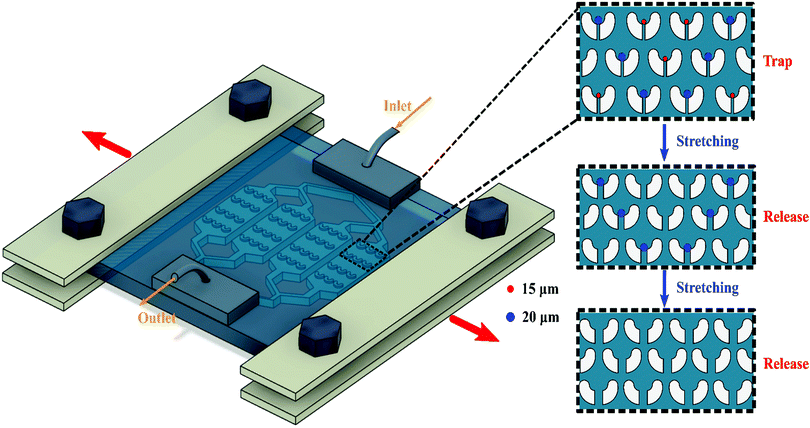
On-demand deterministic release of particles and cells using stretchable microfluidics†
Hedieh
Fallahi
 ,
Haotian
Cha
,
Hossein
Adelnia
,
Yuchen
Dai
,
Hang Thu
Ta
,
Haotian
Cha
,
Hossein
Adelnia
,
Yuchen
Dai
,
Hang Thu
Ta
 ,
Sharda
Yadav
,
Jun
Zhang
,
Sharda
Yadav
,
Jun
Zhang
 * and
Nam-Trung
Nguyen
* and
Nam-Trung
Nguyen
 *
*
Queensland Micro- and Nanotechnology Centre, Griffith University, Nathan, Queensland 4111, Australia. E-mail: nam-trung.nguyen@griffith.edu.au; jun.zhang@griffith.edu.au
First published on 21st February 2022
Abstract
Microfluidic technologies have been widely used for single-cell studies as they provide facile, cost-effective, and high-throughput evaluations of single cells with great accuracy. Capturing single cells has been investigated extensively using various microfluidic techniques. Furthermore, cell retrieval is crucial for the subsequent study of cells in applications such as drug screening. However, there are no robust methods for the facile release of the captured cells. Therefore, we developed a stretchable microfluidic cell trapper for easy on-demand release of cells in a deterministic manner. The stretchable microdevice consists of several U-shaped microstructures to capture single cells. The gap at the bottom edge of the microstructure broadens when the device is stretched along its width. By tuning the horizontal elongation of the device, ample space is provided to release particle/cell sizes of interest. The performance of the stretchable microdevice was evaluated using particles and cells. A deterministic release of particles was demonstrated using a mixture of 15 μm and 20 μm particles. The retrieval of the 15 μm particles and the 20 μm particles was achieved with elongation lengths of 1 mm and 5 mm, respectively. Two different cell lines, T47D breast cancer cells and J774A.1 macrophages, were employed to characterise the cell release capability of the device. The proposed stretchable micro cell trapper provided a deterministic recovery of the captured cells by adjusting the elongation length of the device. We believe that this stretchable microfluidic platform can provide an alternative method to facilely release trapped cells for subsequent evaluation.
New conceptsSingle-cell trapping and analysis allow for the study of cell-to-cell variations within a cell population. Conventional techniques for studying isolated cells based on multi-well plates and flow cytometry are either time consuming or require cell labelling. Microfluidics, the science and technology for handling a tiny amount of fluid, has been used for trapping cells for single-cell studies. However, the bottleneck of the microfluidic techniques is cell retrieval. Retrieval of intact cells after trapping is vital for subsequent studies and analyses. In this study, we introduced the concept of stretchable microfluidics for the convenient release of captured cells in a deterministic manner. Stretchable microfluidics is based on a flexible device, where macroscopic stretching leads to microscale and even nanoscale control of device components such as microchannels and trapping gaps. Macroscopic stretching allows for tuning the gap of a U-shaped trapper, thus allowing the on-demand release of trapped cells. This new concept was first demonstrated with microbeads of various sizes and then tested with breast cancer cells and macrophages. The stretchable cell trapping concept reported here can retrieve 100% of captured cells, a remarkable advancement for the field of single-cell analysis. |
Introduction
Examining cell populations varies considerably from single-cell studies because the cell response analysed in bulk is much different from individual cell responses.1 The reason is that cells are heterogeneous and differ from one another.2 Cellular decision making and cell fate are significantly impacted by cell heterogeneity.3 However, studying cells in bulk can conceal this effect. Thus, cell heterogeneity is of great importance. Analysing cells at a single-cell level allows for the study of cell-to-cell variations within a cell population.4 Consequently, single-cell studies have gained significant attention, and several single-cell analysis methods have been reported.5 Microscopic imaging of isolated cells in multi-well plates is one of the most available methods for studying single cells.6 However, this technique is laborious, requires specialised expertise, and has a low efficiency. Conversely, flow cytometry as the gold standard for cell analysis offers rapid automatic screening of fluorescently labelled cells in a flow with high precision and high throughput.7 Nonetheless, flow cytometry is complex, expensive, cannot monitor cell dynamics and requires a large sample volume.Microfluidics has been developed as an empowering technology for single-cell studies and has demonstrated promising results.8–10 Benefiting from the precise control of fluid flow motion in microchannels, microfluidics has facilitated cell capture and release processes.11–13 Furthermore, microfluidics enables precise control over cell quantity and concentration, leading to higher accuracy than conventional methods. These advanced features allow for the analysis of the morphology and physiology of single cells.14 To date, several microfluidic approaches including passive and active methods have been investigated for single-cell studies.8,9,11,12,14–16
Active methods rely on non-surface contact capturing of cells by employing external force fields such as optical,17–19 acoustic,20,21 magnetic,22 and electrical.23–25 In contrast, passive microfluidic approaches are contact-based techniques which involve chemical or hydrodynamic approaches to capture cells that come into contact with the surface of the device.26 Passive hydrodynamic approaches include micropatterns,27 microwells,28,29 trappers,16 and droplets.15 Hydrodynamic trapping with a microstructure array, designed for capturing single cells, is superior to other passive hydrodynamic methods, as it provides a high throughput and facile operation. The mechanism of hydrodynamic trapping depends on the flow resistance inside the device. That means, the cells follow the route with minimum resistance and avoid obstacles because of the shear stress surrounding them.30–32 While some of these hydrodynamic trapping devices only consist of a dense array of small microstructures to capture cells,33,34 others comprise a bypass channel to regulate the flow resistance to trap cells.30,32
The main drawback of passive hydrodynamic trappers is that cell retrieval is still challenging, and the immobilised cells cannot be easily released.35 In the reported devices, cell retrieval is usually not deterministic and is achieved through backflow,36 multilayer embedded channels,37 and bypass channels.38,39 These techniques require complex design and fabrication procedures as they consist of several layers and valves that need to be prepared using different methods. Also, the operation involves complications as several parameters such as flow rate and pressure need to be delicately regulated.
In the present study, we introduce a stretchable microfluidic device for the easy release of captured particles/cells in a deterministic manner. Stretchable microfluidics, defined as microfluidic devices entirely fabricated from flexible and stretchable materials, has recently been introduced and investigated by our group.40 Elongating the stretchable microdevice allows precise tuning of the microchannel dimensions.41–43 We designed and fabricated a stretchable microfluidic cell trapper that can be stretched along its width. The chip consists of an array of U-shaped microstructures with tiny gaps to trap and release particles and cells. Capturing of particles and cells with this concept is a physical trapping process, where the particles/cells are immobilised without chemical affinity. Elongation of the microchip broadens the gap in the U-shaped microstructures and enables the controllable release of the trapped particles/cells. First, we validated the capture and release capability of the stretchable cell trapper using microbeads. Second, using a mixture of different sized particles, we demonstrated that the particle sizes of interest could be released in a deterministic manner by tuning the elongation of the microchip. Finally, we evaluated the performance of the stretchable cell trapper for capturing and releasing two cell lines, T47D breast cancer cells and J774A.1 macrophages. The results indicated that a deterministic release of the cell sizes of interest as well as a release efficiency as high as 100% of the trapped cells were achieved using the stretchable cell trapper. We believe that the proposed stretchable cell trapper with a capability of easy and deterministic retrieval of 100% of the captured cells is a remarkable advance for subsequent evaluation of the trapped cells.
Materials and methods
Design and fabrication of the stretchable microfluidic cell trapper
The microfluidic channel comprises one inlet and one outlet for the introduction and collection of the samples, respectively, Fig. 1. The inlet channel splits into four and connects to four separate chambers, each comprising 165 U-shaped trapping structures located in 15 rows and 11 columns. The distance between the adjacent columns is 60 μm. Moreover, the adjacent rows have a vertical distance of 60 μm. The small U-shaped structures have a gap of approximately 10 μm used for subsequent releasing of the particles/cells. The channel has a uniform height of approximately 50 μm.A silicon mould comprising the cell trapper design was fabricated using the photolithography technique. In brief, a silicon wafer was spin coated with SU-8 3050 (MicroChem Corp.), followed by soft baking at 95 °C. The microfeatures on a glass chrome mask were transferred to a silicon wafer through UV exposure. Finally, the unexposed photoresist was removed after being immersed in SU8 developer.
The soft lithography technique was employed to fabricate the stretchable microfluidic cell trapper.44 First, poly(dimethylsiloxane) (PDMS) precursor (Sylgard 184, Dow Corning) was mixed with the corresponding curing agent at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After the solution was degassed in a vacuum chamber, it was cast on the silicon mould, degassed, and cured at 70 °C for 2 hours. In addition, another plain layer was prepared using the same casting method. The layers were then peeled off from the substrates, and the one comprising the channel was punched manually to form the inlet and outlet holes. Next, the two thin PDMS layers, one with the channels on it and the other to cover and seal the channel, were plasma-treated using a plasma cleaner (PDC-32G-2, Harrick Plasma) and were bonded followed by thermal treatment at 70 °C for 10 min. The two bonded layers had an overall thickness of approximately 1 mm. Two small blocks of PDMS with a length, a width and a thickness of 8 mm, 5 mm and 5 mm, respectively, were prepared to hold the tubings. After being punched manually, the two small PDMS blocks were bonded on top of the inlet and outlet regions of the previously prepared layers. Fig. S1a (ESI†) shows the fabricated stretchable microfluidic cell trapper. PTFE tubings were then connected to the inlet and outlet for the delivery and collection of the sample.
1. After the solution was degassed in a vacuum chamber, it was cast on the silicon mould, degassed, and cured at 70 °C for 2 hours. In addition, another plain layer was prepared using the same casting method. The layers were then peeled off from the substrates, and the one comprising the channel was punched manually to form the inlet and outlet holes. Next, the two thin PDMS layers, one with the channels on it and the other to cover and seal the channel, were plasma-treated using a plasma cleaner (PDC-32G-2, Harrick Plasma) and were bonded followed by thermal treatment at 70 °C for 10 min. The two bonded layers had an overall thickness of approximately 1 mm. Two small blocks of PDMS with a length, a width and a thickness of 8 mm, 5 mm and 5 mm, respectively, were prepared to hold the tubings. After being punched manually, the two small PDMS blocks were bonded on top of the inlet and outlet regions of the previously prepared layers. Fig. S1a (ESI†) shows the fabricated stretchable microfluidic cell trapper. PTFE tubings were then connected to the inlet and outlet for the delivery and collection of the sample.
Automatic stretching platform
An automated stretching platform was employed to apply an accurate stretch to the micro cell trapper. This platform was custom-made, and the detailed manufacturing procedure has been elaborated in our previous work.43 Briefly, the platform consisted of a linear translation stage connected to a stepper motor (JKM NEMA17 Two Phase 42 mm) controlled by an Arduino board. This motor and linear translation stage were arranged on a poly(methyl methacrylate) (PMMA) sheet that fits into the stage of an inverted microscope. The fabricated stretchable cell trapper device was fixed under the clamps of the stretching platform. The thin stretchable cell trapper was then stretched in its width direction such that the microstructures in the devices were elongated laterally and therefore the gaps in the U-shaped microstructures expanded to release trapped particles/cells. Fig. S1b (ESI†) shows the stretchable platform with the flexible micro cell trapper clamped on it.Experimental setup
The whole setup was mounted on the stage of an inverted microscope (Nikon Eclipse Ti2) equipped with an LED illumination source (pE-4000, CoolLED), Fig. S1c (ESI†). Fluorescence microscopy was conducted to visualise the fluorescently labelled particles and cells. Sample delivery to the microfluidic cell trapper was conducted using a syringe pump (neMESYS, Centoni GmbH). A digital camera (Nikon, DS-Qi2) was connected to the microscope for recording videos and images. NIS-elements imaging software of the microscope was used for processing the images. ImageJ software was used for the quantification of the data acquired from the images. In this paper, retrieval and recovery are used interchangeably to refer to the release of the particles/cells from the microstructures since all the released particles/cells can be retrieved from the outlet. Also, release/retrieval efficiency is defined as the number of released particles/cells in relation to the initial number of particles/cells trapped in the microstructures. The number of released particles/cells was calculated based on the number of captured and remaining particles/cells before and after elongation, respectively.Experimental procedure
Prior to the experiments involving particles, the device was flushed with 70% ethanol to increase the hydrophilicity of the PDMS surface. After that, the particle sample was introduced to the device at a suitable flow rate. Then to flush the particles that were not trapped in the microstructures, a 1% solution of Tween20 (Fisher BioReagents) and DI water was infused into the device for 5 minutes with a flow rate of 4000 μL min−1. The high flow rate was used to remove all the non-immobilised particles stuck in the inlet, channels, and tubings. To release the trapped particles, the thin stretchable device was elongated along the width at 1 mm intervals while a 1% solution of Tween20 and DI water was running through the device with a flow rate of 200 μL min−1. After each 1 mm elongation, the device was kept in that condition for one minute, and both brightfield and fluorescent images were taken to enumerate the released particles. For the experiments with cells, the device was primed with phosphate-buffered saline (PBS) (Thermo scientific) after being flushed with 70% ethanol. The PBS solution was kept inside the channels for 2 hours. Next, the cell suspension was infused into the device at a given flow rate, followed by flushing the untrapped cells using PBS. Finally, the release procedure was performed the same as in the particles but with PBS buffer running through the device.Sample preparation
Three stock solutions of 15 μm, 20 μm and a mixture of 15 μm and 20 μm particles were prepared using a solution of 0.5% Tween20 in DI water. Fluorescently labelled polystyrene particles were purchased from Thermo Fisher. For the cell experiments, we used two cell lines, J774A.1 macrophages and T47D cancer cells from American Type Tissue Culture (ATCC, Manassas, VA, USA).The T47D cell line is a type of human breast cancer. Briefly, the cells were cultured in a 25 cm2 flask in a humidified incubator at 37 °C, with 5% CO2. Dulbecco's modified Eagle medium/Nutrient Mixture F-12 (DEME/F12) containing 10% heat-inactivated fetal bovine serum (FBS), and 1% penicillin/streptomycin were used as the culture medium and were purchased from Gibco-Thermo Fisher Scientific (Waltham, MA, USA). The cancer cells were then detached from the flask and centrifuged to obtain a pellet. Later the pellet was incubated for 30 min in a solution of Mitotracker Green FM (Thermo Fisher Scientific-Eugene, OR, USA) to be stained. Then, the pellet was washed, centrifuged and redispersed in PBS for later use.
Macrophages were grown in a non-treated 100 mm × 20 mm cell culture dish using RPMI (Sigma 1640) supplemented with FBS (10%), penicillin (100 U mL−1), and L-glutamine (1%) at 37 °C with 5% CO2. At confluence, the cells were harvested using cold PBS and treated with the live/dead staining solution for 10 min at room temperature under gentle shaking. The staining solution contained calcein AM (5 μL) and ethidium homodimer-1 (EthD-1) (20 μL) in 10 mL PBS. Finally, the stained cells were washed once (150 × g, 5 min) with PBS, followed by redispersing in PBS.
The as-detached macrophage cell suspension was dispersed in PBS to evaluate viability defined as the number of live cells to the total number of cells. As a reference, half of the cell population was kept at room temperature for 5 min and the other half was immediately introduced into the stretchable device. After the trapping and release processes which were conducted like the cancer cell experiments, the cells were collected from the outlet. The whole experiment took 5 min like the control sample incubated in room temperature. Next, the two samples were immediately stained by calcein/EthD-1 for 10 min (procedure described above), followed by observing the cells under fluorescence microscope. A hemocytometer (BRAND® counting chamber BLAUBRAND® Neubauer improved) was used for cell counting.
Results and discussion
Dimensional changes of the microstructures under elongation
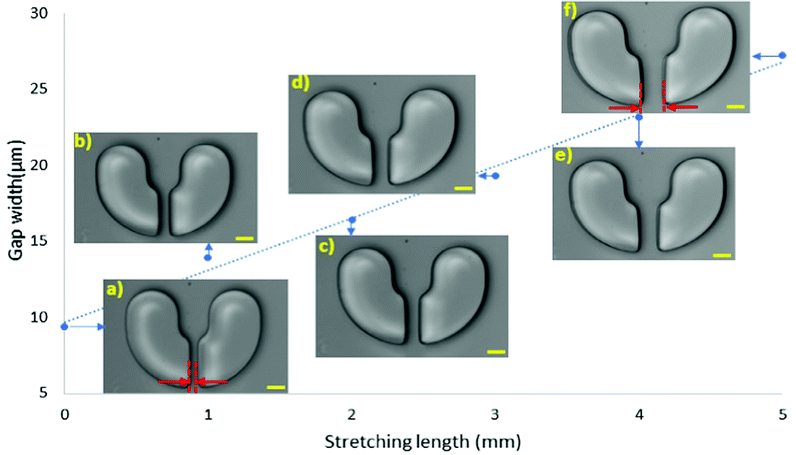
At first, we investigated the effects of stretching on the dimensions of the microstructures. Thus, with an input flow rate of 200 μL min−1 the device was stretched along its width. With 1 mm stretching intervals, the device was elongated up to 5 mm. It was kept constant after each stretch, and images of the same location were captured. Fig. 2 illustrates the dimensional changes of a single microstructure under elongation. Since the device was stretched horizontally (in its width direction), the gap located at the bottom edge of the trapper widened, Fig. 2a–f. The relationship in Fig. 2 exhibits a linear behaviour of the microstructure dimension under elongation with an average increase of about 3 μm in the gap size under each millimetre stretching increase. Thus, the 10 μm gap size widened to around 27 μm after 5 mm elongation. With 5 mm elongation, the device was still in its elastic region, and after removing the stretch, it returned to its initial condition. Therefore, by only 5 mm stretch while still in the elastic region, the gap size increased approximately three times. This variation of gap size under elongation is significant because it should conceivably provide adequate space for the captured particles/cells to pass through.Deterministic trap and release of polystyrene beads
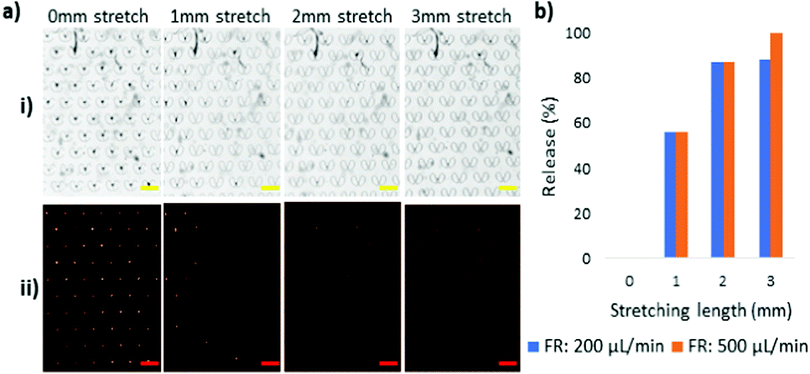
The trap and release performance of the stretchable micro trapper was initially evaluated using spherical polystyrene beads. At the initial no stretch condition, a solution of 15 μm particles was introduced to the device at a flow rate of 200 μL min−1. After flushing the untrapped particles, the immobilised particles were imaged using the microscope. Next, the device was stretched with 1 mm interval while keeping the flow rate constant at 200 μL min−1. Fig. 3 shows the brightfield and fluorescent images of the array of microstructures. At no stretch, the traps were almost occupied with 15 μm particles. With 2 mm stretch, most of the particles were released from the traps as the gap widened to more than 15 μm, which provided the particles with ample space to flow through. The device was stretched for another millimetre to release the few remaining particles in the traps. Three milimetre stretch provided a sufficiently large space for all the 15 μm particles to pass through. Unexpectedly, a few of the particles adhered to the surface of the PDMS, and needed a higher flow rate to flush them out. Thus, all the particles were released by increasing the flow rate to 500 μL min−1. The bar graph in Fig. 3b illustrates the percentage of the particles released by stretching the device. As can be seen, more than 90% were released at 2 mm stretch, and then by increasing the flow rate, all particles were released from the traps.Meanwhile, 20 μm particles were also tested to study the release behaviour of the device. All the experimental conditions were kept the same as for the 15 μm-particles except that a higher flushing flow rate of 1000 μL min−1 was also used for 20 μm particle release since they appeared stickier than the 15 μm particles. Fig. 4 illustrates the trap and release behaviour of 20 μm particles with different flow rate conditions. The 20 μm particles were captured and immobilised in the microstructures at the initial state of the device, followed by being elongated with 1 mm stretching intervals. As shown in Fig. 4a, 20 μm particles need a significantly larger gap size to be released than the 15 μm particles. Therefore, the device was stretched up to 4 mm. A close magnification of the microstructures at 4 mm stretching length reveals that the gap size is sufficiently wide for the 20 μm particles to pass through, Fig. 4e. However, they adhered to the PDMS, making it difficult to retrieve them. Based on this observation, a higher flow rate was required for the sticky particles to flow through the gaps. As such, flow rates of 500 μL min−1 and 1000 μL min−1 were tested, Fig. 4b and c. The quantified data of the experiments are provided in the form of a bar graph, Fig. 4d. At 4 mm stretching length, with the gaps being sufficiently wide for the 20 μm particles to move through, a flow rate of 500 μL min−1 led to the release of 50% of the 20 μm particles while a 100% release of the 20 μm particles was achieved at a 1000 μL min−1 flow rate.
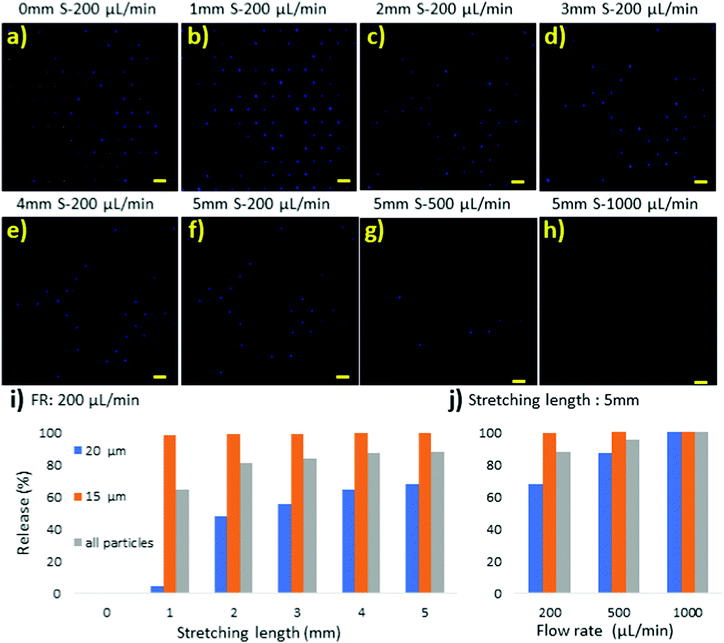
Deterministic release of a mixture of 15 μm and 20 μm particles
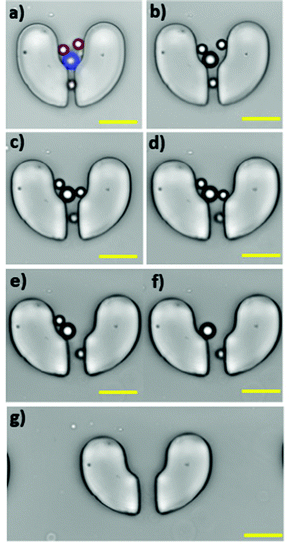
Furthermore, using a mixture of 15 μm and 20 μm particles, we examined the selective release performance of the stretchable microtrapper. Fig. 5a–h shows the fluorescent images of the particles trapped in the device at different stretching lengths and flow rates. The red fluorescent dots represent the 15 μm particles, while the blue dots indicate the 20 μm particles. At the initial no stretch condition, 15 μm and 20 μm particles were captured in the traps, Fig. 5a. Then, the device was stretched horizontally up to 5 mm. By stretching the device for 1 mm under a flow rate of 200 μL min−1, the majority of 15 μm particles (red) were released while most 20 μm particles (blue) were still trapped, Fig. 5b. Next, the device was elongated up to 5 mm where we could retrieve a large number of the 20 μm particles at a flow rate of 200 μL min−1. For the release of the remaining 20 μm particles, the flow rate was increased to 500 μL min−1 and 1000 μL min−1. Finally, a 100% retrieval efficiency of 20 μm particles was obtained at a flow rate of 1000 μL min−1, Fig. 5h. The retrieval efficiency of particles at a flow rate of 200 μL min−1 was also calculated, Fig. 5i. As demonstrated, adjusting the stretching length can achieve a deterministic retrieval of particles. For example, for the release of 15 μm particles, only 1 mm stretch is adequate, but for 20 μm particles, the device needs to be elongated for 5 mm. Fig. 5j shows an increase in release efficiency of 20 μm particles by increasing the flow rate to 1000 μL min−1 at the 5 mm stretching length.In some cases, multiple particles of different sizes were captured in one microstructure. To evaluate the release behaviour of such cases, we closely observed the particle release process from a microstructure with multiple particles trapped in it, Fig. 6. As shown in Fig. 6a, two 15 μm (red) particles as well as one 20 μm (blue) particle were captured in one microstructure. It should be noted that an unwanted particle was stuck and adhered to the bottom of the gap area from the previous experiments, which we do not consider in the discussion. Fig. 6b–e illustrate particles’ movements toward the gap as it widened under elongation of up to 4 mm. At a 4 mm stretch, the 15 μm particle closer to the gap found its way out (Fig. 6e), followed by the other 15 μm particle (Fig. 6f) at a flow rate of 250 μL min−1. Finally, the 20 μm particle was released (Fig. 6g) at a flow rate of 300 μL min−1. The 15 μm particles, due to less resistance around them, found it easier to pass through the narrow gap, while the larger 20 μm particle struggled more to flow through the gap.
Characterisation of cell trap and release in the stretchable microtrapper
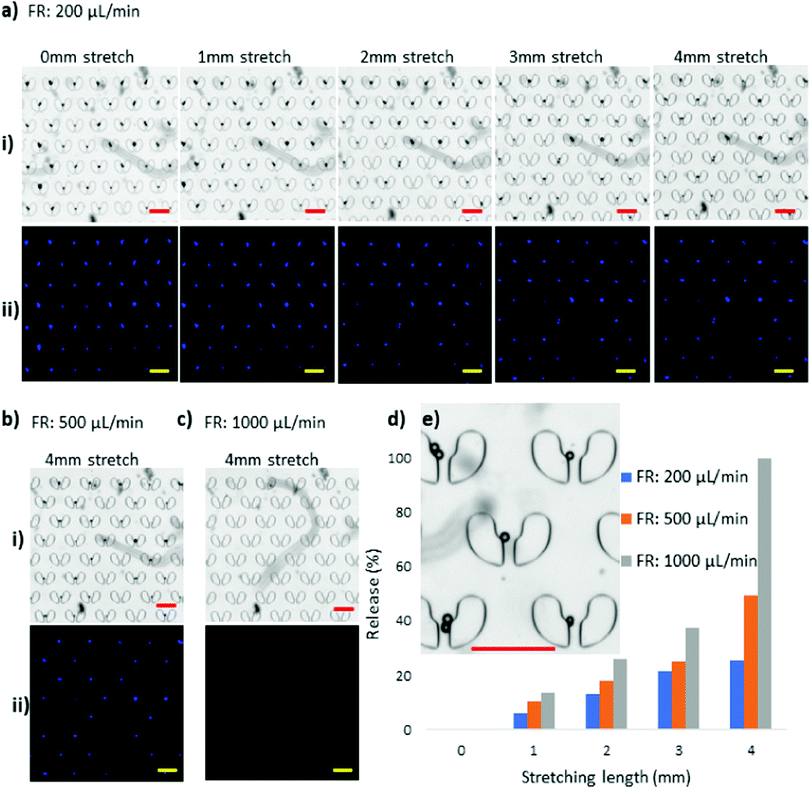
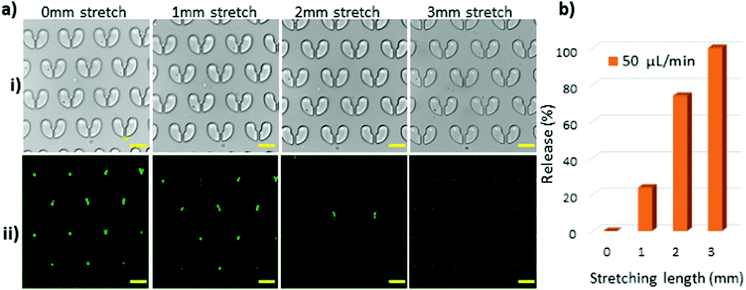
Since the ultimate goal of the stretchable cell trapper is for applications involving biological particles such as cells, we used two cell lines, J774A.1 macrophages and T47D cancer cells, to test the efficiency of the device. We used the relatively gentle flow rate of 50 μL min−1 for cell loading and retrieval to avoid cell deformation and damage caused by high shear. The J774A.1 macrophages had a size distribution of 15–17 μm with almost spherical and regular shapes. They were introduced to the stretchable microdevice and were captured by the U-shaped trappers. Fig. 7a illustrates the brightfield and fluorescent images of the macrophages captured by the stretchable microtrapper. Under the no-stretch condition, the macrophages were immobilised by the microstructures and, as anticipated, stretching the device resulted in their retrieval. The macrophages showed a similar behaviour to that of the 15 μm particles. With a 1 mm stretch, a small percentage of trapped macrophages were released. The majority of the macrophages of 15 and 16 μm diameter were released at a 2 mm stretch. The rest with a larger diameter of about 17 μm were released with a 3 mm stretch, resulting in the full recovery of the cells. Since cells are deformable as compared to particles and cannot withstand high shear stress, we used a much lower flow rate of 50 μL min−1 for cell retrieval. Fig. 7b shows the release efficiency of the macrophages using the stretchable microtrapper.The viability of the released macrophage cells was also evaluated and compared to the reference cell sample. Cells collected from the outlets as well as the reference sample cells being incubated at room temperature were both fully viable after 15 min. Fig. S2 (ESI†) illustrates the viability of the released cells and of the control cells. This result suggests that the device does not lead to cell cytotoxicity. The high viability of the released cells could be attributed to the quick processing time of only 5 min and the condition provided by priming the device with PBS for 2 hours prior to the experiments.
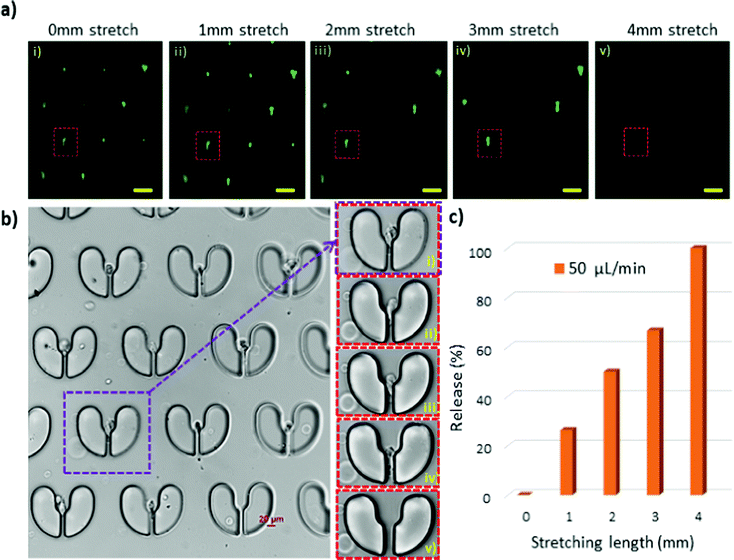
Compared to macrophages, T47D cancer cells have a wider size distribution of 14 μm to 22 μm as well as irregular shapes. Fig. 8a shows the fluorescent images of the T47D cells captured in the cell trapper. By stretching the device, we recovered cells of different sizes under different elongation lengths, where 1, 2, and 3 mm stretching lengths released 14–15, 16–18, and 18–20 μm cells, respectively, and the cells larger than 20 μm were recovered at 4 mm stretch. Despite their large sizes, T47D cells appeared to flow through the gaps easier. We hypothesise that this is due to their spindle-like shapes, which made them easier to slide through the gaps, evident by the image of the trapped T47D cells in Fig. 8b, which shows the details of the release process of a single cell with a size >20 μm. The microstructure of interest is indicated with red squares in Fig. 8a and with a purple square in Fig. 8b. The corresponding enlarged images are provided in Fig. 8b (i–v), illustrating that the single cell is finding its way out of the microstructure during elongation. Fig. 8c exhibits the release efficiency of the T47D cells at different stretching lengths, where a complete retrieval of cells was achieved at a 4 mm stretch. The experiments with both cell types, macrophages and T47D cells, demonstrated that a deterministic release of the trapped cells was achievable by tuning the stretching length of the stretchable microtrapper.
Conclusion
We developed a novel stretchable microfluidic device for facile, on-demand and deterministic release of particles and cells. The hydrodynamic microdevice consisted of dense arrays of U-shaped microstructures designed to capture particles and cells. Simply by stretching the device along its width, the gaps at the bottom edge of the microstructures widened, providing sufficient openings for the particles/cells to pass through. The trap and release performance of the device was validated using particle sizes of 15 μm and 20 μm. Moreover, we demonstrated that by regulating the stretching length of the device, a deterministic release of particle sizes of interest was achieved. For a mixture of 15 μm and 20 μm particles, 15 μm particles were retrieved with 1 mm to 2 mm stretch, while a 4 mm to 5 mm stretch led to the release of 20 μm particles. Two different cell lines were tested to further evaluate the release efficiency of stretchable microfluidics on cells. By stretching the microdevice up to 3 mm, J774A.1 macrophages with a narrow size distribution of 15 to 17 μm were fully recovered. Moreover, adjusting the stretching length from 0 to 4 mm provided a deterministic release of T47D cancer cells with a larger size distribution of 16 to 22 μm. Finally, we believe that this stretchable microfluidic platform offers an alternative method to facilely and deterministically release trapped cells, where a subsequent study of the cells off-chip is necessary.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Hedieh Fallahi acknowledges the support of the higher degree research scholarships from Griffith University. Nam-Trung Nguyen acknowledges the support from the Australian Research Council (ARC) Discovery Project (Grant No. DP180100055) and access to the Queensland node at Griffith of the Australian National Fabrication Facility, a company established under the National Collaborative Research Infrastructure Strategy to provide nano- and microfabrication facilities for Australia's researchers. Jun Zhang acknowledges the support from the Australian Research Council (ARC) DECRA fellowship (Grant No. DE210100692).References
- P. Cluzel, M. Surette and S. Leibler, Science, 2000, 1652–1655 CrossRef CAS PubMed.
- C. V. Rao, D. M. Wolf and A. P. Arkin, Nature, 2002, 420, 231–237 CrossRef CAS PubMed.
- T. J. Perkins and P. S. Swain, Mol. Syst. Biol., 2009, 5, 326 CrossRef PubMed.
- E. Hodzic, Bosnian J. Basic Med. Sci., 2016, 16, 313–314 CrossRef PubMed.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, 1903905 CrossRef CAS PubMed.
- A. Gross, J. Schoendube, S. Zimmermann, M. Steeb, R. Zengerle and P. Koltay, Int. J. Mol. Sci., 2015, 16, 16897–16919 CrossRef CAS PubMed.
- H. M. Davey and D. B. Kell, Microbiol. Rev., 1996, 60, 641–696 CrossRef CAS PubMed.
- R. N. Zare and S. Kim, Annu. Rev. Biomed. Eng., 2010, 12, 187–201 CrossRef CAS PubMed.
- A. Grünberger, W. Wiechert and D. Kohlheyer, Curr. Opin. Biotechnol, 2014, 29, 15–23 CrossRef PubMed.
- V. Lecault, A. K. White, A. Singhal and C. L. Hansen, Curr. Opin. Chem. Biol., 2012, 16, 381–390 CrossRef CAS PubMed.
- B. Deng, H. Wang, Z. Tan and Y. Quan, Micromachines, 2019, 10, 409 CrossRef PubMed.
- H. Yin and D. Marshall, Curr. Opin. Biotechnol, 2012, 23, 110–119 CrossRef CAS PubMed.
- H. Cha, H. Fallahi, Y. Dai, D. Yuan, H. An, N.-T. Nguyen and J. Zhang, Lab Chip, 2021, 22(3), 423–444, 10.1039/D1LC00869B.
- T. Luo, L. Fan, R. Zhu and D. Sun, Micromachines, 2019, 10, 104 CrossRef PubMed.
- H. N. Joensson and H. A. Svahn, Angew. Chem., Int. Ed., 2012, 51, 12176–12192 CrossRef CAS PubMed.
- V. Narayanamurthy, S. Nagarajan, A. A. Y. Firus Khan, F. Samsuri and T. M. Sridhar, Anal. Methods, 2017, 9, 3751–3772 RSC.
- A. Keloth, O. Anderson, D. Risbridger and L. Paterson, Micromachines, 2018, 9, 434 Search PubMed.
- X. Wang, S. Chen, M. Kong, Z. Wang, K. D. Costa, R. A. Li and D. Sun, Lab Chip, 2011, 11, 3656–3662 RSC.
- N.-T. Huang, H.-l. Zhang, M.-T. Chung, J. H. Seo and K. Kurabayashi, Lab Chip, 2014, 14, 1230–1245 RSC.
- D. J. Collins, B. Morahan, J. Garcia-Bustos, C. Doerig, M. Plebanski and A. Neild, Nat. Commun., 2015, 6, 8686 CrossRef CAS PubMed.
- P. Zhang, H. Bachman, A. Ozcelik and T. J. Huang, Annu. Rev. Anal. Chem., 2020, 13, 17–43 CrossRef CAS PubMed.
- R. Khojah, Z. Xiao, M. K. Panduranga, M. Bogumil, Y. Wang, M. Goiriena-Goikoetxea, R. V. Chopdekar, J. Bokor, G. P. Carman, R. N. Candler and D. Di Carlo, Adv. Mater., 2021, 33, 2006651 CrossRef CAS PubMed.
- J. Voldman, Annu. Rev. Biomed. Eng., 2006, 8, 425–454 CrossRef CAS PubMed.
- T. Matsue, N. Matsumoto and I. Uchida, Electrochim. Acta, 1997, 42, 3251–3256 CrossRef CAS.
- P.-E. Thiriet, J. Pezoldt, G. Gambardella, K. Keim, B. Deplancke and C. Guiducci, Micromachines, 2020, 11, 322 CrossRef PubMed.
- S. Takayama, E. Ostuni, P. LeDuc, K. Naruse, D. E. Ingber and G. M. Whitesides, Chem. Biol., 2003, 10, 123–130 CrossRef CAS PubMed.
- L. Lin, Y.-S. Chu, J. P. Thiery, C. T. Lim and I. Rodriguez, Lab Chip, 2013, 13, 714–721 RSC.
- M. C. Park, J. Y. Hur, H. S. Cho, S.-H. Park and K. Y. Suh, Lab Chip, 2011, 11, 79–86 RSC.
- N.-T. Huang, Y.-J. Hwong and R. L. Lai, Microfluid. Nanofluid., 2018, 22, 16 CrossRef.
- W.-H. Tan and S. Takeuchi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1146–1151 CrossRef CAS PubMed.
- J. Chung, Y.-J. Kim and E. Yoon, Appl. Phys. Lett., 2011, 98, 123701 CrossRef PubMed.
- D. Jin, B. Deng, J. X. Li, W. Cai, L. Tu, J. Chen, Q. Wu and W. H. Wang, Biomicrofluidics, 2015, 9, 014101 CrossRef CAS PubMed.
- D. D. Carlo, L. Y. Wu and L. P. Lee, Lab Chip, 2006, 6, 1445–1449 RSC.
- D. Di Carlo, N. Aghdam and L. P. Lee, Anal. Chem., 2006, 78, 4925–4930 CrossRef CAS PubMed.
- S. M. Weiz, M. Medina-Sánchez and O. G. Schmidt, Adv. Biosyst., 2018, 2, 1700193 CrossRef.
- H. S. Kim, T. P. Devarenne and A. Han, Lab Chip, 2015, 15, 2467–2475 RSC.
- C. Lipp, K. Uning, J. Cottet, D. Migliozzi, A. Bertsch and P. Renaud, Lab Chip, 2021, 21, 3686–3694 RSC.
- H. Chai, Y. Feng, F. Liang and W. Wang, Lab Chip, 2021, 21, 2486–2494 RSC.
- A. Adamo and K. F. Jensen, Lab Chip, 2008, 8, 1258–1261 RSC.
- H. Fallahi, J. Zhang, H.-P. Phan and N.-T. Nguyen, Micromachines, 2019, 10, 830 CrossRef PubMed.
- H. Fallahi, J. Zhang, J. Nicholls, H.-P. Phan and N.-T. Nguyen, Anal. Chem., 2020, 92, 12473–12480 CrossRef CAS PubMed.
- H. Fallahi, S. Yadav, H.-P. Phan, H. Ta, J. Zhang and N.-T. Nguyen, Lab Chip, 2021, 21, 2008–2018 RSC.
- H. Fallahi, J. Zhang, J. Nicholls, P. Singha, N.-K. Nguyen, C. H. Ooi and N.-T. Nguyen, Research Square, 2021 DOI:10.21203/rs.3.rs-870684/v2.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nh00679g |
| This journal is © The Royal Society of Chemistry 2022 |