Open Access Article
Open Access ArticleMag-spinner: a next-generation Facile, Affordable, Simple, and porTable (FAST) magnetic separation system†
Sanghoon
Lee
a,
Miseon
Jeong
a,
Soojin
Lee
b,
Sang Hun
Lee
a and
Jin-sil
Choi
 *a
*a
aDept. of Chemical and Biological Engineering, Hanbat National University, 34158 Daejeon, Republic of Korea. E-mail: jinsil.choi@hanbat.ac.kr
bDept. of Microbiology & Molecular Biology, Chungnam National University, 34134 Daejeon, Republic of Korea
First published on 23rd December 2021
Abstract
Mag-spinner, a system in which magnets are combined with a spinner system, is a new type of magnetic separation system for the preprocessing of biological and medical samples. Interference by undesired components restricts the detection accuracy and efficiency. Thus, the development of appropriate separation techniques is required for better detection of the desired targets, to enrich the target analytes and remove the undesired components. The strong response of iron oxide nanoclusters can successfully capture the targets quickly and with high efficiency. As a result, cancer cells can be effectively separated from blood using the developed mag-spinner system. Indeed, this system satisfies the requirements for desirable separation systems, namely (i) fast sorting rates, (ii) high separation efficiency, (iii) the ability to process native biological fluids, (iv) simple operating procedures, (v) low cost, (vi) operational convenience, and (vii) portability. Therefore, this system is widely applicable to sample preparation without limitations on place, cost, and equipment.
Introduction
The development of an efficient separation system is crucial in many fields, including biology, biotechnology, and health/medicinal science.1–5 The separation of target analytes from complex mixtures is generally the first step for the precise detection and analysis of targets. However, while significant advances have been made for establishing effective analytical tools, interference by undesired components restricts the detection accuracy and efficiency of the tools. Therefore, the enrichment of low-abundance target analytes and removal of interfering species by appropriate separation techniques is imperative for better detection of the desired targets.4,6–9 Most current separation systems are designed based on the control of various parameters, such as the filter, inertia, fluid flow, dielectrophoresis, acoustic force, and magnetic attraction.3,5,6,8,10 Most of these processes are performed in laboratories because they require complex instruments and infrastructure. In addition, some analytes such as proteins, DNA, and metabolites are easily degraded in the sample mixture and thus, on-site or quick separation is necessary for accurate detection.5,11,12For the effective detection and analysis of targets, it is important to develop new separation tools that meet the following requirements:4 (i) fast sorting rates, (ii) high separation efficiency, (iii) the ability to process native biological fluids, (iv) simple operating procedures, (v) low cost, (vi) operational convenience, and (vii) portability. Among the various separation techniques, magnetic separation is one of the most effective methods for the high throughput and low-cost isolation of target analytes.13–21 Effective magnetic separation makes it possible to isolate circulating tumor cells in the blood for further analysis, which is crucial in cancer diagnosis and the development of personalized treatments.22–26 Bacteria at very low concentrations can also be purified and concentrated effectively through magnetic separation to monitor food poisoning and infectious disease.27–33 The magnetic activated cell sorting (MACS) system developed by Miltenyi Biotec, is a representative commercialized magnetic separation tool that can separate magnetically tagged targets using a magnetic column. While widely utilized, the MACS system is also designed for laboratory use. Recently, a MACS-based pipette system was reported for high-throughput magnetic separation via simple pipetting.34,35 The MACS tips are designed with permeant magnets and layers of nickel meshes with micropores to generate a magnetic field inside the tip. Although this MACS tip system is applicable in a wider range of settings with high separation efficiency, the cost for micropipettes is a bit high to be equipped in all settings, especially in third world countries, and the tips must be manufactured according to the type of micro-pipette used. Therefore, further investigation is essential to develop the next generation of separation tools.
Herein, we introduce a new type of magnetic separation system, the magnetic spinner (mag-spinner), which is a Facile, Affordable, Simple, and porTable (FAST) separation system of target analytes from complex samples (Scheme 1). Mag-spinner adopts a fidget spinner system so that any unwanted residues can be removed automatically by simple flicking of the spinner. Notably, mag-spinner possesses a compact and simple structure that can be easily produced by 3D printing. Additionally, the separation process employing mag-spinner can be performed without any limitation of place or facility, owing to its hand-held size and simple operation. Iron oxide nanoclusters (NCs), which strongly respond to a magnetic field in a few seconds, are utilized as the magnetic tag. Thus, low-concentration targets can be effectively isolated and concentrated using mag-spinner. Finally, the successful separation of cancer cells from biological fluid using mag-spinner suggests that mag-spinner is a suitable candidate for high-throughput separation methods.
Experimental section
Reagents
All reagents used in the study were used as purchased without further purification. These include iron(III) chloride hexahydrate (FeCl3·6H2O, Sigma-Aldrich), ethylene glycol (99.5%, SAMCHUN), sodium acetate anhydrous (NaOAc, Duksan), polyacrylic acid (PAA, MW: 5000, 50% aqueous solution, Acros Organics), N,N′-dicyclohexylcarbodiimide (DCC, Sigma-Aldrich), 4-dimethylaminopyridine (DMAP, Sigma-Aldrich), biotin (Sigma-Aldrich), dimethyl sulfoxide (DMSO, Sigma-Aldrich), fluorescein isothiocyanate (FITC, Sigma-Aldrich), (3-aminopropyl)trimethoxysilane (APTMS, Sigma-Aldrich), tetraethyl orthosilicate (TEOS, TCI), ethyl alcohol (95%, SAMCHUN), ammonia solution (28–30%, SAMCHUN), potassium chloride (99%, SAMCHUN), egg-white avidin (Sigma-Aldrich), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC, >98%, Acros Organics), N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS, Alfa Aesar), Arg-Gly-Asp (RGD, ≥97%, Sigma-Aldrich) and photopolymer resin (XYZ Print), Lipofectamine™ 3000 Transfection Reagent (Invitrogen), DMEM (high glucose, Simplybiologics). A549 cells were purchased from American Type Culture Collection (USA) and grown in Dulbecco's modified Eagle Medium supplemented with 10% fetal bovine serum. Blood was obtained after CO2 euthanasia of a mouse, and all animal procedures were approved by the Animal Ethics Committee of Chungnam National University.Equipment
The shape and size of the NCs were analyzed by SEM (cold-type FE-SEM, S-4800, Hitachi High Technology, Japan). The hydrodynamic size of the NCs in aqueous solution was measured using a zetasizer (ZSU3200, Malvern PANalytical, England), while their hysteresis curve was obtained by vibrating sample magnetometry (VMS, 7400-S, Lake Shore Cryotronics, USA). The designed mag-spinner was fabricated with a 3D printer (Nobel Superfine, XYZ printing, Taiwan). Finally, fluorescent images of the silica particles and cancer cells captured by mag-spinner were obtained using an Epifluorescence microscope (AE30-31, MOTIC, China).Nanocluster (NC) synthesis
The NCs were synthesized using the solvothermal method.36 In a typical procedure, FeCl3·6H2O (0.34 g, 1.26 mmol), PAA (0.293 g, 0.15 mmol), and NaOAc (3.681 g, 44.87 mmol) were dissolved in 20 mL ethylene glycol. The mixture was vacuumed for several minutes to remove the low-boiling-point impurities and subsequently heated to 160 °C for 1 h under Ar with vigorous stirring. The reaction mixture was then transferred to a Teflon-equipped stainless-steel autoclave and reacted at 200 °C for 10 h. After cooling down to room temperature, the black precipitate was purified by removing the supernatant through centrifugation (3000 rpm) with acetone addition and then dispersed in an aqueous solution.Conjugation of avidin or RGD with NCs
The NCs were conjugated with avidin or RGD for the recognition of targets (biotin-FITC@SiO2 or cancer cells). The NCs (0.2 mg) were dissolved in 200 μL of citrate buffer (10 mM, pH 6) containing EDC (2 mM) and sulfo-NHS (5 mM) and reacted for 15 min at room temperature. Any unreacted reagents were removed using a Zeba™ Spin Desalting Columns (7 K MWCO, ThermoFischer, USA), and the NCs were dispersed in phosphate buffer (10 mM, pH 7.2). Subsequently, the NCs were further incubated with avidin [0.2 mg, 0.003 μmol; 0.015 mM in 200 μL phosphate buffer (pH 7, 10 mM)] or RGD peptide [0.2 mg, 0.58 μmol; 2.5 mM in 200 μL phosphate buffer (pH 7, 10 mM)] for 2 h at room temperature. The avidin/RGD-conjugated NCs were then purified by collecting with a magnet three times.Results and discussion
The details of the developed mag-spinner
The fidget spinner is a toy that is simply rotated by hand; however, it shows applicability to the fluidic chip for point-of-care testing (POCT) in that the centrifugal force can be formed by hand without any equipment.37,38 Therefore, with this in mind, we designed a combined magnet-spinner system as a new type of separation system. The designed mag-spinner was printed out using a 3D printer (Fig. 1a and S1†), and comprises an inlet, magnet-containing separation section, flow path, withdrawal region, and outlet [Fig. 1a(i) and (ii)]. Two magnets (NdFeB, 10 × 10 × 2 mm3) are placed at the center of the magnetic spinner, and the flow paths are located around the magnets (sky blue region). Notably, the compact size of mag-spinner (44 × 31 × 5 mm3) makes it convenient to operate single-handedly (Fig. S1†). The system operates as follows: first, an aqueous solution containing a medical sample (e.g., saliva, urine, feces, and blood) is mixed with magnetic nanoparticles conjugated to target-specific biomolecules and incubated for a few minutes (Fig. 1b(i)). After injecting the mixture solution to the mag-spinner (Fig. 1b(ii)), the magnetically tagged targets are effectively separated with several rotations of the device, and the solution containing the components that are not of analytical interest (waste solution) is drained out to the withdrawal region [Fig. 1b(ii) and (iii)]. Once the waste solution is removed by syringe [Fig. 1b(iv)], the magnetically tagged target is released by removing the magnets from mag-spinner and adding extra saline into the flow path [Fig. 1b(v)]. The medically important targets can then be recovered by the facile spinning of mag-spinner [Fig. 1b(vi) and (vii)] for further analysis. Mag-spinner is easily portable and is operated by the simple flicking of one finger. Therefore, the mag-spinner system satisfies four of the next-generation separation method requirements (i.e., simple operation, operational convenience, low cost, and portability) listed in the Introduction.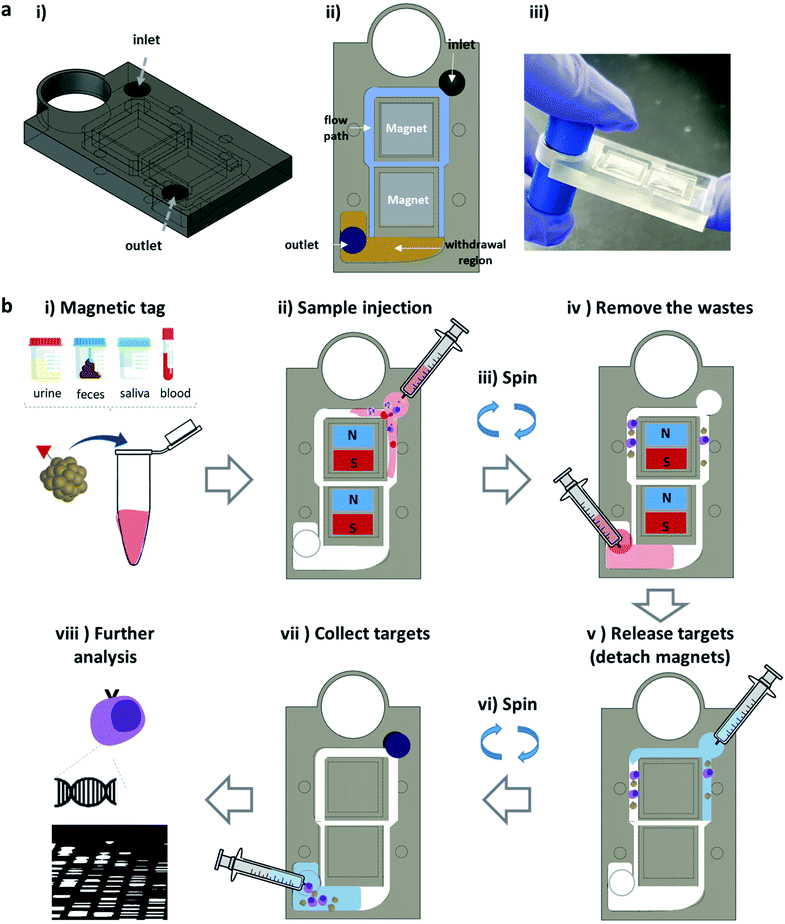 | ||
| Fig. 1 (a) Schematics and picture of the developed mag-spinner fabricated by 3D printing. (b) Schematics of the mag-spinner operation process. | ||
Magnetic tag: iron oxide NCs
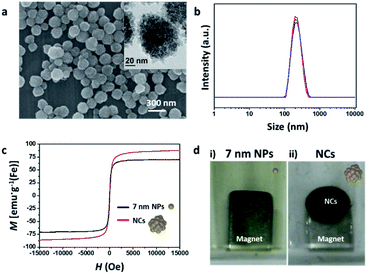
An effective mag-spinner can be realized by using magnetic tags that can sensitively respond to magnets.19 The pulling force of the magnetic nanoparticles by these magnets is proportional to both their saturation magnetization and size.19,20 The magnetic properties of single nanoparticles are improved by controlling their size, shape, and components; however, they still display practical limitations. On the other hand, NCs, which are formed by clustering numerous nanoparticles into well-defined structures, are good candidates as effective magnetic tags because of their superior pulling force over that of single nanoparticles, attained by modulating their magnetic properties and size. In this study, we synthesized iron oxide NCs via a previously reported solvothermal method using PAA, to stabilize the surface of the iron oxide nanoparticles.36 The obtained NCs were spherical and ∼180 nm in size (Fig. 2a). TEM microscopy confirmed that as-synthesized NCs are composed of densely assembled 7 nm nanoparticles (Fig. 2a, inset and Fig. S2†), and their hydrodynamic size was measured as 232 nm (Fig. 2b). The negative charge (−36.31 eV) of the NCs originated from the PAA used to stabilize the NC surface (Fig. S3†). VSM data revealed that the nanoclusters exhibited a saturation magnetization (Msat) of 87 emu g−1 (Fe), which is 139% higher than that of the 7 nm iron oxide nanoparticles (67 emu g−1; Fig. 2c). This increased Msat of the NCs was attributed to the minimized spin-canting effect, owing to close contact between the densely aggregated unit particles.36 When a NdFeB magnet was placed under a solution containing 0.42 mg mL−1 NCs or 7 nm nanoparticles, most of the NCs were pulled down toward the magnet and then gathered around the edge of the magnet within 20 s (dark brown spot in Fig. 2d, right image), while the 7 nm nanoparticles were dispersed in the aqueous solution without any noticeable attracted particles around the magnets (Fig. 2d, left image). Moreover, no recognizable size variation was observed when the collected NCs were redispersed into an aqueous solution (Fig. S4†). Therefore, we supposed that NCs can be applied as adequate magnetic tags to drag targets effectively and quickly to the magnet.Capture of NCs using mag-spinner
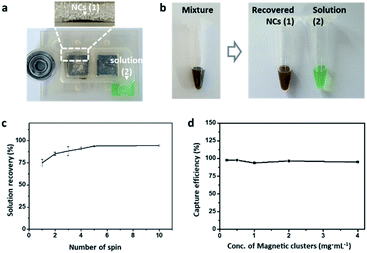
The separation capability of mag-spinner was next examined through the isolation of NCs from the mixture solution (Fig. 3a). The NC-containing aqueous solution was mixed with green food coloring to clearly visualize the movement of the solution in mag-spinner (Fig. 3b). After rotating mag-spinner, the dark brown NCs remained around the magnet, while the green solution moved to the withdrawal region. Notably, >95% (v/v) of the solution successfully migrated to the withdrawal region within five spins (Fig. 3c). The concentration of NCs in an aqueous solution is determined by measuring the absorbance of the solution according to Beer's law (extinction coefficient: 18 mL g−1 cm−1). Based on the NC concentration in the drained-out solution, we confirmed that >98% of the NCs were captured by the magnet in the NC concentration range 0.1–4 mg(Fe) mL−1 (Fig. 3d). On the other hand, the 7 nm Fe3O4 nanoparticles were barely captured by the magnets and thus, the mixture solution retained its brownish-green color, even after separation using mag-spinner (Fig. S5†). Therefore, we concluded that NC-tagged targets can be captured by the magnets with high efficiency in a short time, thereby satisfying another two requirements, namely fast sorting rates and high separation efficiency.Forces applied on magnetic particles in mag-spinner
The theoretical calculations of the force applied to the magnetic particles inside the mag-spinner system help understand the excellent separation effect of the NC particles shown above. In mag-spinner, three major forces are applied to the NCs: a pulling force that attracts the NCs toward the magnets, a drag force that opposes magnetic material movement through the solution, and a centrifugal force that drains the solution down from the spinner (Fig. 4a). The pulling force is determined by the gradient of the magnetic flux density of the magnet and the size and saturation magnetization of the magnetic particles. In the magnetic spinner, the flow path is 1 mm away from the magnet and its width is ∼1 mm. The magnetic flux density over the flow path was calculated based on a simulation using Finite Element Method Magnetics (FEMM) open-source software (Fig. S6†). The pulling force acting on the NCs in the flow path was calculated to be in the range 1–2.5 pN (top of aligned magnets, red line) or 4.7–10 pN (side of aligned magnets, blue line), depending on the location of the NCs (Fig. 4b). In addition, the drag force was calculated as 27.3 fN, based on the distance traveled for a certain period (Fig. S7†), a value that was considered negligible compared to the calculated pulling force. The centrifugal force was determined from the angular speed of rotation of the spinner and the distance from the spinning center. The average angular speed was measured as ∼240 rpm, and the distance that the sample moved in mag-spinner by centrifugal force was measured as 1–4 cm from the spinning center (Fig. S8†). Thus, the centrifugal force of the NCs was calculated as 0.1–0.5 fN (Fig. 4c). Because the magnetic pulling force was higher than the drag and centrifugal forces around the injection site, where the particles were mainly attached (Fig. 4a), the NCs were effectively captured by the magnets, and any undesired components were easily removed. Notably, the magnetic pulling force applied to the 7 nm iron oxide nanoparticles was significantly smaller (aN level) than that applied to the NCs (Fig. S9†). These results revealed that NCs can be effectively captured by the mag-spinner magnets, overcoming the centrifugal force.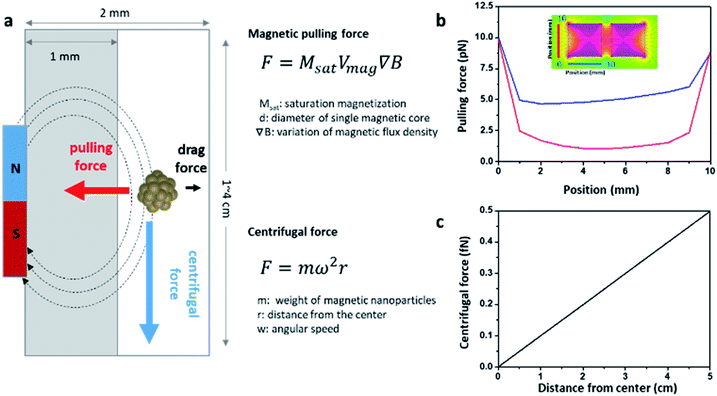 | ||
| Fig. 4 (a) Schematics of the forces applied on the NCs across the mag-spinner flow path (top view). Calculated (b) magnetic pulling force and (c) centrifugal force. | ||
Capture of fluorescent silica particles using mag-spinner
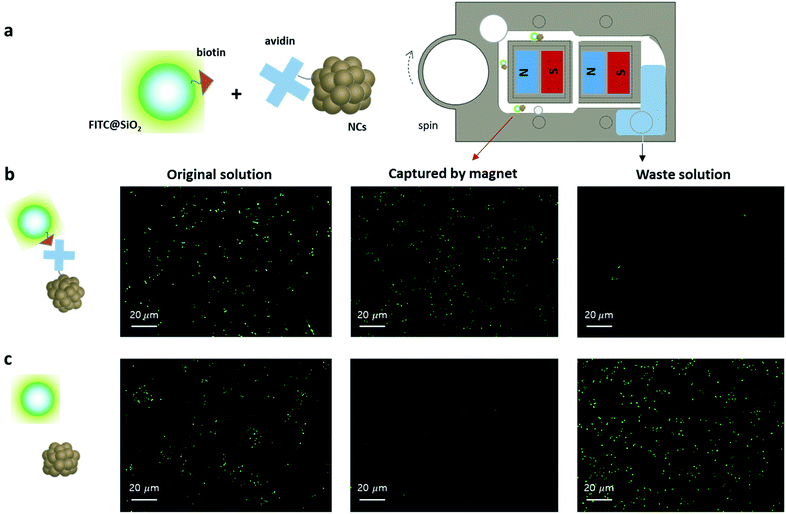
Once the effective NC capture capability of the developed mag-spinner was confirmed, we proceeded to apply the system to separate specific targets, e.g., fluorescent silica particles (FITC-SiO2, Fig. S10†), from an aqueous solution to evaluate its applicability to magnetic separation (Fig. 5a). Thus, NCs and FITC-SiO2 were conjugated to avidin and biotin, respectively, to achieve biotin–avidin recognition, whereby the biotin–avidin interaction is the strongest non-covalent interaction between a protein and monovalent ligand (Ka = 1015 M−1).39–41 After mixing the biotin–FITC@SiO2 and avidin–NC conjugates for 10 min, FITC@SiO2 was isolated following the method illustrated in Fig. 1b. As a control, pristine FITC@SiO2 and NCs, without biotin or avidin conjugation, were tested following the same separation procedures as those of their corresponding conjugated forms. The presence of FITC@SiO2 in each solution was confirmed by the green fluorescence in the microscopic images. Indeed, strong green fluorescence originating from FITC@SiO2 was observed in the recovered NC solution for the biotin–FITC@SiO2 and avidin–NC mixture, while negligible green fluorescence was observed in the waste solution (Fig. 5b). On the other hand, in the control group, strong green fluorescence was observed in the waste solution, while no recognizable green fluorescence was detected in the recovered NC solution (Fig. 5c). These results confirmed that biotin–FITC@SiO2 can be efficiently separated by the mag-spinner–NC system.Separation of cancer cell from blood using mag-spinner
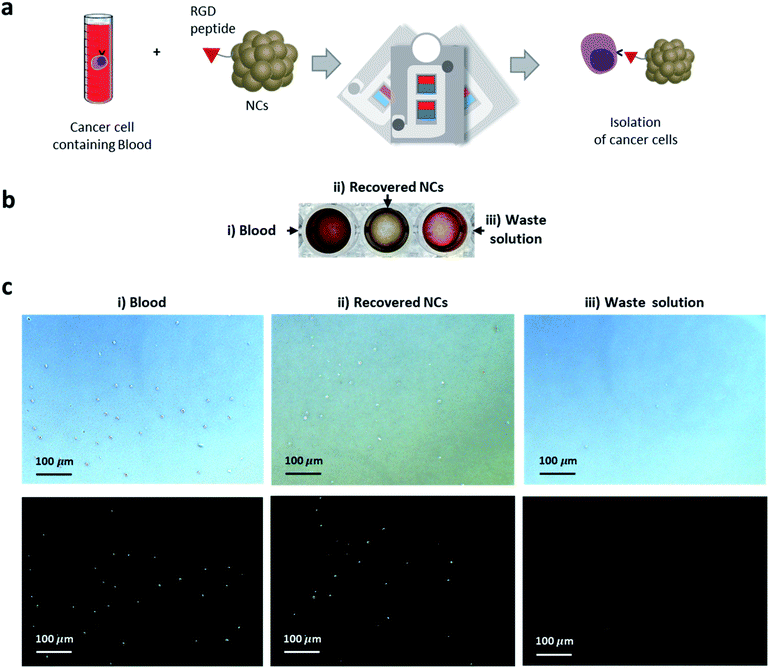
The applicability of mag-spinner for the preprocessing of medical samples was next examined through the separation of biological targets from a biological fluid. Circulating tumor cells (CTCs) are cells that have shed into the vasculature or lymphatics from a primary tumor and are carried around the body in the blood circulation, causing cancer metastasis.42–44 Therefore, separation of CTCs is important because their existence serves as an indicator to determine the status of a cancer, and the same biological composition of the CTC-to-mother tumor tissue can be helpful to develop personalized treatments.44,45 However, CTC population in blood is very rare and thus, the development of an effective separation method is highly desirable.46,47 In this study, separation of A549, an adenocarcinoma human alveolar basal epithelial cell, was performed using mag-spinner. RGD peptides, which can recognize integrin overexpressed on the surface of A549 were conjugated on the NC surface (Fig. 6a). The green fluorescent protein (GFP) was expressed in A549 cells to enable the cells to be identified with green fluorescence in fluorescence microscopic images. A549 cells expressing GFP were incubated with RGD–NCs and subsequently separated using mag-spinner. After spinning the mag-spinner, all the blood solution moved to the withdrawal site and the recovered NC solution turned brown (Fig. 6b). In the fluorescent images (Fig. 6c), vivid green fluorescent signals from the A549 cells were observed in the recovered NC solution, which matched those displayed in the bright-field images. On the other hand, no green fluorescence was observed in the blood solution. In addition, no recognizable cell growth was observed when the waste solution was cultured for 4 d (Fig. S11†). Moreover, the cell damage caused by mag-spinner physical stimulation was deemed negligible (Fig. S12†). Therefore, the mag-spinner system shows great potential as a new type of separation method. In addition, the concentration of the separated samples can be adjusted according to the demand of the next analysis step, so that low-abundance samples can be concentrated effectively to overcome the detection limits of analytical tools.Conclusions
Briefly, successful target separation and concentration were performed using a new type of FAST magnetic separation system, which we named mag-spinner. The developed system satisfies the suggested requirements for next-generation separation methods. Although mag-spinner is a prototype, further study to optimize the fluidics and magnetic force in the spinner system can be performed to enable pretreatment of more complex systems than those performed in this study. The effective preprocessing of CTC using mag-spinner can increase the detection sensitivity and accuracy, thus enabling diagnosis in the early or early metastasis stages of cancer and the development of personalized treatment. Mag-spinner is finger operated and the separation process is simple and fast. Moreover, the structure of the mag-spinner device is simple and easily produced by 3D printings. These advantages of the mag-spinner system diminish the limits of the separation system in terms of space, time, and facility. Thus, the developed mag-spinner is expected to be applied widely for on-site separation for further analysis or in third counties which do not possess well-facilitated laboratories.Author contributions
Investigation, formal analysis: SL, MJ; conceptualization, funding acquisition, project administration: JC; methodology, resources: SJL, SHL; validation, visualization, supervision, and writing – original draft: JC; writing – review & editing: JC, SJL, SHL.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (2020R1C1C1011863 and 2020R1A5A8017671).References
- M. K. Barman and A. Patra, J. Photochem. Photobiol., C, 2018, 37, 1–22 CrossRef CAS.
- D. Pappas and K. Wang, Anal. Chim. Acta, 2007, 601, 26–35 CrossRef CAS PubMed.
- J. Chen, J. Li and Y. Sun, Lab Chip, 2012, 12, 1753 RSC.
- C. Wyatt Shields IV, C. D. Reyes and G. P. López, Lab Chip, 2015, 15, 1230–1249 RSC.
- Y. Gao, W. Li and D. Pappas, Analyst, 2013, 138, 4714 RSC.
- L. Yu, S. R. Ng, Y. Xu, H. Dong, Y. J. Wang and C. M. Li, Lab Chip, 2013, 13, 3163 RSC.
- J. E. Petch, P. Gurnani, G. Yilmaz, F. Mastrotto, C. Alexander, S. Heeb, M. Cámara and G. Mantovani, ACS Appl. Mater. Interfaces, 2021, 13, 19230–19243 CrossRef CAS PubMed.
- P. Shinde, L. Mohan, A. Kumar, K. Dey, A. Maddi, A. Patananan, F.-G. Tseng, H.-Y. Chang, M. Nagai and T. Santra, Int. J. Mater. Sci., 2018, 19, 3143 Search PubMed.
- J. Cheong, H. Yu, C. Y. Lee, J. Lee, H.-J. Choi, J.-H. Lee, H. Lee and J. Cheon, Nat. Biomed. Eng., 2020, 4, 1159–1167 CrossRef CAS PubMed.
- A. Dalili, E. Samiei and M. Hoorfar, Analyst, 2019, 144, 87–113 RSC.
- M. Zhang, M. Wei, Z. Dong, H. Duan, S. Mao, S. Feng, W. Li, Z. Sun, J. Li, K. Yan, H. Liu, X. Meng and H. Ge, BMC Biotechnol., 2019, 19, 99 CrossRef CAS PubMed.
- C.-H. Lee, Endocrinol. Metab., 2017, 32, 18 CrossRef CAS PubMed.
- J. Xu, J. Sun, Y. Wang, J. Sheng, F. Wang and M. Sun, Molecules, 2014, 19, 11465–11486 CrossRef PubMed.
- M. Hejazian, W. Li and N.-T. Nguyen, Lab Chip, 2015, 15, 959–970 RSC.
- J. He, M. Huang, D. Wang, Z. Zhang and G. Li, J. Pharm. Biomed., 2014, 101, 84–101 CrossRef CAS PubMed.
- L. Borlido, A. M. Azevedo, A. C. A. Roque and M. R. Aires-Barros, Biotechnol. Adv., 2013, 31, 1374–1385 CrossRef CAS PubMed.
- I. Safarik and M. Safarikova, BioMag. Res. Technol., 2004, 2, 7 CrossRef PubMed.
- M. A. Witek, I. M. Freed and S. A. Soper, Anal. Chem., 2020, 92, 105–131 CrossRef CAS PubMed.
- S. S. Leong, Z. Ahmad, S. C. Low, J. Camacho, J. Faraudo and J. Lim, Langmuir, 2020, 36, 8033–8055 CrossRef CAS PubMed.
- S. S. Leong, S. P. Yeap and J. Lim, Interface Focus, 2016, 6, 20160048 CrossRef PubMed.
- O. Olsvik, T. Popovic, E. Skjerve, K. S. Cudjoe, E. Hornes, J. Ugelstad and M. Uhlén, Clin. Microbiol. Rev., 1994, 7, 12 CrossRef PubMed.
- W. Zhao, Y. Liu, B. D. Jenkins, R. Cheng, B. N. Harris, W. Zhang, J. Xie, J. R. Murrow, J. Hodgson, M. Egan, A. Bankey, P. G. Nikolinakos, H. Y. Ali, K. Meichner, L. A. Newman, M. B. Davis and L. Mao, Lab Chip, 2019, 19, 1860–1876 RSC.
- Z. Wang, N. Sun, H. Liu, C. Chen, P. Ding, X. Yue, H. Zou, C. Xing and R. Pei, ACS Appl. Mater. Interfaces, 2019, 11, 39586–39593 CrossRef CAS PubMed.
- R. Nasiri, A. Shamloo, S. Ahadian, L. Amirifar, J. Akbari, M. J. Goudie, K. J. Lee, N. Ashammakhi, M. R. Dokmeci, D. Di Carlo and A. Khademhosseini, Small, 2020, 16, 2000171 CrossRef CAS PubMed.
- L. Rao, Q.-F. Meng, Q. Huang, Z. Wang, G.-T. Yu, A. Li, W. Ma, N. Zhang, S.-S. Guo, X.-Z. Zhao, K. Liu, Y. Yuan and W. Liu, Adv. Funct. Mater., 2018, 28, 1803531 CrossRef.
- S. Yaman, M. Anil-Inevi, E. Ozcivici and H. C. Tekin, Front. Bioeng. Biotechnol., 2018, 6, 192 CrossRef PubMed.
- T. Bu, X. Yao, L. Huang, L. Dou, B. Zhao, B. Yang, T. Li, J. Wang and D. Zhang, Talanta, 2020, 206, 120204 CrossRef CAS PubMed.
- F. Schaumburg, C. S. Carrell and C. S. Henry, Anal. Chem., 2019, 91, 9623–9630 CrossRef CAS PubMed.
- A. Abo Markeb, J. Llimós-Turet, I. Ferrer, P. Blánquez, A. Alonso, A. Sánchez, J. Moral-Vico and X. Font, Water Res., 2019, 159, 490–500 CrossRef CAS PubMed.
- J. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Chem. Soc. Rev., 2017, 46, 1272–1283 RSC.
- W. Qi, L. Wang, N. Rong, X. Huo, Y. Li, M. Liao and J. Lin, Talanta, 2022, 239, 123095 CrossRef CAS PubMed.
- M. Rashid, M. A. Rabbi, T. Ara, M. M. Hossain, M. S. Islam, A. Elaissari, H. Ahmad and Md. M. Rahman, RSC Adv., 2021, 11, 36319–36328 RSC.
- H. Han, B. Sohn, J. Choi and S. Jeon, Biomed. Eng. Lett., 2021, 11, 297–307 CrossRef PubMed.
- S. Oh, S. H. Jung, H. Seo, M.-K. Min, B. Kim, Y. K. Hahn, J. H. Kang and S. Choi, Sens. Actuators, B, 2018, 272, 324–330 CrossRef CAS.
- J. Y. Noh, S.-W. Yoon, Y. Kim, T. V. Lo, M.-J. Ahn, M.-C. Jung, T. B. Le, W. Na, D. Song, V. P. Le, S. Haam, D. G. Jeong and H. K. Kim, Sci. Rep., 2019, 9, 16661 CrossRef PubMed.
- M. Jung, S. Lee, D. Y. Song, S. Kang, T. Shin and J. Choi, ACS Omega, 2021, 6, 31161–31167 CrossRef PubMed.
- C.-H. Liu, C.-A. Chen, S.-J. Chen, T.-T. Tsai, C.-C. Chu, C.-C. Chang and C.-F. Chen, Anal. Chem., 2019, 91, 1247–1253 CrossRef CAS PubMed.
- I. Michael, D. Kim, O. Gulenko, S. Kumar, S. Kumar, J. Clara, D. Y. Ki, J. Park, H. Y. Jeong, T. S. Kim, S. Kwon and Y.-K. Cho, Nat. Biomed. Eng., 2020, 4, 591–600 CrossRef CAS PubMed.
- R. P. Haugland and W. W. You, Methods Mol. Biol., 2008, 418, 13–24 CAS.
- X. Duan, Y. Li, N. K. Rajan, D. A. Routenberg, Y. Modis and M. A. Reed, Nat. Nanotechnol., 2012, 7, 401–407 CrossRef CAS.
- M. De Odrowaz Piramowicz, P. Czuba, M. Targosz, K. Burda and M. Szymoński, Acta Biochim. Pol., 2006, 53, 93–100 CrossRef PubMed.
- I. Krol, F. D. Schwab, R. Carbone, M. Ritter, S. Picocci, M. L. De Marni, G. Stepien, G. M. Franchi, A. Zanardi, M. D. Rissoglio, A. Covelli, G. Guidi, D. Scarinci, F. Castro-Giner, L. Mazzarella, C. Doglioni, F. Borghi, P. Milani, C. Kurzeder, W. P. Weber and N. Aceto, Br. J. Cancer, 2021, 125, 23–27 CrossRef CAS PubMed.
- N. Aceto, A. Bardia, D. T. Miyamoto, M. C. Donaldson, B. S. Wittner, J. A. Spencer, M. Yu, A. Pely, A. Engstrom, H. Zhu, B. W. Brannigan, R. Kapur, S. L. Stott, T. Shioda, S. Ramaswamy, D. T. Ting, C. P. Lin, M. Toner, D. A. Haber and S. Maheswaran, Cell, 2014, 158, 1110–1122 CrossRef CAS PubMed.
- R. Vaidyanathan, R. H. Soon, P. Zhang, K. Jiang and C. T. Lim, Lab Chip, 2019, 19, 11–34 CAS.
- L. Keller and K. Pantel, Nat. Rev. Cancer, 2019, 19, 553–567 CrossRef CAS.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Nature, 2007, 450, 1235–1239 CrossRef CAS PubMed.
- C. Alix-Panabières and K. Pantel, Nat. Rev. Cancer, 2014, 14, 623–631 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details: manufacture of mag-spinner using a 3D-printer, synthetic method of 7 nm iron oxide, iron oxide nanocluster (NC) zeta-potentials, comparison of hydrodynamic size of NCs pulled by magnets, test for capture efficiency of 7 nm iron oxide by mag-spinner, simulation of magnetic flux density around magnets, calculation of the drag force of NCs near magnets, and cell viability test after spinning in mag-spinner. See DOI: 10.1039/d1na00791b |
| This journal is © The Royal Society of Chemistry 2022 |