Intelligent and robust DNA robots capable of swarming into leakless nonlinear amplification in response to a trigger†
Shaofei
Li
abc,
Yizhuang
Cheng
ac,
Miao
Qin
ac,
Guoliang
Zhou
ac,
Pan
Li
 a and
Liangbao
Yang
a and
Liangbao
Yang
 *ad
*ad
aInstitute of Health and Medical Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei 230031, Anhui, China. E-mail: lbyang@iim.ac.cn
bSchool of Life Science, Anhui University, Hefei 230601, Anhui, China
cUniversity of Science & Technology of China, Hefei 230026, Anhui, China
dHefei Cancer Hospital, Chinese Academy of Sciences, Hefei 230031, Anhui, China
First published on 22nd April 2022
Abstract
Nonlinear DNA signal amplification with an enzyme-free isothermal self-assembly process is uniquely useful in nanotechnology and nanomedicine. However, progress in this direction is hampered by the lack of effective design models of leak-resistant DNA building blocks. Here, we propose two conceptual models of intelligent and robust DNA robots to perform a leakless nonlinear signal amplification in response to a trigger. Two conceptual models are based on super-hairpin nanostructures, which are designed by innovating novel principles in methodology and codifying them into embedded programs. The dynamical and thermodynamical analyses reveal the critical elements and leak-resistant mechanisms of the designed models, and the leak-resistant behaviors of the intelligent DNA robots and morphologies of swarming into nonlinear amplification are separately verified. The applications of the designed models are also illustrated in specific signal amplification and targeted payload enrichment via integration with an aptamer, a fluorescent molecule and surface-enhanced Raman spectroscopy. This work has the potential to serve as design guidelines of intelligent and robust DNA robots and leakless nonlinear DNA amplification, and also as the design blueprint of cargo delivery robots with the performance of swarming into nonlinear amplification in response to a target automatically, facilitating their future applications in biosensing, bioimaging and biomedicine.
New conceptsTwo conceptual models of intelligent and robust DNA robots based on super-hairpin nanostructures are demonstrated for the first time, which are capable of swarming into leakless nonlinear amplification by the embedded programs in response to a trigger. The two models are composed of multifunctional manipulators with optional attachments (drugs, labels, grippers, and so on), trigger validators, smart leak-resistant path controllers and automatic assembly motors. Manipulators can capture triggers to validators and also actively dock with swarmed partners in assembly. After the validators finish validating the triggers, the controllers are spontaneously opened to provide the docking locations for the partners. And then, the motors are activated to run for assembly with the result that two controllers are further opened. Next, a nonlinear cascaded amplification will be executed, in which two intelligent robot species continue to assemble in a two-to-one ratio with the controllers and the motors working alternately. By innovating the design principles and codifying the novel embedded programs, this study breaks through the limitation of DNA building blocks in terms of leakage and also overturns the current design patterns of nonlinear DNA amplification devices in methodology. Thus, this work offers new design blueprints for advanced signal amplifiers and intelligent targeted drug delivery platforms, facilitating their future applications in nanotechnology and nanomedicine. |
Introduction
DNA, a traditional genetic molecule with excellent sequence programmability, has become a versatile building block for synthesizing static nanostructures and dynamic nanodevices in nanotechnology.1,2 DNA-based nanoscale devices have the capability to undergo controlled motion and reconfiguration via reaction networks in response to environmental stimulation.3,4 DNA-based nonlinear signal amplification devices with enzyme-free isothermal self-assembly processes hold great promise in sensing and imaging,1 which are not possible with the most popular method, polymerase chain reaction (PCR), because of strict thermocycling.5,6 Moreover, they have the potential to become promising platforms of nanoscale drug carriers with reduced drug toxicity and enhanced drug targeting for precise treatments.7–9Some approaches have been reported for the programming of nonlinear signal amplification devices based on the hybridization chain reaction (HCR) by designing building blocks of more DNA oligonucleotide species.10–13 The HCR involves a series of hybridization events during which DNA hairpins open alternately via toehold-mediated strand displacements when exposed to the DNA trigger and can assemble into long DNA nanostructures.14,15 Multiple DNA oligonucleotides, including hairpin species, single-stranded assistants and double-stranded substrates, are used in the HCR-based strategies, with single-stranded assistants being critical to sustaining assembly.12,13 However, because of spontaneous breathing of the duplex strand and end fraying, single-stranded DNA can inevitably initiate strand displacement and cause system leakage, even in the absence of a DNA trigger, resulting in high background signals and false-positive results.16–19 Therefore, system leakage is an insurmountable limitation in conventional DNA building blocks of programming nonlinear amplification devices. In addition, it can be assumed that more DNA oligonucleotide species are not convenient in assembly for in vivo imaging and targeted therapy. The new design strategies should probably renounce the conventional HCR-based ones in methodology or avoid using single-stranded assistants. To date, effective methods have remained challenging tasks.20–22
Toehold-mediated strand displacement is central to the dynamic assembly of DNA nanodevices3,23 and involves three general steps: toehold binding, branch migration and strand dissociation.16,24 Toehold binding is both the initiation step of strand displacement and the rate-limiting step, which involves forming an intermediate complex between two short complementary single-stranded domains.16,25 Branch migration, which includes three-way branch migration and four-way branch migration, is a walking process with nucleotide dissociation and hybridization steps just like a motor.25–28 Three-way branch migration involves a single strand and a duplex strand such as HCR,16,24 whereas four-way branch migration involves exchanging two duplexes at a mobile four-way junction.27–29 Finally, strand dissociation is carried out to complete the exchange.16
By introducing the basic idea of four-branch migration instead of HCR-based strategies and further innovating the anti-leakage mechanism, here we proposed two conceptual models of intelligent and robust DNA robots based on two super-hairpin nanostructures to perform leakless nonlinear signal amplification by the embedded programs in response to a trigger. According to three general steps of toehold-mediated strand displacement reaction, the critical elements of the designed modality were first explored individually by extracting the relevant functional motifs from two super-hairpin nanostructures. The validity of intelligent and robust DNA robots was then confirmed by examining the molecular weight and morphology of swarming into nonlinear amplification. The applications were also illustrated for specific signal amplification and loading cargo via integration with an aptamer, a fluorescent molecule and surface-enhanced Raman spectroscopy (SERS). Our study confirmed that the two conceptual models of the intelligent DNA robots had the potential to serve as guidelines and also offered new design patterns for programming nonlinear DNA amplification devices, facilitating their future applications in DNA nanotechnology and nanomedicine.
Results and discussion
Models of intelligent and robust DNA robots
Two conceptual models (R1 and R2) are shown in Fig. 1. R1 and R2 were composed of multifunctional manipulators with optional attachments (drugs, signal labels, multiple grippers, and so on), trigger validators, smart leak-resistant path controllers and automatic assembly motors. The sub-sequences were specified with letters. Letters marked with an asterisk denote the strand segments that were complementary to the corresponding unmarked segments. Arrows indicate 3′-ends. Two end overhangs of each super-hairpin species were used as multifunctional manipulators with optional attachments. The stems mainly contain two species domains (b/b* and d/d*) to form stable duplexes, which were used to carry out four-way branch migration as automatic assembly motors. The bulged loops in the middle of the duplexes along with the adjacent base pairs that were sequestered by complementarity in the stem domains were specially designed to control the assembly path and resist the system leakage. Moreover, the stems next to the manipulators were also used to validate the triggers. The hairpin loop (g) acted as a flexible linker between connecting sub-sequences, allowing for different spatial arrangements.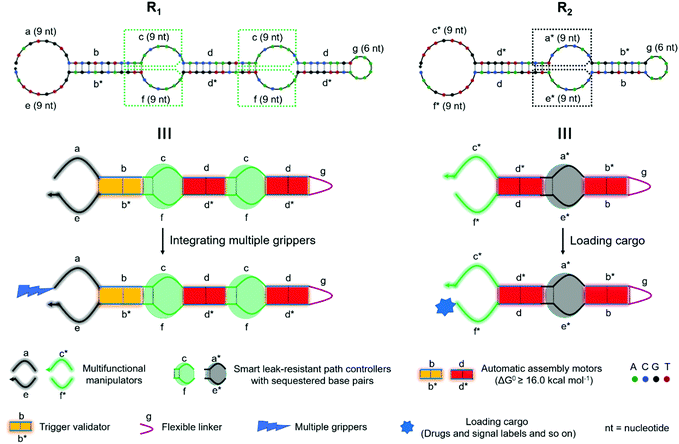 | ||
| Fig. 1 Schematic illustration for two conceptual models based on super-hairpin nanostructures. Two models are illustrated at the sequence level and domain level. | ||
Pathway and pattern of leakless nonlinear amplification
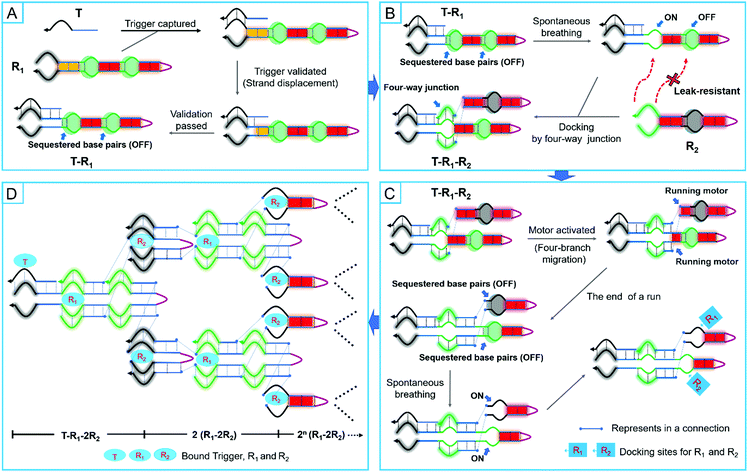
Under the designed models, R1 and R2 would stably coexist due to the constraints of the smart leak-resistant path controllers. When they were exposed to a correct trigger, a cascade event would be initiated which resulted in swarming into nonlinear amplification by the embedded programs. In the first step, a multifunctional manipulator of R1 captured the trigger (T) to the validator for further validation by toehold-mediated strand displacement. Unlike the conventional toehold-mediated strand displacement, some base pairs were sequestered by complementarity in the stem domains, which would act as the switch of a smart leak-resistant path controller (Fig. 2A). After the validator finished validating the trigger by three-branch migration, the sequestered base pairs of the first smart leak-resistant path controller would be opened spontaneously by breathing fluctuations. Thus, the only opened controller would provide suitable locations for two manipulators of R2 to dock by a four-branch junction (Fig. 2B). And then, two automatic assembly motors would be automatically activated to run along with four-branch migration. After two automatic assembly motors finished running, the sequestered base pairs in two different smart leak-resistant path controllers were not exchanged equally but remained by complementarity in the stem domains. Next, the two controllers would be further opened by breathing fluctuations, which could provide suitable locations for R1 and R2 to dock (Fig. 2C). Finally, a nonlinear cascaded amplification would be executed, in which two intelligent robot species continued to assemble in a two-to-one ratio with the controllers and motors working alternately (Fig. 2D). In the whole process of leakless nonlinear amplification, the smart leak-resistant path controllers were the pivots for docking and assembly between two intelligent robots.Dynamical investigation of the designed models
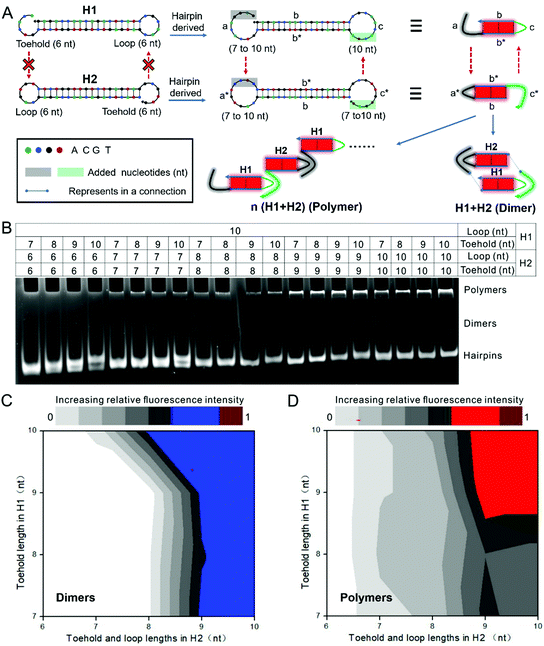
In order to perform the leakless nonlinear signal amplification, four-branch migration was utilized in the automatic assembly motors instead of HCR-based strategies, and multifunctional manipulators and smart leak-resistant path controllers were specially designed to fulfil the initiation of four-branch migration by a four-way junction. The critical elements of multifunctional manipulators and smart leak-resistant path controllers were firstly revealed by the dynamics analysis.Toehold binding is an initiation and rate-limiting step.16 The rate constant of the strand displacement increases with the toehold length and reaches saturation when the toehold is approximately 6 nucleotides (nt) long.17 Therefore, a 6-nt toehold and a 6-nt loop were usually used in the traditional hairpin with a stem-loop structure,16,30 and also in which the 6-nt loop can execute topological constraints to inhibit the hybridization with a complementary toehold.31–33 Moreover, our previous studies have also shown that a 6-nt toehold could not even bind with the trapped loops that extend to 18 nt.34
To achieve the designed models, we firstly derived a series from two complementary hairpins (H1 and H2) by extending the lengths of the toeholds and loops, with the expectation of increasing the binding dynamics between the toehold and loop and thus overcoming the topological constraint of the loops. Accordingly, the complementary toeholds and loops should form dimers and polymers with “kissed” structures (Fig. 3A and Fig. S1, S2A, B, ESI†). Each pair of derived hairpins was incubated in the reaction buffer for 2 hours at room temperature. Native polyacrylamide gel electrophoresis analysis revealed hybridization events. The hybrid reactions developed gradually as the toeholds and loops were extended synchronously. The longer polymers were blocked in the gel loading well. Moreover, relatively short products, which should be dimer complexes, were also observed (Fig. 3B and Fig. S2C, D, ESI†). When the toehold and loop lengths were both extended to more than 8 nt, hybrid products clearly appeared based on the relative fluorescence intensity (Fig. 3C and D). This means that the exposed toehold with lengths of 9 nt can bind with the complementary loop with lengths of 9 nt to form complexes.
Then, considering the sequence dependence, some other hairpins were systematically investigated, using toeholds and loops of varying sequences and lengths, as well as different stem sequences. In these cases, consistent results also showed that the hybrid products clearly formed when the lengths of the toeholds and the loops were extended to 9 nt (Fig. S3–S6 and Table S1, ESI†). The results indicated that the lengths of the toehold and loop were critical for increasing the binding dynamics between the toehold and loop. The extended loop and toehold may increase the likelihood of binding by overcoming the topological constraint.
One further point must be discussed. In a DNA hairpin, the base pairing of the stem is involved in enthalpy, whereas the loop is involved in entropy.35 The free energy or the stability of a hairpin can decrease as the loop size increases.36 Long products may also form due to leakage hybridization if the stem opens spontaneously (Fig. S7, ESI†). However, because each hairpin species of H1 has a loop with a length of 10 nt in Fig. 3B, the leakage should occur in each pair of complementary hairpins if the added loop entropy contributes to unzipping the stem. This is obviously a contradiction. Indeed, there was a slight difference in the overall free energy between hairpins with extended loops, and the change was less than 1 kcal mol−1, as estimated by the Nucleic Acids Package (NUPACK) (Fig. S1, S3 and S4, ESI†). Moreover, the free energy of each derived hairpin met a testing benchmark of −16.0 kcal mol−1 to resist leakage.34 Therefore, the added loop entropy cannot also theoretically contribute to leakage. More results also ruled out the possibility of leakage, as shown in Fig. S3 and S4 (ESI†).
Thus, based on the traditional hairpin species with a stem-loop structure, the dynamics revealed that a toehold with lengths of 9 nt can bind with a complementary loop with lengths of 9 nt, but it is unable to associate with the complementary loop with a length of 6 nt. Therefore, switching the length of the complementary loop between 9 nt and 6 nt can control the toehold binding step. These will serve as the dynamical foundation of the critical elements of the designed models including multifunctional manipulators and smart leak-resistant path controllers.
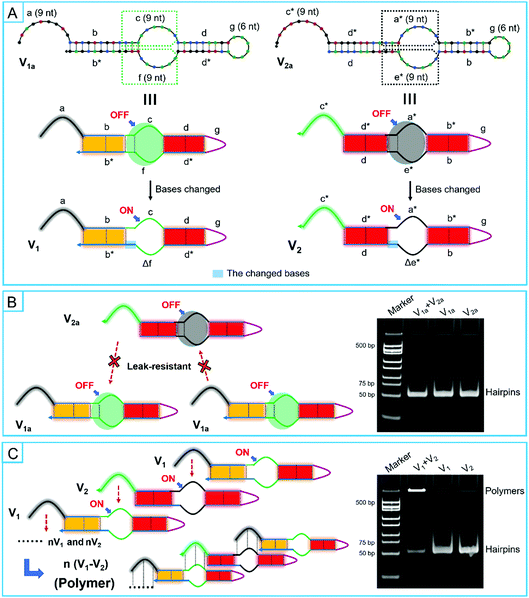
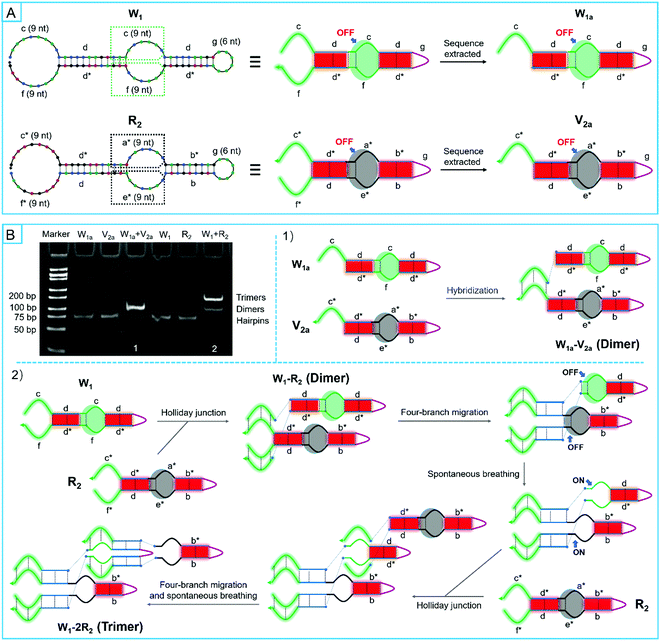
Unlike the traditional hairpin species, the bulged loops were specially designed in super-hairpin species to control the assembly path and resist system leakage. In order to investigate the design principles of smart leak-resistant path controllers as well as multifunctional manipulators, V1a and V2a were firstly extracted from motifs of R1 and R2 by discarding some sub-sequences. They had a complete smart leak-resistant path controller. Then, V1 and V2 were further derived from V1a and V2a, in which smart leak-resistant path controllers were opened by changing the sequestered bases (Fig. 4A and Table S2, ESI†). Next, the two pairs of super-hairpins were incubated in the reaction buffer for 2 hours at room temperature separately. A native polyacrylamide gel electrophoresis analysis showed that V1a and V2a could stably coexist (Fig. 4B), while V1 and V2 formed longer polymers (Fig. 4C). These results suggested that the above dynamical parameters were appropriate for the super-hairpins. Therefore, smart leak-resistant path controllers can work by switching the length of the complementary loop when the length of the multifunctional manipulator is 9 nt, and the sequestered bases in the stem domains can act as leak-resistant switching elements.
In addition, based on the relative fluorescence intensity of electrophoresis analysis, dimers between V1 and V2 were not observed. This result might have occurred because the two super-hairpins might not be structurally suited for the formation of dimers (Fig. S8, ESI†). In fact, this may be beneficial for the efficient use of hairpin monomers and the formation of long polymers.
Thermodynamical investigation of the designed models
Smart leak-resistant path controllers were the pivots for docking and assembly between two intelligent robots. The sequestered bases in the stem domains were designed as switching elements of the smart leak-resistant path controllers. The new G–C pairs contribute −1.5 kcal mol−1 of stabilization on average, and the A–T pairs add −0.8 kcal mol−1.37 However, according to the intuitive energy landscape model, the sequestered bases will suffer the free energy penalty of branch migration in terms of thermodynamics.25,38–41 Moreover, under the designed models as previously stated, the sequestered base pairs would be opened spontaneously by breathing fluctuations in the process of the leakless nonlinear amplification. Since switching the length of the complementary loop between 9 nt and 6 nt can control the toehold binding step, the sequestered bases in our design were adopted as lengths of 3 nt to accommodate the thermodynamics and dynamics. In order to evaluate the validity, the thermodynamics analysis was done.Firstly, three base pairs of the smart leak-resistant path controller would be retained by complementarity in the stem domains after the validator finished validating the correct trigger by three-branch migration. Based on the traditional hairpin species with a stem-loop structure, our previous research has shown that the remaining double chains could spontaneously melt within 6 or 7 bp due to breathing fluctuations after the trigger performed the displacement by three-branch migration.34 Therefore, this thermodynamical mechanism was used in the open situation of a smart leak-resistant path controller after the validator finished validating the trigger.
Then, automatic assembly motors were designed to run by four-branch migration so that the smart path controllers could be further opened. Because the sequestered bases of 3 nt in sequences were different between two hairpins, two stem domains of the four-branch migration do not exchange equally along complementary regions. Therefore, it may be possible to inhibit four-branch migration due to the thermodynamic potential barrier. According to the dynamics analysis of toehold binding, 9 nt lengths were available to overcome the topological constraint. To confirm the validity in thermodynamics, W1 and W1a were extracted from motifs of R1 by discarding some sub-sequences (Fig. 5A and Table S3, ESI†). Moreover, R2 and V2a that have been used above were also prepared. As shown in Fig. 5B, based on the complementary sequences of these super-hairpin species, forming trimers would act as an expected barometer for the completion of four-branch migration and the exposure of the switching elements. Next, the two pairs of super-hairpin species were incubated separately in the reaction buffer for 2 hours at room temperature. A native polyacrylamide gel electrophoresis analysis showed the reaction products. The results showed that trimers obviously formed between W1 and R2. Therefore, this confirmed that the four-branch migration occurred to expose the switching elements, resulting in the formation of more trimers between W1 and R2. In contrast, only dimers formed between W1a and V2a, which indicated that the switching elements were not exposed; if they had been, trimers would have formed (Fig. 5B). Compared with W1 and R2, W1a and V2a have only one multifunctional manipulator each. Based on these findings, two multifunctional manipulators of each super-hairpin species were found to be necessary for four-branch migration. Furthermore, 9 nt lengths of multifunctional manipulators were available to overcome the thermodynamic potential barrier of the sequestered bases by the Holiday junction. That was probably because the added free energy resulting from two pairs of multifunctional manipulators was thermodynamically in favor of driving the reaction forward by the enthalpy net gain of base pairs due to the Holiday junction. To further confirm this reasoning, a series of super-hairpins were derived from W1 and R2 by varying the lengths of one multifunctional manipulator of each super-hairpin species (Fig. S9A and Table S3, ESI†). In the view of the thermodynamics, the probability that a resting microstate would form a complex between the derived hairpins was estimated by NUPACK. The results suggested that four-branch migration could be carried out to expose the next sequestered bases when the length of the varied manipulator was not less than 6 nt (Fig. S9B, ESI†). Considering the sequence dependence, lots of other hairpins were also estimated by NUPACK, and similar results were obtained (data not shown). Hence, combining dynamical and thermodynamical analyses, each manipulator of 9nt was appropriate in length.
Validity and application of the designed models
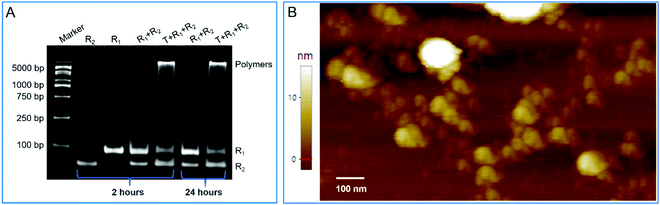
The dynamical and thermodynamical analyses confirmed the critical elements of and leak-resistant mechanism of the designed models. Based on the designed models, we fabricated two intelligent DNA robots (R1 and R2) in response to a trigger (T) (Table S4, ESI†). For these components, as well as the primary assembly complexes among them, the minimum free energy (MFE) structures were estimated by NUPACK and agreed with the expected results (Fig. S10, ESI†). Then, R1, R2 and T were incubated in the reaction buffer at room temperature. Native polyacrylamide gel electrophoresis analysis showed the reaction products. As shown in Fig. 6A, the longer assembly products were blocked in the gel loading well. However, no assembly products formed in the absence of a trigger. The results indicated that an assembly event was carried out, and two intelligent DNA robots were leak-resistant. Moreover, more details on the running steps for assembly were also explored (Fig. S11 and S12, ESI†).Next, atomic force microscopy (AFM) further confirmed the native polyacrylamide gel electrophoresis results and directly revealed the expected morphology of nonlinear amplification. In the presence of a trigger, large nonlinear assembly products were observed. In contrast, in the absence of a trigger, only some tiny yellow spots that represented the unassembled super-hairpins were observed (Fig. S13, ESI†). The partially enlarged details of the atomic force microscopy images for nonlinear amplification are shown in Fig. 6B. These results suggested that the two intelligent DNA robots were capable of swarming into leakless nonlinear amplification by the embedded programs in response to a trigger.
In order to preliminarily evaluate the applicability of the designed models, intelligent and robust DNA robots based on different super-hairpin species were illustrated for specific signal amplification and loading cargo via integration with aptamers, fluorescent molecules and SERS.
Has-let-7d-5p (Tb) and Hsa-miR-18a-5p (Tc) were randomly selected as the representative miRNA triggers, and the corresponding super-hairpin species were designed for specific signal amplification in biosensing (Table S4, ESI†). For these components, as well as the primary assembly complexes among them, the minimum free energy (MFE) structures were estimated by NUPACK and agreed with the expected results (Fig. S14 and S15, ESI†). And then, they were incubated in the reaction buffer for 2 hours at room temperature. The readouts of the leakless nonlinear amplification signals were provided by label-free surface-enhanced Raman spectroscopy (SERS). More details are described in Fig. S16 (ESI†). In the presence of triggers, SERS readouts of leakless nonlinear amplifications showed the marker band of DNA at ∼732 cm−1 (Fig. 7A-a1), which has been assigned to the ring breathing mode of adenine.34 Native polyacrylamide gels electrophoresis analysis with ethidium bromide further confirmed the results of SERS (Fig. 7A-a2).
Exosomes as small extracellular vesicles have offered promise as drug delivery systems for biological payloads. Taking surface protein CD63 of exosomes as a target,42 we evaluated the performance of the designed models in bioimaging as well as cargo delivery and enrichment by integrating the aptamer and a fluorescent molecule. The aptamer was used to bind CD63,43 and the fluorescent molecule was used as a label for bioimaging or as a loading cargo. Components including R1, target strand T1a containing aptamer CD63, and R2a containing the fluorescent dye were prepared. For these components, as well as the primary assembly complexes among them, the minimum free energy (MFE) structures were estimated by NUPACK and agreed with the expected results (Fig. S17 and Table S4, ESI†). Then, these components were used to target exosomes. T was used as a control instead of T1a to eliminate false positives caused by nonspecific adsorption of the target strand. Finally, fluorescence assays and dynamic light scattering were performed. The results showed that exosomes exhibited distinct green fluorescence signals in the presence of all components of T1a, R1 and R2a but not in controls (Fig. 7B-b1 and B-b2). Under dynamic light scattering, the size distribution of imaged exosomes was relatively large, which may be consistent with nonlinear amplification (Fig. 7B-b3 and B-b4). Moreover, Has-let-7d-5p in exosomes was also detected by label-free surface-enhanced Raman spectroscopy (SERS). More details are described in the supporting methods and Fig. S18 (ESI†).
Therefore, these results suggested that intelligent and robust DNA robots had good adaptability and applicability, and were capable of swarming into leakless nonlinear amplification in response to triggers in biosensing, bioimaging and targeted cargo enrichment.
Conclusions
Two conceptual models of intelligent and robust DNA robots based on super-hairpin nanostructures were confirmed to perform leakless nonlinear signal amplification in response to a trigger through detailed experimental and theoretical analyses of the toehold-mediated strand displacement reaction. Two models were composed of multifunctional manipulators with optional attachments, a trigger validator, smart leak-resistant path controllers and automatic assembly motors. The critical elements of the designed models were revealed in light of dynamics and thermodynamics. We demonstrated that the toehold binding could occur to overcome the topological constraints in the loop when the lengths of the toehold and loop were extended to 9 nt. On that basis, the design principle of a smart leak-resistant path controller was established by switching the length of the complementary loop between 9 nt and 6 nt when the length of the multifunctional manipulator was 9 nt. To overcome the thermodynamic potential barrier of the switches, 9 nt lengths of two multifunctional manipulators in each super-hairpin were considered to drive the four-branch migration of the automatic assembly motors by the enthalpy net gain of base pairs due to the Holiday junction. Therefore, this work improved the understanding of the toehold-mediated strand displacement dynamics and thermodynamics while also innovating the leak-resistant mechanism and providing a new pathway and pattern of leakless nonlinear amplification. The designed models can also be combined with signal readout platforms and biological payloads to offer a wide range of applications in biosensing, bioimaging and targeted cargo enrichment. Future work will focus on optimizing the assembly efficiency and in-depth evaluation of the application performance. We envision that this work has the potential to serve as design guidelines of intelligent and robust DNA robots and leakless nonlinear DNA amplification and also as the design blueprint of cargo delivery robots with the performance of swarming into nonlinear amplification in response to environmental stimulation automatically, facilitating their future applications in DNA nanotechnology and nanomedicine.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation of China (No. 21974142), the Natural Science Research Project of Universities in Anhui Province (No. K120438026) and the Nature Science Research Project of Anhui Province (No. 1908085QB65).References
- F. Li, J. Li, B. Dong, F. Wang, C. Fan and X. Zuo, Chem. Soc. Rev., 2021, 50, 5650–5667 RSC.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, Chem. Rev., 2020, 120, 9420–9481 CrossRef CAS PubMed.
- M. DeLuca, Z. Shi, C. E. Castro and G. Arya, Nanoscale Horiz., 2020, 5, 182–201 RSC.
- Y. Zhang, V. Pan, X. Li, X. Yang, H. Li, P. Wang and Y. Ke, Small, 2019, 15, e1900228 CrossRef PubMed.
- Y. Duan, R. Glazier, A. Bazrafshan, Y. Hu, S. A. Rashid, B. G. Petrich, Y. Ke and K. Salaita, Angew. Chem., Int. Ed., 2021, 60, 19974–19981 CrossRef CAS PubMed.
- C. Zhang, J. Chen, R. Sun, Z. Huang, Z. Luo, C. Zhou, M. Wu, Y. Duan and Y. Li, ACS Sens., 2020, 5, 2977–3000 CrossRef CAS PubMed.
- Y. Zhang, J. Tu, D. Wang, H. Zhu, S. K. Maity, X. Qu, B. Bogaert, H. Pei and H. Zhang, Adv. Mater., 2018, 30, e1703658 CrossRef PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- A. Keller and V. Linko, Angew. Chem., Int. Ed., 2020, 59, 15818–15833 CrossRef CAS PubMed.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- F. Baneyx and J. K. Park, Biotechnol. J., 2013, 8, 158–159 CrossRef CAS PubMed.
- F. Xuan and I. M. Hsing, J. Am. Chem. Soc., 2014, 136, 9810–9813 CrossRef CAS PubMed.
- S. Bi, M. Chen, X. Jia, Y. Dong and Z. Wang, Angew. Chem., Int. Ed., 2015, 54, 8144–8148 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha and N. A. Pierce, Development, 2018, 145, dev156869 CrossRef PubMed.
- F. C. Simmel, B. Yurke and H. R. Singh, Chem. Rev., 2019, 119, 6326–6369 CrossRef CAS PubMed.
- D. Y. Z. A. E. Winfree, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef PubMed.
- C. Li, Y. Li, Y. Chen, R. Lin, T. Li, F. Liu and N. Li, RSC Adv., 2016, 6, 74913–74916 RSC.
- A. J. Genot, D. Y. Zhang, J. Bath and A. J. Turberfield, J. Am. Chem. Soc., 2011, 133, 2177–2182 CrossRef CAS PubMed.
- Z. Zeng, R. Zhou, R. Sun, X. Zhang, Z. Cheng, C. Chen and Q. Zhu, Biosens. Bioelectron., 2020, 173, 112814 CrossRef PubMed.
- R. Duan, X. Lou and F. Xia, Chem. Soc. Rev., 2016, 45, 1738–1749 RSC.
- J. Wu, J. Lv, X. Zheng and Z. S. Wu, Talanta, 2021, 234, 122637 CrossRef CAS PubMed.
- Z. Wang, Y. Hu and L. Pan, Angew. Chem., Int. Ed., 2020, 59, 14979–14985 CrossRef CAS PubMed.
- N. Srinivas, T. E. Ouldridge, P. Sulc, J. M. Schaeffer, B. Yurke, A. A. Louis, J. P. Doye and E. Winfree, Nucleic Acids Res., 2013, 41, 10641–10658 CrossRef CAS PubMed.
- R. R. Machinek, T. E. Ouldridge, N. E. Haley, J. Bath and A. J. Turberfield, Nat. Commun., 2014, 5, 5324 CrossRef CAS PubMed.
- S. Kotani and W. L. Hughes, J. Am. Chem. Soc., 2017, 139, 6363–6368 CrossRef CAS PubMed.
- R. Shah Punatar, M. J. Martin, H. D. Wyatt, Y. W. Chan and S. C. West, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 443–450 CrossRef PubMed.
- W. Meng, R. A. Muscat, M. L. McKee, P. J. Milnes, A. H. El-Sagheer, J. Bath, B. G. Davis, T. Brown, R. K. O'Reilly and A. J. Turberfield, Nat. Chem., 2016, 8, 542–548 CrossRef CAS PubMed.
- D. Ren, C. Sun, Z. Huang, Z. Luo, C. Zhou and Y. Li, Sens. Actuators, B, 2019, 296, 126577 CrossRef CAS.
- D. Zhao, Q. Yin, Y. Chang and M. Liu, TrAC, Trends Anal. Chem., 2020, 122, 115706 CrossRef CAS.
- R. M. D. A. N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef PubMed.
- J. S. Bois, Nucleic Acids Res., 2005, 33, 4090–4095 CrossRef CAS PubMed.
- S. J. Green, D. Lubrich and A. J. Turberfield, Biophys. J., 2006, 91, 2966–2975 CrossRef CAS PubMed.
- S. Li, P. Li, M. Ge, H. Wang, Y. Cheng, G. Li, Q. Huang, H. He, C. Cao, D. Lin and L. Yang, Nucleic Acids Res., 2020, 48, 2220–2231 CrossRef CAS PubMed.
- G. Mishra, D. Giri, M. S. Li and S. Kumar, J. Chem. Phys., 2011, 135, 035102 CrossRef PubMed.
- T. Tran and B. Cannon, Biochemistry, 2017, 56, 6448–6459 CrossRef CAS PubMed.
- K. Oertell, E. M. Harcourt, M. G. Mohsen, J. Petruska, E. T. Kool and M. F. Goodman, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2277–E2285 CrossRef CAS PubMed.
- C. A. Figg, P. H. Winegar, O. G. Hayes and C. A. Mirkin, J. Am. Chem. Soc., 2020, 142, 8596–8601 CrossRef CAS PubMed.
- N. E. C. Haley, T. E. Ouldridge, I. Mullor Ruiz, A. Geraldini, A. A. Louis, J. Bath and A. J. Turberfield, Nat. Commun., 2020, 11, 2562 CrossRef CAS PubMed.
- D. Y. Zhang, S. X. Chen and P. Yin, Nat. Chem., 2012, 4, 208–214 CrossRef CAS PubMed.
- S. X. Chen and G. Seelig, J. Am. Chem. Soc., 2016, 138, 5076–5086 CrossRef CAS PubMed.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz, N. Rahbari, C. Reissfelder, C. Pilarsky, M. F. Fraga, D. Piwnica-Worms and R. Kalluri, Nature, 2015, 523, 177–182 CrossRef CAS PubMed.
- Z. Wang, S. Zong, Y. Wang, N. Li, L. Li, J. Lu, Z. Wang, B. Chen and Y. Cui, Nanoscale, 2018, 10, 9053–9062 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nh00018k |
| This journal is © The Royal Society of Chemistry 2022 |

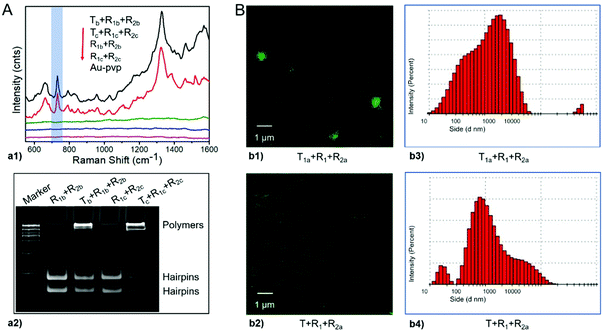
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. (B) Bioimaging and drug delivery for targeted exosomes. b1 and b2 show the laser confocal microscopy fluorescence images, and b3 and b4 show the size distributions by dynamic light scattering.
000. (B) Bioimaging and drug delivery for targeted exosomes. b1 and b2 show the laser confocal microscopy fluorescence images, and b3 and b4 show the size distributions by dynamic light scattering.