Open Access Article
Open Access ArticleThe protein corona reduces the anticancer effect of graphene oxide in HER-2-positive cancer cells†
Lishan
Cui‡
 a,
Erica
Quagliarini‡
b,
Siyao
Xiao
a,
Erica
Quagliarini‡
b,
Siyao
Xiao
 b,
Francesca
Giulimondi
b,
Serena
Renzi
b,
Luca
Digiacomo
b,
Giulio
Caracciolo
b,
Francesca
Giulimondi
b,
Serena
Renzi
b,
Luca
Digiacomo
b,
Giulio
Caracciolo
 b,
Junbiao
Wang
a,
Augusto
Amici
a,
Cristina
Marchini
*a and
Daniela
Pozzi
b,
Junbiao
Wang
a,
Augusto
Amici
a,
Cristina
Marchini
*a and
Daniela
Pozzi
 *b
*b
aSchool of Biosciences and Veterinary Medicine, University of Camerino, 62032 Camerino, Italy. E-mail: cristina.marchini@unicam.it
bNanoDelivery Lab, Department of Molecular Medicine, Sapienza University of Rome, Viale Regina Elena 291, 00161 Rome, Italy. E-mail: daniela.pozzi@uniroma1.it
First published on 24th August 2022
Abstract
In the last decade, graphene oxide (GO)-based nanomaterials have attracted much attention for their potential anti-cancer properties against various cancer cell types. However, while in vitro studies are promising, following in vivo investigations fail to show any relevant efficacy. Recent research has clarified that the wide gap between benchtop discoveries and clinical practice is due to our limited knowledge about the physical–chemical transformation of nanomaterials in vivo. In physiological environments, nanomaterials are quickly coated by a complex dress of biological molecules referred to as the protein corona. Mediating the interaction between the pristine material and the biological system the protein corona controls the mechanisms of action of nanomaterials up to the sub-cellular level. Here we investigate the anticancer ability of GO in SK-BR-3 human breast cancer cells over-expressing the human epidermal growth factor receptor 2 (HER-2), which is functionally implicated in the cell growth and proliferation through the activation of downstream pathways, including the PI3K/AKT/mTOR and MAPK/ERK signaling cascades. Western blot analysis demonstrated that GO treatment resulted in a marked decrease in total HER-2, associated with a down-regulation of the expression and activation of protein kinase B (AKT) and extracellular signal-regulated kinase (ERK) thus indicating that GO may act as a potent HER-2 inhibitor. On the other side, the protein corona reverted the effects of GO on HER-2 expression and molecular downstream events to the control level. Our findings may suggest a mechanistic explanation of the reduced anticancer properties of GO-based nanomaterials in vivo. These results may also represent a good prediction strategy for the anticancer activity of nanomaterials designed for biomedical purposes, reaffirming the necessity of exploring their effectiveness under physiologically relevant conditions before moving on to the next in vivo studies.
Introduction
To predict the effectiveness of a nanosystem designed for biological applications, it must be considered how it behaves within a biological system. First, its biological identity is governed by the interaction between the nanosystem and biological fluids.1,2 In fact, due to their high surface free energy bare nanoparticles once immersed in biological media, are passed by several biomolecules that immediately interact with their surface, alter their synthetic identity and form a protein-enriched shell that is commonly referred to as a ‘protein corona’.3–5 This surface biotransformation has a strong impact on the pharmacological and toxicological profile of nanosystems in an unpredictable manner. As an example, it has been observed that only 0.7% of nanosystem doses reached the target tissue since the bio-nano interactions might perturb their primary function and influence cellular recognition and uptake.6 With the growing application of nanomaterials in biomedicine and considering the increase in human exposure to nano-based therapies and treatments, this aspect is becoming a priority. Especially in the field of cancer therapy, the commercialization of nanoparticle-based anticancer therapeutics is increasing considerably with a rise in the number of available products on the market.7 These include polymeric carriers8,9 (e.g., hydrogels, polymersomes, dendrimers, and nanofibers), lipid-based vehicles10–12 (e.g., liposomes, solid lipid nanoparticles, and micelles), metallic nanoparticles13 (e.g., gold, silver, and titanium), carbon structures (e.g., nanotubes, nanohorns, nanodiamonds), and graphene.14,15 However, less than 10% of such nanotherapeutics get translated into clinical applications, remaining with a large proportion of promising, but clinically ineffective, experimental therapies.16 This makes translational research a long and expensive enterprise which significantly increases health care costs. On the other hand, cancer continues to be one of the leading causes of death worldwide.17 The fact that nanomedicine has opened the door to potential effective therapies prompts many researchers to ask about the underlying cause of clinical failures. Thus, driven by the craving to understand the reason beyond such limitations, we exposed our comprehensive study developing an in vitro evaluation on the impact that a mimetic physiological environment may have on graphene oxide (GO) nanosheets' anticancer capabilities. GO-based nanodevices have always shown great potential in cancer therapy.18,19 One of the key parameters that make GO nanosheet highly fascinating for biological purposes is its high aspect ratio. A sheet of derivatized graphene with oxygen-containing functional group. More particularly, it is the result of graphite's oxidation under acidic conditions. The oxidation process results in the presence of different oxidized functional groups on its surface such as carboxyl groups (–OOH) on the edge of its structure, as well as epoxy (–O) and hydroxyl (–OH) groups on the basal plane. Although GO, like graphene, is a 2D carbon material, its properties are extremely far from that of graphene.20 In fact, the presence of the hydrophilic functional groups favours a more reliable aqueous dispersibility and colloidal stability of both single and few-layered GO. Moreover, the oxygen groups confer to the GO surface a negative charge that allows an easier functionalization with several molecules. Thus, its physical–chemical characteristics can be adjusted to the need of a variety of biomedical applications ranging from gene therapy or delivery of chemotherapeutics to the detection of biomarkers for cancer diagnosis.21 However, if on one hand, the higher GO affinity towards biomolecules allows high bioavailability and versatility, on the other it can turn out to be a double-edged sword since biomolecules present in the physiological environment may cause to a greater extent the alteration of its anticancer capacity. Thus, motivated by the necessity of developing the most reliable anticancer therapeutics, our work was defined to fulfill two requirements: (i) validating the anticancer capacity of GO in both its synthetic and biological forms (ii) understanding the molecular mechanisms underlying the GO anticancer potential. Regarding the first point, we used three different cancer cell lines frequently used in life science research, i.e., U-87 human glioblastoma multiforme (GBM) cell line, HeLa human cervical carcinoma cell line, and CasKi human cervical epidermoid carcinoma cell line, that we treated with naked GO and GO exposed to increasing human plasma (HP) concentrations. As for the second point, we chose SK-BR-3 cells as a model system for the HER-2-positive human breast cancer, one of the most aggressive breast cancer subtypes.22 HER-2 is a transmembrane receptor with tyrosine kinase activity that plays a role in regulating cell growth, survival, differentiation, and tumor proliferation through downstream activation of the PI3K/AKT and MAPK/ERK pathways. When HER-2 is overexpressed at the cell membrane, the downstream pathways are activated constitutively.23 Thus, we validated the ability of GO and human plasma protein-coated GO to inhibit HER-2 driven oncogenic signaling pathways.Materials & methods
Preparation of GO–protein complexes
Graphene oxide (GO) water dispersion was purchased from Graphenea (San Sebastián, Spain). GO nanosheets exhibited a typical one-atom thickness (Fig. S1 in the ESI†).24 Human plasma (HP) was purchased from Sigma-Aldrich, Inc. (Merk KGaA, Darmstadt, Germany). GO–protein complexes were obtained by incubating a GO solution (0.25 mg mL−1) with different percentages of HP (i.e., 2.5% vol., 5% vol., 10% vol., 20% vol., 30% vol., and 50% vol.) for 1 h at 37 °C.Size and zeta potential experiments
Dynamic light scattering (DLS) and zeta-potential analysis were performed to evaluate the size and surface charge of GO in the absence and presence of HP at room temperature using Zetasizer Nano ZS (Malvern Panalytical, Malvern, UK). The measured data are the average of at least 10 runs. The average size and mean zeta potential of each sample were computed using the software provided by the manufacturer.1D SDS-PAGE experiments
GO–protein complexes were subjected to centrifugation at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 RCF for 15 min at 4 °C. Next, the pellets were washed with ultrapure water three times to eliminate free proteins. After the washing step, the pellet was resuspended in 20 μL of Laemmli loading buffer 1×, boiled at 100 °C for 10 min, and centrifuged at 18
620 RCF for 15 min at 4 °C. Next, the pellets were washed with ultrapure water three times to eliminate free proteins. After the washing step, the pellet was resuspended in 20 μL of Laemmli loading buffer 1×, boiled at 100 °C for 10 min, and centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 RCF for 15 min at 4 °C. Finally, 10 μL of supernatants were recovered and loaded on a stain-free gradient polyacrylamide gel (4–20% TGX precast gels, BioRad, Hercules, CA, United States) and run at 100 V for about 150 min. Gel images were obtained with a ChemiDocTM imaging system (Bio-Rad, Hercules, CA, United States) and were processed by ImageLab Software and custom Matlab (MathWorks, Natick, MA, United States) scripts to evaluate the one-dimensional intensity distribution function of each sample and obtain the corresponding one-dimensional molecular weight (MW) distribution. Further details can be found elsewhere.25
620 RCF for 15 min at 4 °C. Finally, 10 μL of supernatants were recovered and loaded on a stain-free gradient polyacrylamide gel (4–20% TGX precast gels, BioRad, Hercules, CA, United States) and run at 100 V for about 150 min. Gel images were obtained with a ChemiDocTM imaging system (Bio-Rad, Hercules, CA, United States) and were processed by ImageLab Software and custom Matlab (MathWorks, Natick, MA, United States) scripts to evaluate the one-dimensional intensity distribution function of each sample and obtain the corresponding one-dimensional molecular weight (MW) distribution. Further details can be found elsewhere.25
Cell culture
U87 human glioblastoma cells, CaSki human cervical epidermoid carcinoma, and HeLa human cervical carcinoma cells were purchased from the American Type Culture Collection (ATTC, Manassas, VA, USA). U87 cells and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, EuroClone), 2% penicillin-streptomycin (Sigma-Aldrich) and 2% L-glutamine (Sigma-Aldrich). CaSki cells were maintained in RPMI medium (Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum, and 2% penicillin-streptomycin, and 2% L-glutamine. SK-BR-3 (ER−/PR−/HER2+) cells were obtained from American Type Culture Collection (Rockville, MD) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco, Life Technologies) and 1% penicillin-streptomycin (Gibco, Life Technologies). Cells were cultivated in T75 flasks and kept at 37 °C in 5% CO2 humidity.Cell viability assay
Cell viability of U87, CaSki, and HeLa cells was evaluated by 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (TXT assay, cell proliferation Kit II, Roche). Cells were seeded on 96-well plates (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and, after 24 h, were incubated with 7.8125, 15.625, 31.25, 62.5, 125, and 250 μg mL−1 concentrations of GO in DMEM plus 10% FBS for U87 and HeLa cells and RPMI plus 10% FBS for CaSki. The cells were incubated for 48 h at 37 °C. Then, 50 μL of TXT solution, previously prepared as indicated in the kit protocol, was added to each well, and cells were incubated at 37 °C for 3 h. After 3 h, the absorbance of each well was measured with Glomax Discover System (Promega, Madison, WI, USA), a detection multi-mode instrument with high performance. As for the SK-BR-3 cells, 105 cells were seeded in each well of the 96-well plate in a complete DMEM medium and incubated overnight at 37 °C with 5% CO2. After 24 h, SK-BR-3 cells were also treated with a series of 7.8125, 15.625, 31.25, 62.5, 125, and 250 μg mL−1 concentration of GO in DMEM supplemented with 2% FBS. After 24 h or 72 h of incubation, 10 μL of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide Sigma Aldrich, St. Louis, MO] was added and the cultures were further incubated for 4 h. After incubation, MTT was aspirated and 100 μL of DMSO (Sigma-Aldrich) was added to dissolve the formazan crystals. The absorbance was measured at 540 nm in the Multiskan Ascent 96/384 Plate Reader. Finally, for all the cell lines 125 and 250 μg mL−1 were chosen as effective concentrations against cancer cell line co-cultured with different percentages (from 2.5% vol. to 50% vol.) of human plasma protein to investigate protein corona's effect.
000 cells per well) and, after 24 h, were incubated with 7.8125, 15.625, 31.25, 62.5, 125, and 250 μg mL−1 concentrations of GO in DMEM plus 10% FBS for U87 and HeLa cells and RPMI plus 10% FBS for CaSki. The cells were incubated for 48 h at 37 °C. Then, 50 μL of TXT solution, previously prepared as indicated in the kit protocol, was added to each well, and cells were incubated at 37 °C for 3 h. After 3 h, the absorbance of each well was measured with Glomax Discover System (Promega, Madison, WI, USA), a detection multi-mode instrument with high performance. As for the SK-BR-3 cells, 105 cells were seeded in each well of the 96-well plate in a complete DMEM medium and incubated overnight at 37 °C with 5% CO2. After 24 h, SK-BR-3 cells were also treated with a series of 7.8125, 15.625, 31.25, 62.5, 125, and 250 μg mL−1 concentration of GO in DMEM supplemented with 2% FBS. After 24 h or 72 h of incubation, 10 μL of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide Sigma Aldrich, St. Louis, MO] was added and the cultures were further incubated for 4 h. After incubation, MTT was aspirated and 100 μL of DMSO (Sigma-Aldrich) was added to dissolve the formazan crystals. The absorbance was measured at 540 nm in the Multiskan Ascent 96/384 Plate Reader. Finally, for all the cell lines 125 and 250 μg mL−1 were chosen as effective concentrations against cancer cell line co-cultured with different percentages (from 2.5% vol. to 50% vol.) of human plasma protein to investigate protein corona's effect.
Western blot assay
After 48 h, treated cells were homogenized in RIPA buffer (0.1% SDS, 1% NP40, 0.5% CHAPS) supplemented with protease inhibitors aprotinin, sodium orthovanadate, and phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO). After 30 minutes of incubation on ice, cell lysates were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C, for 20 minutes. The supernatant was collected, and proteins were quantified via Bradford assay (Bio-Rad). For western blot analysis, equal amounts of protein lysates were separated onto Criterion™ TGX™ precast gels (Bio-Rad) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilion P, Millipore) using Criterion™ Blotter (Bio-Rad). After blocking in EveryBlot Blocking Buffer (Bio-Rad) for 15 minutes, membranes were incubated with primary antibodies (HER-2, pHER-2, ERK, pERK, AKT, pAKT, β-actin, all from Cell Signaling Technology 1
000 rpm, 4 °C, for 20 minutes. The supernatant was collected, and proteins were quantified via Bradford assay (Bio-Rad). For western blot analysis, equal amounts of protein lysates were separated onto Criterion™ TGX™ precast gels (Bio-Rad) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilion P, Millipore) using Criterion™ Blotter (Bio-Rad). After blocking in EveryBlot Blocking Buffer (Bio-Rad) for 15 minutes, membranes were incubated with primary antibodies (HER-2, pHER-2, ERK, pERK, AKT, pAKT, β-actin, all from Cell Signaling Technology 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) at 4 °C overnight. The membranes were washed three times and incubated with secondary antibodies (Sigma-Aldrich, 1
1000) at 4 °C overnight. The membranes were washed three times and incubated with secondary antibodies (Sigma-Aldrich, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20000) at room temperature for 1 h. After TBS-T washing, protein bands were incubated with LiteAblot PLUS reagent (Euroclone) and detected via ChemiDoc™ XRS+ System (Bio-Rad). Densitometry analysis was performed through ImageJ software.
20000) at room temperature for 1 h. After TBS-T washing, protein bands were incubated with LiteAblot PLUS reagent (Euroclone) and detected via ChemiDoc™ XRS+ System (Bio-Rad). Densitometry analysis was performed through ImageJ software.
Data and statistical analysis
Quantitative data are presented as means ± SEM from three independent experiments. The significance of differences was evaluated with a two-tailed Student's t-test or one-way ANOVA followed by Bonferroni or Tukey's multiple comparison post-test. Statistical analysis was executed with GraphPad Prism9 software (San Diego, CA, USA), using p < 0.05 as the critical level of significance (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001).Results and discussion
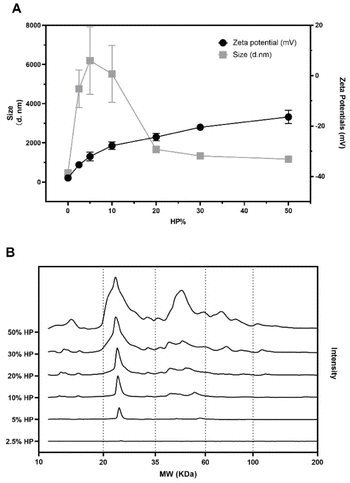
We characterized graphene oxide (GO) nanoflakes in terms of size, and surface charge. We found homogenously dispersed GO nanosheets with a diameter around 200 nm and a zeta potential around −40 mV (see Fig. 1A). However, exposure to biological media alters the physicochemical properties of GO.26 Thus, we investigated the effect of protein concentration on the size and zeta potential of GO nanoflakes exposed to the increasing amount of human plasma (HP). As shown in Fig. 1A, with the increasing HP concentration, zeta potential values passed from −40 mV to around −20 mV at the highest plasma concentration (i.e., 50% HP). A characteristic pattern for the size of GO–protein complexes was also observed. At lower HP concentrations the GO–protein corona complexes were large, meaning that a rapid particle clustering occurred.A further increase of HP concentration resulted in a marked size decrease until a plateau was reached at around 30% HP exposure. To investigate protein binding to GO nanoflakes, 1D SDS-PAGE was performed. A representative gel image and the corresponding data analysis are reported in Fig. S2.† The one-dimensional (1D) absolute protein patterns are shown in Fig. 1B.
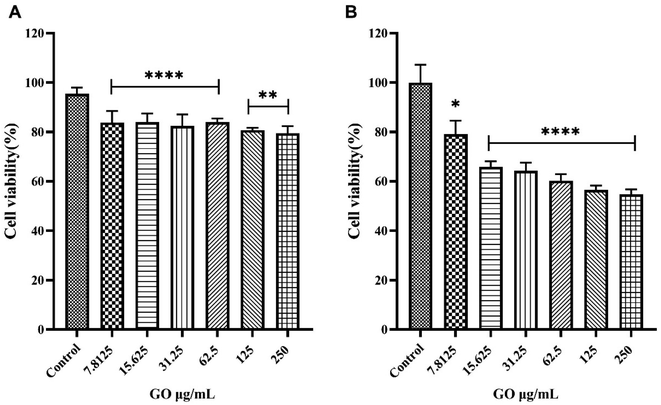
At the lowest protein concentration (i.e., 2.5% HP) the intensity profile exhibited a barely visible single peak around 25 kDa, which becomes more intense at HP = 5%. The complexity of the protein pattern increased with increasing HP percentage, with the appearance of peaks in the molecular weight (MW) intervals from 35 kDa to 60 kDa at 10% HP and from 60 kDa to 100 kDa at 30% HP. Globally these results show that as the HP percentage increased, more protein bound to the GO, and the complexity of the PC itself became greater. As a next step, the anticancer activity of naked GO was evaluated using an XTT assay. To this end, we employed three different models of cancer cell lines typically used in the field of cancer therapy and drug testing, i.e., U-87 human glioblastoma multiforme (GBM) cell line, HeLa human cervical carcinoma cell line, and CasKi human cervical epidermoid carcinoma cell line. After 48 h treatments, a remarkable decrease in cell viability occurred as a function of GO concentration until a minimum was reached at a GO concentration of 250 μg mL−1 with cell viability of around 40% for all the three cell lines (see Fig. S3 in the ESI†).
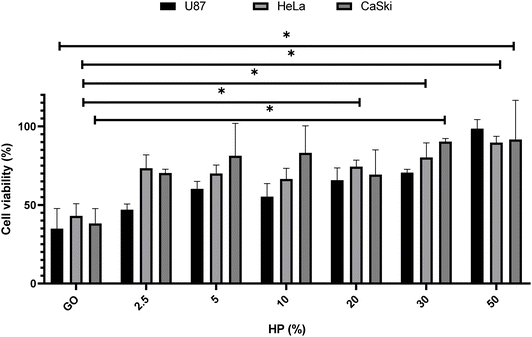
This step was mandatory for choosing the optimal GO concentration for protein corona studies, i.e., the one that produced the largest impact on cell viability. Thus, GO at 250 μg mL−1 was incubated with different percentages of HP (i.e., from 2.5% to 50%) (Fig. 2).
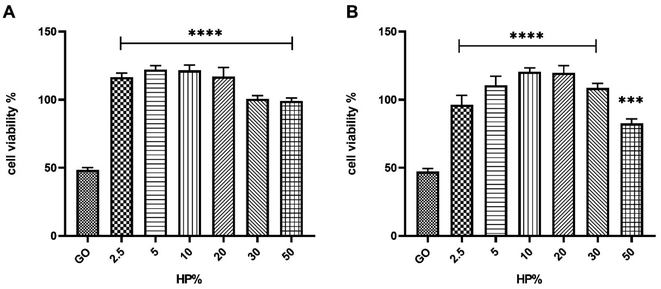
As shown in Fig. 2, for all three cell lines a significant boost in cell viability with increasing HP percentage was observed. At the highest plasma concentration, the cell viability was close to 90% for all three cell lines. Results displayed in Fig. 2 are in line with previous findings27,28 and suggest that the anti-cancer effect of GO may be drastically impaired in a protein-enriched physiological environment likely due to the reduction in cell penetration. This phenomenon has been studied for a wide variety of nanoparticles and cell systems.29 For instance, it has been demonstrated that the uptake levels change when particles are exposed to cells in a serum-free medium or with a preformed protein coating.30 In particular, even though the underlying uptake mechanisms are mostly uncertain, it has been noted that in serum-free ambient the nanoparticle uptake is higher than the one in the presence of serum31 and may lead to higher cytotoxicity. Thus, this behavior could have repercussions on the toxicity levels of the system. To deepen into this aspect and, therefore, clarify the molecular mechanisms underlying the anticancer potential of GO under biologically relevant conditions, we explored the effect of coronated GO on the viability of HER-2 positive cancer cells and used pristine GO as a reference. As a model of HER-2 positive cancer cells, we used SK-BR-3 cancer cells that over-express HER-2 and tend to grow and spread faster than other breast cancer subtypes. Several pharmacological treatments have been developed to target specifically HER-2 protein and inhibit its function, such as trastuzumab, pertuzumab, lapatinib, and ado-trastuzumab emtansine (T-DM1).32 Although HER-2 targeted drugs have significantly improved survival outcomes for HER-2-positive breast cancer patients, some of them may show no response or develop drug resistance after a period of treatment. Thus, there is still a need to develop new anti HER-2 anticancer agents and understand their modes of action. As a first step, SK-BR-3 cells were incubated with increasing concentrations of GO for 48 hours (Fig. 3A) or 72 hours (Fig. 3B). As Fig. 3 clearly shows, GO decreased the cell viability of SK-BR-3 cells in a dose-dependent manner with the GO concentrations of 125 and 250 μg mL−1 exhibiting a remarkable efficacy after 72 hours of treatment.
For both 125 μg mL−1 and 250 μg mL−1 of GO concentrations and the longest incubation time (i.e., t = 72 hours), the impact on the cell viability was weakened in the presence of the protein corona (Fig. 4).
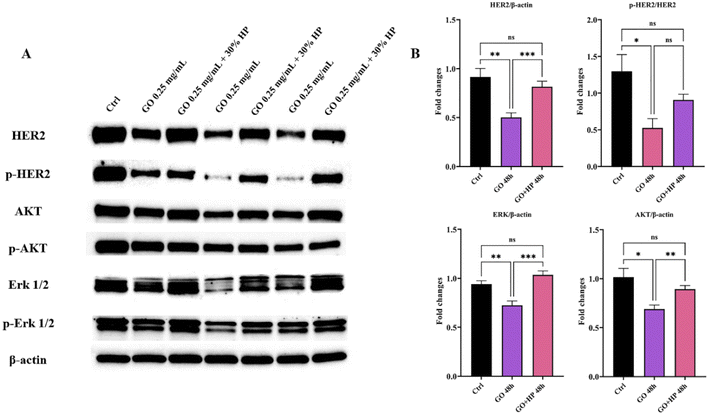
Similar results were obtained for the largest GO concentration and the lowest incubation time (i.e., t = 48 hours) (Fig. S4 in the ESI†). Considering the greater efficacy displayed by 30% HP with respect to the others, we selected it for further studies. To provide mechanistic insights on the anticancer potential of GO, we performed a western blot analysis on SK-BR-3 cells, that exhibit an enhanced activation of HER-2-driven signaling pathways such as phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathways, which mediate cancer cell survival and proliferation. We observed that GO treatment resulted in a marked decrease in total HER-2, in a drastic reduction of the level of phosphorylated HER-2, indicating that GO may act as a potent HER-2 inhibitor, in association with a down-regulation of the expression and activation of ERK and AKT (Fig. 5). Interestingly, the presence of 30% HP significantly reduced the anticancer activity of GO, showing that the expression of HER-2 and its molecular downstream events reverted to the control level. Similar results were obtained using 50% HP (Fig. S5 in the ESI†). The ability of HP to override GO effect was likely caused by interdicted GO physical interaction with HER-2 exposed to cell membrane.
Conclusion
The protein corona that covers nanomaterials in a physiological environment is the molecular interface that determines their physiological response. The limited knowledge of the role of the protein corona is believed to be the main factor limiting the translation of nanomaterials into clinical practice. Previous studies have shown that the protein corona reduces interaction with the plasma membrane by endowing the nanomaterials with stealth properties. This aspect is particularly relevant when the protein coating allows preventing interaction with some cell types (e.g., immune cells). In this scenario, several strategies have been delineated to reduce the impact of protein corona on the effectiveness of nanomaterials. These include the use of different ligands (such as peptides, hydrophobin, etc.) or stealth polymers (e.g., polyethylene glycol (PEG), polyvinylpyrrolidone (PVP), dextran, etc.) to avoid the involving of opsonin proteins in the corona, guaranteeing the maintenance of the targeting efficiency of nanomedicines and the escape from the mononuclear phagocyte system.33 Moreover, besides ligand and stealth polymer-based approach, recently the artificial protein corona has greatly emerged, namely by pre-coating the nanomaterials surface with antibodies or proteins to modulate the protein corona formation and induce an active cancer-targeting aimed at ensuring the specific therapeutic action of the system.34 On the other hand, few studies have investigated how the altered interaction with the plasma membrane caused by the protein corona can influence the anticancer properties of nanomaterials. In this study, we used SK-BR-3 human breast cancer cells as model system of HER-2 positive cancer cells and GO as a model of HER-2 inhibitor. We have shown that the GO treatment resulted in a marked decrease in total HER-2, associated with a down-regulation of the expression and activation of protein kinase B (AKT) and extracellular signal-regulated kinase (ERK). On the other side, the protein corona reverted the effects of GO on HER-2 expression and molecular downstream events to the control level. Moreover, this work reaffirms the absolute need to conduct in vitro studies on anti-tumor properties of nanomaterials under experimental conditions that accurately mimic the physiological environment.Author contributions
Conceptualization, G. C., D. P. A. A. and C. M.; data curation, L. D.; formal analysis, L. D., funding acquisition, G. C. and D. P.; investigation, L. C., E. Q., S. X., F. G., S. R. and J. W.; project administration, D. P. and C. M.; resources, D. P. and C. M.; software, L. D.; supervision, G. C., D. P., A. A and C. M.; validation, L. C. and E. Q.; visualization, L. D.; writing – original draft, L. C., E. Q.; S. X. and G. C.; writing – review & editing, G. C., D. P. and C. M.Conflicts of interest
There are no conflicts to declare.References
- G. Caracciolo, O. C. Farokhzad and M. Mahmoudi, Biological identity of nanoparticles in vivo: clinical implications of the protein corona, Trends Biotechnol., 2017, 35(3), 257–264 CrossRef CAS PubMed.
- L. Digiacomo, Impact of the protein corona on nanomaterial immune response and targeting ability, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12(4), e1615 CAS.
- P. C. Ke, A decade of the protein corona, ACS Nano, 2017, 11(12), 11773–11776 CrossRef CAS PubMed.
- L. Treuel, Impact of protein modification on the protein corona on nanoparticles and nanoparticle–cell interactions, ACS Nano, 2014, 8(1), 503–513 CrossRef CAS PubMed.
- G. Caracciolo, The protein corona effect for targeted drug delivery, Bioinspired, Biomimetic Nanobiomater., 2013, 2(1), 54–57 CrossRef CAS.
- S. Wilhelm, Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater., 2016, 1(5), 1–12 Search PubMed.
- Y. Yao, Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance, Front. Mol. Biosci., 2020, 193 CrossRef CAS PubMed.
- J. Ding, Polymer-mediated penetration-independent cancer therapy, Biomacromolecules, 2019, 20(12), 4258–4271 CrossRef CAS PubMed.
- H. Yang, Targeted nanosystems: advances in targeted dendrimers for cancer therapy, Nanomedicine, 2016, 12(2), 309–316 CrossRef CAS PubMed.
- A. Arcella, Brain targeting by liposome–biomolecular corona boosts anticancer efficacy of temozolomide in glioblastoma cells, ACS Chem. Neurosci., 2018, 9(12), 3166–3174 CrossRef CAS PubMed.
- G. Perini, Inhibiting the Growth of 3D Brain Cancer Models with Bio-Coronated Liposomal Temozolomide, Pharmaceutics, 2021, 13(3), 378 CrossRef CAS PubMed.
- E. Quagliarini, Microfluidic formulation of DNA-loaded multicomponent lipid nanoparticles for gene delivery, Pharmaceutics, 2021, 13(8), 1292 CrossRef CAS PubMed.
- A. Sharma, A. K. Goyal and G. Rath, Recent advances in metal nanoparticles in cancer therapy, J. Drug Targeting, 2018, 26(8), 617–632 CrossRef CAS PubMed.
- E. Quagliarini, Effect of protein corona on the transfection efficiency of lipid-coated graphene oxide-based cell transfection reagents, Pharmaceutics, 2020, 12(2), 113 CrossRef CAS PubMed.
- Y. Wang, Emerging 2D material-based nanocarrier for cancer therapy beyond graphene, Coord. Chem. Rev., 2019, 400, 213041 CrossRef CAS.
- P. Satalkar, Challenges of clinical translation in nanomedicine: a qualitative study, Nanomedicine, 2016, 12(4), 893–900 CrossRef CAS PubMed.
- R. L. Siegel, Cancer statistics, 2022, Ca-Cancer J. Clin., 2022, 72, 7–33 CrossRef PubMed.
- A. Shafiee, S. Iravani and R. S. Varma, Graphene and graphene oxide with anticancer applications: challenges and future perspectives, MedComm, 2022, 3(1), e118 CrossRef CAS PubMed.
- E. Quagliarini, Mechanistic insights into the release of doxorubicin from graphene oxide in cancer cells, Nanomaterials, 2020, 10(8), 1482 CrossRef CAS PubMed.
- S. Priyadarsini, Graphene and graphene oxide as nanomaterials for medicine and biology application, J. Nanostruct. Chem., 2018, 8(2), 123–137 CrossRef CAS.
- C. Chung, Biomedical applications of graphene and graphene oxide, Acc. Chem. Res., 2013, 46(10), 2211–2224 CrossRef CAS PubMed.
- T. Sørlie, Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(19), 10869–10874 CrossRef PubMed.
- Y. Yarden and M. X. Sliwkowski, Untangling the ErbB signalling network, Nat. Rev. Mol. Cell Biol., 2001, 2(2), 127–137 CrossRef CAS PubMed.
- V. Palmieri, Graphene oxide coatings prevent Candida albicans biofilm formation with a controlled release of curcumin-loaded nanocomposites, Nanomedicine, 2018, 13(22), 2867–2879 CrossRef CAS PubMed.
- L. Digiacomo, The biomolecular corona of gold nanoparticles in a controlled microfluidic environment, Lab Chip, 2019, 19(15), 2557–2567 RSC.
- M. Papi, Plasma protein corona reduces the haemolytic activity of graphene oxide nano and micro flakes, RSC Adv., 2015, 5(99), 81638–81641 RSC.
- G. Duan, Protein corona mitigates the cytotoxicity of graphene oxide by reducing its physical interaction with cell membrane, Nanoscale, 2015, 7(37), 15214–15224 RSC.
- Y. Yang, Protein corona reduced graphene oxide cytotoxicity by inhibiting endocytosis, Colloid Interface Sci. Commun., 2021, 45, 100514 CrossRef.
- A. Lesniak, Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency, J. Am. Chem. Soc., 2013, 135(4), 1438–1444 CrossRef CAS PubMed.
- Y. Yan, Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines, ACS Nano, 2013, 7(12), 10960–10970 CrossRef CAS PubMed.
- A. Lesniak, Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells, ACS Nano, 2012, 6(7), 5845–5857 CrossRef CAS PubMed.
- X. Zhang, Efficacy and Safety of Anti-HER2 Agents in Combination with Chemotherapy for Metastatic HER2-Positive Breast Cancer Patient: A Network Meta-Analysis, Front. Oncol., 2021, 3183 Search PubMed.
- Y. Zou, Polyglycerol grafting shields nanoparticles from protein corona formation to avoid macrophage uptake, ACS Nano, 2020, 14(6), 7216–7226 CrossRef CAS PubMed.
- F. Giulimondi, Opsonin-deficient nucleoproteic corona endows unPEGylated liposomes with stealth properties in vivo, ACS Nano, 2022, 16(2), 2088–2100 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2na00308b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |