flagella (ΔflaA–flaH, HCB137), motile E. coli without functional chemotactic receptors (ΔcheA–cheZ, HCB437), and E. coli with an incessant tumbling motion (ΔcheZ, HCB1394) are used as model bacterial strains for these studies.The least adhesive bacteria, tumbling E. coli ΔcheZ, did not infiltrate or adhere to either pillar or pore substrates as demonstrated by PRISM (Fig. 3A), as they possibly do not have a long enough residence time to adhere to the surface.72 Minor infiltration was also observed for E. coli ΔflaA–flaH without flagella. Analysis of intensity changes for these strains (Fig. 3C) reveals a minor interaction of these cells with the different surfaces in total, as the low intensity changes indicate a minor adherence of the cells to the upper plane of the microstructures. These results suggest a crucial role of cell motility in the adhesion process of the cells to the surfaces as the resulted available surface contact area for bacterial binding differs.21 Contrarily, the non-chemotactic strain of E. coli behaves similar to the WT strain, confirming that the movement of bacteria into the microstructures during the PRISM assays is not directed by chemotaxis.
The motility of the E. coli WT, E. coli K-12, and S. epidermidis strains are measured in a semi-soft agar assay, demonstrating that E. coli WT swims faster than E. coli K-12, while S. epidermidis proves to be immotile (Fig. 3D). This suggests that infiltration and adherence into pore microstructures are achieved by active accumulation of motile cells, whereas E. coli WT exhibits a higher degree of accumulation, possibly due to the faster rate of swimming. Differences in swimming speeds between the two E. coli strains could possibly be explained by the varying number of flagella per cell as seen in optical microscope images (Fig. 3D) after Ryu flagellar staining.73,74 On the contrary, non-motile cells passively “fall” into the microstructures as the result of Brownian motion and a gravitational force.75 Though S. epidermidis and E. coli ΔflaA–flaH are both non-motile strains, S. epidermidis infiltrates and adheres more to OX pillar structures than E. coli ΔflaA–flaH does (see Fig. 2B, left panel). This may be explained by the significant difference in cell size between E. coli and S. epidermidis; we speculate that the larger rod-shaped E. coli are unable to easily fit within the pillars as the smaller and spherical Staphylococci. Additional substantial difference is observed in the intensity signal of these non-motile strains (Fig. 3C) indicating that motility itself cannot be solely accounted for cell adherence preferences.
Cell collisions with the microstructures are evidently crucial for attachment, though to an extent, the uncontrolled motility of bacteria may work against adhesion, as seen with E. coli ΔcheZ.76 In summary, while flagellar motors are not crucial for bacterial adhesion to the microstructured surface, they certainly assist in the process,77 particularly to pore topologies deficient in wetting and especially for larger sized cells that cannot easily fit within the confines of the pillars. These results also suggest that cells can passively diffuse between the pillar topology but must actively swim into the pores to attach.
Electrostatic forces promote cell adhesion at the surface
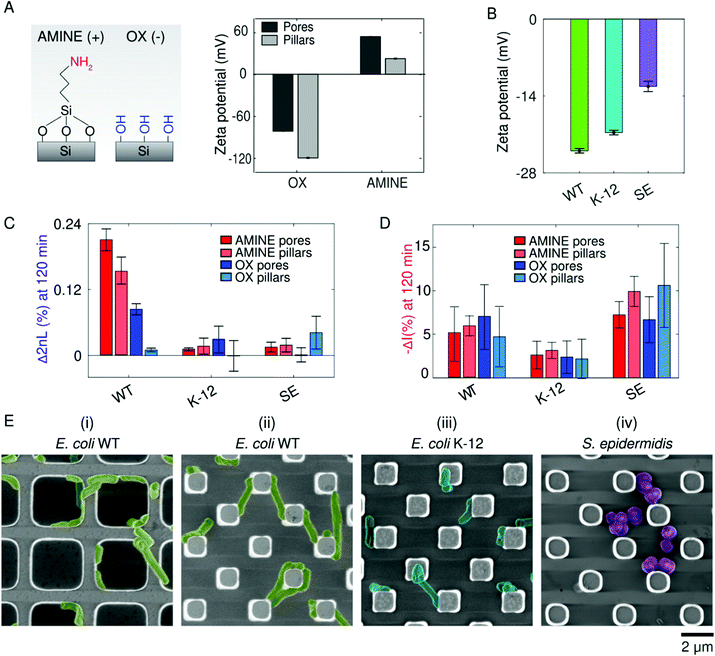
Once within the vicinity of the microstructures, bacterial adhesion is achieved by cellular appendages and secretions, but is also thought to be promoted by physiochemical forces.78 In particular, when bacteria reach a certain distance from the surface, they can then undergo reversible and irreversible attachment.75 Many studies try to draw parallels between the attraction of colloidal particles based on surface charge (described by DLVO theory) with bacteria cells adhering to abiotic surface.78 To test this hypothesis, we compare bacterial adhesion to positively and negatively charged substrates using PRISM (Fig. 4). Fig. 4A presents zeta potential measurements of the different OX and AMINE Si microstructures in pH-neutral MB, demonstrating that OX Si surfaces exhibit an overall negative charge due to the presence of hydroxyl groups, while AMINE surfaces exhibit an overall positive charge in pH-neutral MB.79 Furthermore, for the same surface chemistry, we observe differences in the charge between the pore and pillar structures,80 where OX pillar microstructures are the most negatively charged (−118 mV) and the AMINE pores are the most positive (53 mV). It is important to note that the differently charged surfaces also affect the contact angle values, as illustrated in Fig. 2A, but to a lower extent.81,82 The overall surface charge of each of the studied bacterial strains is also measured as a function of electrophoretic mobility by zeta potential measurements in MB (Fig. 4B).While generally all bacteria exhibit a negative cell surface charge,78,83 of the bacteria screened in our assays in accordance with the zeta potential measurements (Fig. 4B), E. coli WT exhibits the most negative surface charge, while S. epidermidis is the least negatively charged. These differences may arise due to differences in the peptidoglycan layer, the presence and structure of lipopolysaccharides, and/or the presence of appendages such as flagella (insinuating that charge is not homogenous throughout a cell).78,84 Differences in measured cell charges may also result from inhomogeneity within the cell population depending on the life cycle stage of the cells.85 Interestingly, even within the same species, E. coli WT was statistically more negatively charged than the K-12 strain. These differences in bacterial cell charge can be seen in the attained average Δ2nL (%) values after 120 min (Fig. 4C). The most prominent effect was realized for E. coli WT, which exhibited a ∼16 times and ∼3 times greater average Δ2nL (%) values on positively charged pillar and pore surfaces, respectively, than on the corresponding negatively charged surfaces. On the contrary, no preference was observed for the other strains, which are less motile and less negatively charged. This is supported by the intensity changes for the different strains, as depicted in Fig. 4D. It is interesting to note that for E. coli WT, a smaller intensity change and a larger 2nL change were observed in AMINE pores, compared to the other surfaces, suggesting that the cells prefer to infiltrate into the pores. Besides the difference of surface charge of the pore and pillar structures, the preference of the strains to the different structures is also dictated by the cell charge, motility, and size. Particularly, the lower preference of E. coli WT towards the pillars is attributed to the cell size, which can less conveniently reside between the pillars, compared to its infiltration into the larger pores. Yet, it should be noted that the amine modification of the pillars is sufficient to induce the E. coli WT infiltration and adhesion onto the pillar substrate, demonstrating the significant effect of the surface charge for this species.
Additional insight is obtained from complementary electron microscopy studies and Fig. 4E presents micrographs of the different bacterial species on AMINE surfaces. E. coli WT cells are observed to be elongated and stretch from pillar to pillar or wrap around the pore walls (Fig. 4E(ii and i), respectively), perhaps as a way to maximize surface coverage or possibly as the beginning of a filamentation stage that may occur during the initial stages of biofilm formation.86 Furthermore, optical microscopy of E. coli WT on amine-functionalized planar substrates (see Movie S1, ESI†) reveals cells spinning in circles while attached to the surface, suggesting that the AMINE surfaces promote tethering of cells to the surface and by particular locations of the cell body; specifically, the spinning of all the cells in a counterclockwise direction suggests tethering of the flagella to the surface72,87,88 and incidentally with the bacterial strain containing the most flagella per cell, namely E. coli WT (as observed in Fig. 3D).
The similar optical response on OX and AMINE surfaces found for the non-motile S. epidermidis strain, which is also the least negatively charged bacteria, may suggest that motility is a crucial factor for infiltration and adhesion, compared to surface charge. Yet, an effect of surface charge was apparent in the SEM micrographs, where the S. epidermidis cells were found to cluster on the AMINE surfaces and to be scattered on the OX surfaces (see Fig. 4E-right and Fig. 2C-right, respectively). The impact of motility over charge is also supported by the adhesion behaviour of the non-motile mutant E. coli ΔflaA–flaH, where no difference was observed in its adhesion to positively charged AMINE surface (see Fig. S3, ESI†).
Cell-to-cell communication promotes cohesion of bacterial communities
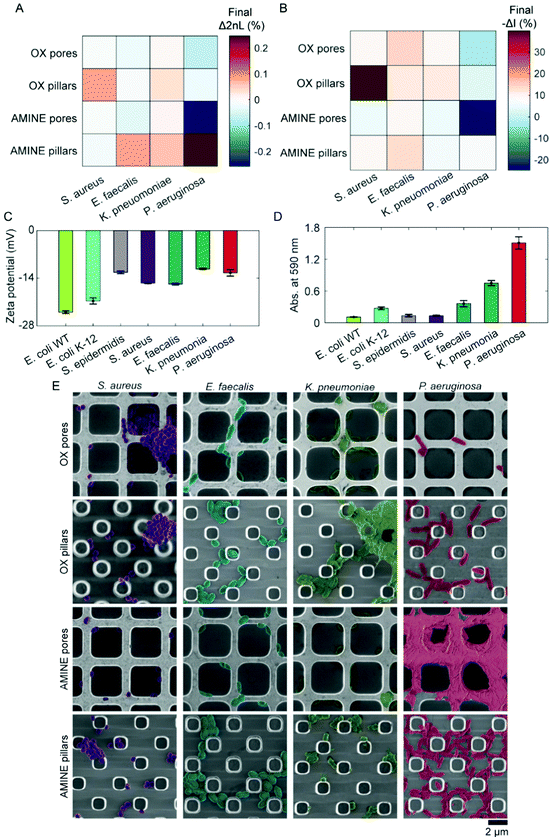
After bacterial cells colonize a surface, often pathogenic strains will proceed to form intertwined networks of communities in the process of biofilm formation.89,90 Thus, we lastly investigate the role of cell-to-cell adhesion in the accumulation of cells on a substrate. For these studies, a variety of pathogenic bacterial strains were employed, including S. aureus, Enterococcus faecalis (E. faecalis), Klebsiella pneumoniae (K. pneumoniae), and Pseudomonas aeruginosa (P. aeruginosa). While all these strains are clinical isolates that have been maintained in a lab environment, it is interesting to note that the P. aeruginosa was isolated from an intravenous coronary stent of a patient at a neighbouring medical center (Bnai Zion Medical Center, Israel). The attachment behaviours of these four clinically relevant isolates are depicted in Fig. 5A and B as a heat map in which more saturated colours represent more bacterial attachment after 180 min of accumulation in MB. However, unlike the non-pathogenic bacterial strains, in the testing of the clinical isolates, negative values of Δ2nL (%) and −ΔI (%) are surprisingly observed for some of the pathogenic bacteria, particularly in the case of P. aeruginosa to AMINE pores.To understand this behaviour of these bacterial strains in-depth, cell surface zeta potentials are measured (n = 3), see Fig. 5C. Unlike the E. coli strains, cell surface charge did not directly correlate with accumulation on oppositely charged surfaces as measured by PRISM, suggesting that surface heterogeneity on individual cells and also phenotypic heterogeneity within populations may play an even greater role in pathogenic bacteria.37,85,91,92 Using a standard crystal violet biofilm formation assay,93,94 we were able to measure the ability of bacterial strains to develop biofilms by the staining of secreted exopolysaccharides and cells adhered to the bottom of a 96-well plate after the removal of planktonic cells (Fig. 5D). As expected, the clinically isolated, highly pathogenic P. aeruginosa exhibited the highest biofilm forming abilities as seen in Fig. 5D, which corresponds well to the extreme PRISM values (Fig. 5A and B). Furthermore, SEM images of the substrates during the PRISM assay after 180 min of cell accumulation reveal interesting interactions of both cells with each other and cells with the substrate. For example, S. aureus formed clusters of cells, while E. faecalis arranged into chains of cells. K. pneumoniae appeared to develop multiple phenotypes with excreted substances, including some cells with numerous fimbriae. P. aeruginosa also acquired multiple phenotypes of cells, as well as the initiation of a notable biofilm formation, during which it appears as if the cells migrate from inside to the top of the pore walls. Such changes in phenotype are known to occur when cells contact a surface.1 Additionally, the formation of the biofilm on top of the AMINE pores, as opposed to inside the pores, suggests that the excessive presence of cells attached outside the pores can lead to negative values of Δ2nL (%) that are only observed with the biofilm-forming strains. The precise mathematical reasoning for this decrease in terms of physics and optical path differences remains unknown. In contrast, a negative −ΔI (%) value, which indicates an increase in the intensity of the reflected light, may be attributed to the secretion of extracellular matrix and proteins, causing Rayleigh scattering.95 Interestingly, from the observation of these bacteria, we were able to conclude that cells that work together in biofilm formation tend to form the largest, thriving communities (e.g., P. aeruginosa)96 on Si surfaces. Thus, in a relatively short time of 2 to 3 hours, PRISM can be used to investigate both initial bacterial adhesion preferences and biofilm formation, respectively. This is significantly faster than the current methods used, such as direct colony forming units counting, crystal violet staining, mass measurements, and flow cytometry of fully formed biofilms, and does not require labelling as does microscopy often does. Additionally, employing PRISM as a method for early detection of biofilm formation would also prove useful for studying the susceptibility of biofilms to antimicrobials/antibiotics.97
Addressing the statistical significance
The statistical significance of the presented results is investigated with a full factorial multiway analysis of variance (ANOVA) to evaluate the effects of surface topology, chemistry, and bacterial species on the mean values of Δ2nL (%) and −ΔI (%) acquired after 120 minutes, see Tables S1 and S2 (ESI†), respectively. An effect is considered significant with a P value <0.05. The analysis indicates that the 2nL changes are significantly influenced by each of the parameters investigated, where the bacteria (species or strain) had the most pronounced effect (P value <0.0001), followed by the surface topology. Moreover, whereas surface chemistry alone has less influence compared to the other parameters, it is found to be dependent on the bacteria as the interaction of both these parameters is found to have a substantial influence on the 2nL values. Thus, statistical analysis indicates that the species or strain of the bacteria is the most important parameter that will dictate cell adhesion to the surface, while the surface parameters investigated will have a secondary effect. The intensity changes are also most significantly dependent on the bacteria. Yet, the intensity is found to be a less sensitive parameter compared to the 2nL, supporting its use as a secondary and complementary analysis to the 2nL in this study. ANOVA analysis supports the complexity of the process of bacteria attachment on the surface and allows us to elucidate the contribution of the multiple parameters investigated in this study.Conclusions
The intricacies of bacterial colonization and biofilm formation have become a topic of high importance for microbiologists and materials engineers. However, finding methods and platforms to investigate these topics in a quantitative and label-free way remains challenging. Thus, we devised a unique platform for real-time monitoring of bacterial attachment in a controlled environment without the use of fluorescent labels or imaging. This platform is based on periodic arrays of microstructures, resulting in photonic diffractive gratings. With these materials, an extensive study of bacteria behaviour can be performed, enabling the differentiation of top-surface adhesion, infiltration, and bottom-surface adhesion.We found that substrate wetting, which is dependent on surface topology, can critically affect the potential of cells to reach a surface. Cell motility will guide cells to the surface if the wetting of the substrate permits. Furthermore, the charge of the surface can play a role in cell attachment once the cells have reached the surface. However, cell surface charge is not homogenous and may not be homogenous within populations, leading to a more complex cell–surface interfaces than opposite charges will attract. Lastly, the cooperation of cells working as a team to develop complex communities will have a direct effect on the accumulation of cells on a microstructured surface. While many studies have focused on anti-biofouling materials based on observations of non-pathogenic materials, these studies demonstrate that pathogenic bacteria can have extremely different responses. While colloidal theories can compare cells to particles, these theories overlook the complex networks that the cells develop as communities to thrive under various conditions.
While we focus on factors such as surface charge, substrate topology, and substrate wetting, there are still infinite other factors that can be studied with these materials and platform. Additional mechanical factors, such as the possession of bacterial fimbriae, and the role of the surrounding fluid (e.g., temperature, ionic strength, pH, and flow) are a few other factors that can be extensively monitored with this platform, while further application of machine learning algorithms will enable generalization of the procedure. However, we demonstrate that both the nature of the bacteria themselves and the characteristics of the environment around them, specifically the abiotic surface, is what results (or dissuades) bacteria from sticking and forming larger communities on the material. By simple modifications of materials, we have demonstrated that the maturation of cells and development of bacterial communities is the combined result of the bacteria cells’ nature and substrates’ nurturing environment.
In summary, the presented platform and the PRISM assay enable real-time quantification of bacteria–surface interactions in a relatively short time of up to 3 hours. It allows to elucidate the effect of various characteristics of the bacteria, surface, media, and flow on the bacteria attachment to a surface and predict biofilm formation. This is of crucial importance for the investigation and control of biofouling and bacterial infections in various medical and environmental fields.
Experimental
Reagents and solutions
Phenol, tannic acid, crystal violet, sodium lactate, L-methionine, ethylenediaminetetraacetic acid (EDTA), 3-(aminopropyl)triethoxysilane (APTES), glutaraldehyde, and tryptic soy broth (TSB) were purchased from Sigma-Aldrich, Israel. Acetone and methanol were supplied by Gadot, Israel. Agar was supplied by Difco. Aluminum potassium sulfate dodecahydrate was supplied by Scharlau, Spain. All motility buffer (MB) salts and absolute ethanol were supplied by Merck, Germany. Acetic acid was supplied by Bio-Lab Ltd, Israel. All aqueous solutions were prepared in Milli-Q water (18.2 MΩ cm). MB consisted of 6.2 mM K2HPO4, 3.8 mM KH2PO4, 67 mM NaCl, 0.1 mM EDTA, 1 μM L-methionine, and 10 mM sodium lactate.Fabrication of photonic chips
Si photonic chips containing either lamellar arrays of micropillars or pores were fabricated using standard photolithography and reactive ion etching techniques at the Wolfson Microelectronics Center (Micro–Nano-Fabrication and Printing Unit, Technion – Israel Institute of Technology). Developed whole wafers containing pores were thermally oxidized at 850 °C for 21 min in a tube diffusion furnace (BDF-4 RTRI-878, Bruce Technologies Inc, USA). Each developed wafer was then coated with photoresist to protect the microstructures and diced into 4 mm × 4 mm chips by an automated dicing saw (DAD3350; Disco, Japan). Chips were cleaned in acetone. Wafers containing micropillars were diced, followed by thermal oxidation in a tube furnace (Lindberg/Blue M 1200 °C Split-Hinge; Thermo Scientific, USA) at 800 °C for 1 h in ambient air. The resulting oxidized samples are termed “OX.” Amine-modified chips (termed “AMINE”) were generated by incubation of the oxidized samples in 2% v/v APTES in 50% methanol for 1 h, after which they were washed with ethanol and dried under nitrogen.Bacterial strains and growth conditions
Five different bacterial species were evaluated, including Escherichia coli (including the strains E. coli ATCC 25922, K-12, HCB437, HCB1394, and HCB137), Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis ATCC 14990, Klebsiella pneumoniae ATCC 700603, Enterococcus faecalis ATCC 29212, and Pseudomonas aeruginosa (clinical isolate collected by Bnai Zion Medical Center; Haifa, Israel). All ATCC bacterial strains were obtained from American Type Culture Collection, USA. HCB strains were kindly donated by Dr Karen Fahrner and Prof. Howard Berg (Harvard University; Cambridge, MA). E. coli K-12 was generously donated by Prof. Sima Yaron (Technion – Israel Institute of Technology). All cultures were maintained on tryptic soy agar (TSA) at 4 °C. Each bacterial strain was cultured individually in tryptic soy broth (TSB) overnight at 37 °C, followed by sub-culturing in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution until an optical density measured at a wavelength of 600 nm (OD600) value of ∼0.5. All HCB strains were cultured at 30 °C.
100 dilution until an optical density measured at a wavelength of 600 nm (OD600) value of ∼0.5. All HCB strains were cultured at 30 °C.
Adhesion assays using PRISM
Bacterial cultures were centrifuged at 2575 g for 5 min at room temperature. The resulting pellet was resuspended in filtered MB and adjusted to a concentration of 107 cells mL−1 as determined by calculated OD600 curves. For each adhesion trial, one PRISM chip was fixed in a polycarbonate flow cell at room temperature. A broadband light source (LS1, Ocean Optics) connected to a bifurcated fiber fitter with a collimated lens was focused normal to the surface of the chip. Reflectance spectra were collected by a USB4000 spectrometer (Ocean Optics) every 2 min. Bacterial suspensions were introduced into the flow chamber at a speed of 20 μL min−1. PRISM was expressed as Δ2nL (%) or intensity changes [−ΔI (%)] and calculated as follows:where 2nL0 and I0 refer to the values of 2nL and intensity at t = 0 (of the bacteria accumulation time), respectively. For each assay, 70% ethanol was briefly injected into the flow cell, followed by continuous flow of MB until stable values of 2nL were achieved, after which the solution was changed to the bacterial suspension. Please note that bacteria adhesion results mostly in intensity decrease due to light scattering effects. However, for convenience, intensity changes are presented as negative values to correlate a positive signal to bacterial adhesion.
Assays of pathogenic bacteria
Due to the pathogenicity of the strains and desire to observe faster differences in adhesion preferences, assays of pathogenic bacteria, namely E. faecalis, S. aureus, P. aeruginosa, and K. pneumoniae, were modified. In these assays, bacteria were plated on TSA overnight. Colonies were selected from freshly incubated agar plates and suspended in MB. The suspension was diluted until achieving and OD600 of 0.1.Crystal violet biofilm formation assay
Aliquots of 10 μL of overnight bacterial cultures in TSB were seeded in a 96-well polystyrene plate. Cells were overlaid with 190 μL of TSB and incubated for 24 h at room temperature. The plate was submerged in ddH2O for 5 min to remove planktonic cells, followed by inversion of the plate to remove remaining water drops. Wells were incubated for 10 min with 0.1% crystal violet solution, after which the plate was submerged in ddH2O for 5 min to remove excess crystal violet. The plate was again inverted to remove large water droplets containing crystal violet. After drying for 20 min, 200 μL of 96% ethanol was added to each well and incubated for 5 min to solubilized crystal violet contained within the biofilms. 180 μL of the solubilized crystal violet was transferred to a new 96-well plate, where absorbance at 590 nm was measured by a microplate reader (Synergy HT, BioTek Instruments). The average absorbance value of wells inoculated with only growth medium was subtracted from the absorbance values of bacteria-containing wells to calibrate for any excess crystal violet adhered to the well plate.Motility assay in semi-solid agar
Semi-solid agar was prepared by adding 0.25% agar to TSB. Following autoclaving, ∼20 mL of agar was poured into each plate. Agar plates remained at room temperature overnight. A bacterial suspension of 10 μL was spotted in the middle of each plate and incubated at room temperature to mimic conditions of the optical experiments. Agar plates were imaged, and the diameter of each radial growth area was measured using ImageJ software.Contact angle measurements
Contact angles of 5 μL drops of MB on various substrates were measured with an Attension Theta Lite optical tensiometer (Biolin Scientific, Sweden). A CCD camera acquired digital images of the droplet on each substrate over time. Contact angles were calculated by OneAttension Software using the Young–Laplace equation. Single measurements were performed on three different chips of the same type as the droplet covered the majority of the substrate surface.Zeta potential measurements
Zeta potential measurements of different bacterial strains were performed using a ZetaSizer ZSP (Malvern Instruments, UK) in triplicate samples. Bacterial suspensions taken from freshly incubated plates were prepared to an OD600 0.1 in MB (pH = 6.9). Zeta potential measurements of oxidized or amine-modified Si microstructures were conducted using SurPass electrokinetic analyzer (Anton Paar GmbH, Graz, Austria), in a clamping cell. An asymmetric sample mounting strategy was applied, using polypropylene film as a reference, placed at a distance of 100 μm from the measured sample in the clamping cell. pH titration was performed automatically by the instrument from pH 6 to 9 using a 1 mM NaCl electrolyte solution with a target ramp pressure of 450 mbar. The pH was controlled with 0.1 M HCl or 0.05 M NaOH solutions. Two cycles of pressure ramping were conducted in each direction and the zeta potential were computed from the streaming current based on all four cycles. The calculation was performed using the Helmholtz–Smoluchowski equation:98| ζ = (dI/dp) × (η/ε × ε0) × (L/A) |
| ζsurface = 2ζmeasured − ζreference |
Imaging
All PRISM chips were fixed and stored at 4 °C in 2.5% glutaraldehyde. For scanning electron microscopy (SEM), samples were washed in subsequent 50–70–100% ethanolic solutions, each for 5 min and then dried under a stream of nitrogen. Samples were analysed at 1 keV using a high-resolution SEM (Ultra Zeiss Plus). To visualize flagella, a Ryu flagellar stain was performed. Briefly, bacterial cells were grown overnight on tryptic soy agar. Drops of motile cells were prepared in sterile water by touching a loopful of water to the colony margin on the agar and then suspending the bacteria by touching a drop of water on a glass slide, which was covered with a cover slip and examined for motile cells. Then, two drops of Ryu stain were applied to the edge of the cover slip and allowed to diffuse by capillary action. The Ryu stain is composed of a filtered 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mixture of two solutions, in which the first solution contains 5% phenol and 20% tannic acid in a saturated aqueous solution of aluminium potassium sulphate dodecahydrate and the second solution contains 12% crystal violet in 95% ethanolic solution. Bacterial cells were examined for flagella after 10 min using a Zeiss Axio Scope A1 microscope. Images were obtained by an Axio Cam MRc (Zeiss) camera.
10 mixture of two solutions, in which the first solution contains 5% phenol and 20% tannic acid in a saturated aqueous solution of aluminium potassium sulphate dodecahydrate and the second solution contains 12% crystal violet in 95% ethanolic solution. Bacterial cells were examined for flagella after 10 min using a Zeiss Axio Scope A1 microscope. Images were obtained by an Axio Cam MRc (Zeiss) camera.
Statistical analysis
Full factorial multiway analysis of variance (ANOVA) of the data was performed with JMP® software (Pro15, SAS Institute) with a significant level of p < 0.05.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Funding sources
This research was supported by the ISRAEL SCIENCE FOUNDATION (grant no. 2458/21).Conflicts of interest
There are no conflicts to declare.Acknowledgements
We dedicate this work to the late Prof. Howard Berg, who termed our silicon pores “blind alleys.” We thank him and Dr Karen Fahrner of Harvard University for donating E. coli mutant strains and for their useful discussions and insight. We also thank Dima Peselev, Orna Ternyak, and the rest of the staff at the Micro Nano-Fabrication and Printing Unit (MNFPU) at the Technion – Israel Institute of Technology for their efforts in the design and fabrication of the diffraction gratings. We grateful to Prof. Yechezkel Kashi for hosting our experiments in his lab and for fruitful discussions. We thank Prof. Boaz Pokroy and Beni Rich for guidance with contact angle measurements.References
- H. H. Tuson and D. B. Weibel, Soft Matter, 2013, 9, 4368–4380 RSC.
- P. S. Stewart and J. William Costerton, Lancet, 2001, 358, 135–138 CrossRef CAS.
- A. Penesyan, I. T. Paulsen, S. Kjelleberg and M. R. Gillings, npj Biofilms Microbiomes, 2021, 7, 80 CrossRef CAS PubMed.
- L. Hall-Stoodley and P. Stoodley, Cell. Microbiol., 2009, 11, 1034–1043 CrossRef CAS PubMed.
- M. E. Davey and G. A. O'toole, Microbiol. Mol. Biol. Rev., 2000, 64, 847–867 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- C. Liao, X. Liang, M. L. Soupir and L. R. Jarboe, J. Appl. Microbiol., 2015, 119, 315–330 CrossRef CAS PubMed.
- H. Leonard, R. Colodner, S. Halachmi and E. Segal, ACS Sens., 2018, 3, 2202–2217 CrossRef CAS PubMed.
- J. S. Biteen, P. C. Blainey, Z. G. Cardon, M. Chun, G. M. Church, P. C. Dorrestein, S. E. Fraser, J. A. Gilbert, J. K. Jansson, R. Knight, J. F. Miller, A. Ozcan, K. A. Prather, S. R. Quake, E. G. Ruby, P. A. Silver, S. Taha, G. van den Engh, P. S. Weiss, G. C.-L. Wong, A. T. Wright and T. D. Young, ACS Nano, 2016, 10, 6–37 CrossRef CAS PubMed.
- M. Ding, H.-Y. Shiu, S.-L. Li, C. K. Lee, G. Wang, H. Wu, N. O. Weiss, T. D. Young, P. S. Weiss, G. C.-L. Wong, K. H. Nealson, Y. Huang and X. Duan, ACS Nano, 2016, 10, 9919–9926 CrossRef CAS PubMed.
- A. Muras, A. Parga, C. Mayer and A. Otero, Mar. Drugs, 2021, 19, 74 CrossRef CAS PubMed.
- C. Berne, C. K. Ellison, A. Ducret and Y. V. Brun, Nat. Rev. Microbiol., 2018, 16, 616–627 CrossRef CAS PubMed.
- O. E. Petrova and K. Sauer, J. Bacteriol., 2012, 194, 2413–2425 CrossRef CAS PubMed.
- M. Schaffner, P. A. Rühs, F. Coulter, S. Kilcher and A. R. Studart, Sci. Adv., 2017, 3, eaao6804 CrossRef PubMed.
- E. Sgobba and V. F. Wendisch, Curr. Opin. Biotechnol, 2020, 62, 72–79 CrossRef CAS PubMed.
- S. Achinas, N. Charalampogiannis and G. J.-W. Euverink, Appl. Sci., 2019, 9, 2801 CrossRef CAS.
- X.-Q. Dou, D. Zhang, C. Feng and L. Jiang, ACS Nano, 2015, 9, 10664–10672 CrossRef CAS PubMed.
- S. M. Imani, R. Maclachlan, K. Rachwalski, Y. Chan, B. Lee, M. McInnes, K. Grandfield, E. D. Brown, T. F. Didar and L. Soleymani, ACS Nano, 2020, 14, 454–465 CrossRef CAS PubMed.
- N. Encinas, C.-Y. Yang, F. Geyer, A. Kaltbeitzel, P. Baumli, J. Reinholz, V. Mailänder, H.-J. Butt and D. Vollmer, ACS Appl. Mater. Interfaces, 2020, 12, 21192–21200 CrossRef CAS PubMed.
- S. W. Lee, K. S. Phillips, H. Gu, M. Kazemzadeh-Narbat and D. Ren, Biomaterials, 2021, 268, 120595 CrossRef CAS PubMed.
- R. S. Friedlander, H. Vlamakis, P. Kim, M. Khan, R. Kolter and J. Aizenberg, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5624–5629 CrossRef CAS PubMed.
- A. I. Hochbaum and J. Aizenberg, Nano Lett., 2010, 10, 3717–3721 CrossRef CAS PubMed.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Appl. Environ. Microbiol., 2013, 79, 2703–2712 CrossRef CAS PubMed.
- K. W. Kolewe, S. Kalasin, M. Shave, J. D. Schiffman and M. M. Santore, ACS Appl. Mater. Interfaces, 2019, 11, 320–330 CrossRef CAS PubMed.
- G. Feng, Y. Cheng, S.-Y. Wang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, npj Biofilms Microbiomes, 2015, 1, 15022 CrossRef CAS PubMed.
- G. B. Hwang, K. Page, A. Patir, S. P. Nair, E. Allan and I. P. Parkin, ACS Nano, 2018, 12, 6050–6058 CrossRef CAS PubMed.
- Y. Cao, B. Su, S. Chinnaraj, S. Jana, L. Bowen, S. Charlton, P. Duan, N. S. Jakubovics and J. Chen, Sci. Rep., 2018, 8, 1071 CrossRef PubMed.
- W. Wang, Y. Lu, H. Zhu and Z. Cao, Adv. Mater., 2017, 29, 1606506 CrossRef PubMed.
- N. Massad-Ivanir, G. Shtenberg, T. Zeidman and E. Segal, Adv. Funct. Mater., 2010, 20, 2269–2277 CrossRef CAS.
- G. Bodelón, V. Montes-García, V. López-Puente, E. H. Hill, C. Hamon, M. N. Sanz-Ortiz, S. Rodal-Cedeira, C. Costas, S. Celiksoy, I. Pérez-Juste, L. Scarabelli, A. La Porta, J. Pérez-Juste, I. Pastoriza-Santos and L. M. Liz-Marzán, Nat. Mater., 2016, 15, 1203–1211 CrossRef PubMed.
- C. C. Liao, H. K. Huang, Y. Z. Chen and N. T. Huang, Lab Chip, 2020, 20, 2520–2528 RSC.
- I. M. Marcus, M. Herzberg, S. L. Walker and V. Freger, Langmuir, 2012, 28, 6396–6402 CrossRef CAS PubMed.
- Y. Shan, L. Liu, Y. Liu, H. Harms and L. Y. Wick, Environ. Sci. Technol., 2020, 54, 14036–14045 CrossRef CAS PubMed.
- K. K.-W. Wong, A. L.-J. Olsson, B. Asadishad, B. van der Bruggen and N. Tufenkji, Langmuir, 2017, 33, 4066–4075 CrossRef CAS PubMed.
- M. A.-S. Vigeant, R. M. Ford, M. Wagner and L. K. Tamm, Appl. Environ. Microbiol., 2002, 68, 2794–2801 CrossRef CAS PubMed.
- N. Cermak, S. Olcum, F. F. Delgado, S. C. Wasserman, K. R. Payer, M. A. Murakami, S. M. Knudsen, R. J. Kimmerling, M. M. Stevens, Y. Kikuchi, A. Sandikci, M. Ogawa, V. Agache, F. Baléras, D. M. Weinstock and S. R. Manalis, Nat. Biotechnol., 2016, 34, 1052–1059 CrossRef CAS PubMed.
- B. Ferdman, L. E. Weiss, O. Alalouf, Y. Haimovich and Y. Shechtman, ACS Nano, 2018, 12, 11892–11898 CrossRef CAS PubMed.
- T. Vissers, A. T. Brown, N. Koumakis, A. Dawson, M. Hermes, J. Schwarz-Linek, A. B. Schofield, J. M. French, V. Koutsos, J. Arlt, V. A. Martinez and W. C.-K. Poon, Sci. Adv., 2018, 4, eaao1170 CrossRef PubMed.
- T. R. Garrett, M. Bhakoo and Z. Zhang, Prog. Nat. Sci., 2008, 18, 1049–1056 CrossRef CAS.
- J. de Anda, E. Y. Lee, C. K. Lee, R. R. Bennett, X. Ji, S. Soltani, M. C. Harrison, A. E. Baker, Y. Luo, T. Chou, G. A. O’Toole, A. M. Armani, R. Golestanian and G. C.-L. Wong, ACS Nano, 2017, 11, 9340–9351 CrossRef CAS PubMed.
- J. Deng, C. Zhao, J. P. Spatz and Q. Wei, ACS Nano, 2017, 11, 8282–8291 CrossRef CAS PubMed.
- J. Fang, Y.-Y. Hsueh, J. Soto, W. Sun, J. Wang, Z. Gu, A. Khademhosseini and S. Li, ACS Nano, 2020, 14, 1296–1318 CrossRef CAS PubMed.
- F. J. Harding, S. Surdo, B. Delalat, C. Cozzi, R. Elnathan, S. Gronthos, N. H. Voelcker and G. Barillaro, ACS Appl. Mater. Interfaces, 2016, 8, 29197–29202 CrossRef CAS PubMed.
- Y. Chen, J. Wang, X. Li, N. Hu, N. H. Voelcker, X. Xie and R. Elnathan, Adv. Mater., 2020, 32, 2001668 CrossRef CAS PubMed.
- H. Leonard, S. Halachmi, N. Ben-Dov, O. Nativ and E. Segal, ACS Nano, 2017, 11, 6167–6177 CrossRef CAS PubMed.
- C. Heuer, H. Leonard, N. Nitzan, A. Lavy-Alperovitch, N. Massad-Ivanir, T. Scheper and E. Segal, ACS Infect. Dis., 2020, 6, 2560–2566 CrossRef CAS PubMed.
- H. Leonard, C. Heuer, D. Weizman, N. Massad-Ivanir, S. Halachmi, R. Colodner and E. Segal, Rapid diagnostic susceptibility testing of bacteria and fungi from clinical samples using silicon gratings, SPIE, 2019.
- Y. Mirsky, A. Nahor, E. Edrei, N. Massad-Ivanir, L. M. Bonanno, E. Segal and A. Sa'ar, Appl. Phys. Lett., 2013, 103, 033702 CrossRef.
- N. Massad-Ivanir, Y. Mirsky, A. Nahor, E. Edrei, L. M. Bonanno-Young, N. Ben Dov, A. Sa'ar and E. Segal, Analyst, 2014, 139, 3885–3894 RSC.
- C. Ude, N. Ben-Dov, A. Jochums, Z. Li, E. Segal, T. Scheper and S. Beutel, Appl. Microbiol. Biotechnol., 2016, 100, 4147–4159 CrossRef CAS PubMed.
- T. Davidov, N. Granik, S. Zahran, H. Leonard, I. Adir, O. Elul, T. Fried, A. Gil, B. Mayo, S. Ohayon, S. Sarig, N. Shasha, S. Tsedef, S. Weiner, M. Brunwasser-Meirom, A. Ereskovsky, N. Katz, B. Kaufmann, Y. Haimov, E. Segal and R. Amit, ACS Biomater. Sci. Eng., 2019, 5, 603–612 CrossRef CAS PubMed.
- S. Arshavsky-Graham, N. Massad-Ivanir, E. Segal and S. Weiss, Anal. Chem., 2019, 91, 441–467 CrossRef CAS PubMed.
- M. Hermansson, Colloids Surf., B, 1999, 14, 105–119 CrossRef CAS.
- S. Al-Amshawee, M. Y.-B. M. Yunus, J. G. Lynam, W. H. Lee, F. Dai and I. H. Dakhil, Environ. Technol. Innovation, 2021, 21, 101233 CrossRef.
- S. Utech, K. Bley, J. Aizenberg and N. Vogel, J. Mater. Chem. A, 2016, 4, 6853–6859 RSC.
- O. Rzhepishevska, S. Hakobyan, R. Ruhal, J. Gautrot, D. Barbero and M. Ramstedt, Biomater. Sci., 2013, 1, 589–602 RSC.
- M. N. Chandraprabha and K. A. Natarajan, Miner. Process. Extr. Metall. Rev., 2009, 31, 1–29 CrossRef.
- C. Graf, Q. Gao, I. Schütz, C. N. Noufele, W. Ruan, U. Posselt, E. Korotianskiy, D. Nordmeyer, F. Rancan, S. Hadam, A. Vogt, J. Lademann, V. Haucke and E. Rühl, Langmuir, 2012, 28, 7598–7613 CrossRef CAS PubMed.
- M. Kaasalainen, E. Mäkilä, J. Riikonen, M. Kovalainen, K. Järvinen, K.-H. Herzig, V.-P. Lehto and J. Salonen, Int. J. Pharm., 2012, 431, 230–236 CrossRef CAS PubMed.
- A. Nogalska, A. Trojanowska, B. Tylkowski and R. Garcia-Valls, Phys. Sci. Rev., 2020, 5, 20190083 Search PubMed.
- P. Formentín and L. F. Marsal, Nanomaterials, 2021, 11, 670 CrossRef PubMed.
- G. Whyman, E. Bormashenko and T. Stein, Chem. Phys. Lett., 2008, 450, 355–359 CrossRef CAS.
- B.-X. Zhang, S.-L. Wang and X.-D. Wang, Langmuir, 2019, 35, 662–670 CrossRef CAS PubMed.
- N. S.-K. Gunda, M. Singh, L. Norman, K. Kaur and S. K. Mitra, Appl. Surf. Sci., 2014, 305, 522–530 CrossRef CAS.
- P. P. Lele, A. Shrivastava, T. Roland and H. C. Berg, Sci. Adv., 2015, 1, e1500299 CrossRef PubMed.
- M. Fletcher and D. C. Savage, Bacterial adhesion: mechanisms and physiological significance, Springer Science & Business Media, 2013 Search PubMed.
- G. Li, L.-K. Tam and J. X. Tang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18355–18359 CrossRef CAS PubMed.
- A. van Belkum, C. Almeida, B. Bardiaux, S. V. Barrass, S. J. Butcher, T. Çaykara, S. Chowdhury, R. Datar, I. Eastwood, A. Goldman, M. Goyal, L. Happonen, N. Izadi-Pruneyre, T. Jacobsen, P. H. Johnson, V. A.-J. Kempf, A. Kiessling, J. L. Bueno, A. Malik, J. Malmström, I. Meuskens, P. A. Milner, M. Nilges, N. Pamme, S. A. Peyman, L. R. Rodrigues, P. Rodriguez-Mateos, M. G. Sande, C. J. Silva, A. C. Stasiak, T. Stehle, A. Thibau, D. J. Vaca and D. Linke, Diagnostics, 2021, 11, 1259 CrossRef CAS PubMed.
- K. Yang, J. Shi, L. Wang, Y. Chen, C. Liang, L. Yang and L.-N. Wang, J. Mater. Sci. Technol., 2022, 99, 82–100 CrossRef.
- M. Bossù, L. Selan, M. Artini, M. Relucenti, G. Familiari, R. Papa, G. Vrenna, P. Spigaglia, F. Barbanti, A. Salucci, G. Di Giorgio, J. V. Rau and A. Polimeni, Microorganisms, 2020, 8, 807 CrossRef PubMed.
- N. Lu, W. Zhang, Y. Weng, X. Chen, Y. Cheng and P. Zhou, Food Control, 2016, 68, 344–351 CrossRef CAS.
- M. D. Manson, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11151–11152 CrossRef CAS PubMed.
- H. Kodaka, A. Y. Armfield, G. L. Lombard and V. R. Dowell, J. Clin. Microbiol., 1982, 16, 948–952 CrossRef CAS PubMed.
- M. E. Heimbrook, W. L. Wang and G. Campbell, J. Clin. Microbiol., 1989, 27, 2612–2615 CrossRef CAS PubMed.
- Y. H. An and R. J. Friedman, J. Biomed. Mater. Res., 1998, 43, 338–348 CrossRef CAS PubMed.
- M. Qi, X. Gong, B. Wu and G. Zhang, Langmuir, 2017, 33, 3525–3533 CrossRef CAS PubMed.
- C. K. Lee, A. J. Kim, G. S. Santos, P. Y. Lai, S. Y. Lee, D. F. Qiao, J. D. Anda, T. D. Young, Y. Chen, A. R. Rowe, K. H. Nealson, P. S. Weiss and G. C.-L. Wong, ACS Nano, 2016, 10, 9183–9192 CrossRef CAS PubMed.
- A. T. Poortinga, R. Bos, W. Norde and H. J. Busscher, Surf. Sci. Rep., 2002, 47, 1–32 CrossRef CAS.
- D. V. Okhrimenko, A. Budi, M. Ceccato, M. Cárdenas, D. B. Johansson, D. Lybye, K. Bechgaard, M. P. Andersson and S. L.-S. Stipp, ACS Appl. Mater. Interfaces, 2017, 9, 8344–8353 CrossRef CAS PubMed.
- A. Drechsler, A. Caspari and A. Synytska, Surf. Interface Anal., 2020, 52, 991–995 CrossRef CAS.
- L. De Stefano, G. Oliviero, J. Amato, N. Borbone, G. Piccialli, L. Mayol, I. Rendina, M. Terracciano and I. Rea, J. R. Soc., Interface, 2013, 10, 20130160 CrossRef PubMed.
- M. Oggianu, C. Figus, S. Ashoka-Sahadevan, N. Monni, D. Marongiu, M. Saba, A. Mura, G. Bongiovanni, C. Caltagirone, V. Lippolis, C. Cannas, E. Cadoni, M. L. Mercuri and F. Quochi, RSC Adv., 2021, 11, 15557–15564 RSC.
- S. Zheng, M. Bawazir, A. Dhall, H.-E. Kim, L. He, J. Heo and G. Hwang, Front. Bioeng. Biotechnol., 2021, 9, 643722 CrossRef PubMed.
- Y.-L. Ong, A. Razatos, G. Georgiou and M. M. Sharma, Langmuir, 1999, 15, 2719–2725 CrossRef CAS.
- H. C. van der Mei and H. J. Busscher, PLoS Pathog., 2012, 8, e1002821 CrossRef CAS PubMed.
- J. Möller, P. Emge, I. A. Vizcarra, P. Kollmannsberger and V. Vogel, New J. Phys., 2013, 15, 125016 CrossRef.
- M. Silverman and M. Simon, Nature, 1974, 249, 73–74 CrossRef CAS PubMed.
- H. C. Berg, Nature, 1974, 249, 77–79 CrossRef CAS PubMed.
- L. Hall-Stoodley and P. Stoodley, Trends Microbiol., 2005, 13, 7–10 CrossRef CAS PubMed.
- W. Lei, J. Bruchmann, J. L. Rüping, P. A. Levkin and T. Schwartz, Adv. Sci., 2019, 6, 1900519 CrossRef PubMed.
- R. Avraham, N. Haseley, D. Brown, C. Penaranda, H. B. Jijon, J. J. Trombetta, R. Satija, A. K. Shalek, R. J. Xavier, A. Regev and D. T. Hung, Cell, 2015, 162, 1309–1321 CrossRef CAS PubMed.
- P. S. Stewart and M. J. Franklin, Nat. Rev. Microbiol., 2008, 6, 199–210 CrossRef CAS PubMed.
- G. A.-A. O’Toole, JoVE, 2011, 2437 Search PubMed.
- J. H. Merritt, D. E. Kadouri and G. A. O'Toole, Curr. Protoc. Microbiol., 2011, 22, 1B.1.1–1B.1.18 Search PubMed.
- C. Pacholski, C. Yu, G. M. Miskelly, D. Godin and M. J. Sailor, J. Am. Chem. Soc., 2006, 128, 4250–4252 CrossRef CAS PubMed.
- C. K. Lee, J. Vachier, J.-d Anda, K. Zhao, A. E. Baker, R. R. Bennett, C. R. Armbruster, K. A. Lewis, R. L. Tarnopol, C. J. Lomba, D. A. Hogan, M. R. Parsek, G. A. O’Toole, R. Golestanian, G. C.-L. Wong and F. M. Ausubel, mBio, 2020, 11, e02644-02619 Search PubMed.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS PubMed.
- M. Elimelech, W. H. Chen and J. J. Waypa, Desalination, 1994, 95, 269–286 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Plan view SEM images of bare Si microtopologies and in-depth statistical analysis (PDF) and a movie of E. coli WT on an amine-functionalized surface (AVI). See DOI: https://doi.org/10.1039/d2nh00130f |
| This journal is © The Royal Society of Chemistry 2022 |