Open Access Article
Open Access ArticleEpigallocatechin-gallate tailors the cell adhesivity of fibronectin coatings in oxidation and concentration-dependent manner
Beatrix
Peter
 *a,
Nicolett
Kanyo
a,
Inna
Szekacs
*a,
Nicolett
Kanyo
a,
Inna
Szekacs
 a,
Antal
Csampai
b,
Szilvia
Bosze
cd and
Robert
Horvath
a,
Antal
Csampai
b,
Szilvia
Bosze
cd and
Robert
Horvath
 a
a
aNanobiosensorics Laboratory, Research Centre for Natural Sciences, Institute for Technical Physics and Materials Science, Konkoly-Thege u, 29-33, Budapest, H-1120, Hungary. E-mail: peter.beatrix@ek-cer.hu
bInstitute of Chemistry, Eötvös Loránd University, 112, POB 32, Budapest, H-1518, Hungary
cELKH-ELTE Research Group of Peptide Chemistry, Eötvös Loránd Research Network (ELKH), Eötvös Loránd University, 112, POB 32, Budapest, H-1518, Hungary
dNational Public Health Center, 1097, Budapest, Hungary
First published on 5th October 2022
Abstract
Fibronectin is an extracellular matrix component that plays a significant role in many physiological processes, such as cell adhesion, growth, differentiation, and migration. In this study, we revealed the interaction between this important protein and the widely studied natural active substance green tea polyphenol epigallocatechin-gallate (EGCG) and its oxidized form. Furthermore, we investigated the kinetics of cancer cell adhesion on the polyphenol-treated fibronectin coatings. We applied a high-throughput, label-free optical biosensor capable of monitoring the above processes in real time with an excellent signal-to-noise ratio. Our results show that EGCG and its oxidized form bind to fibronectin in a concentration-dependent manner and can form multilayers as well. Furthermore, both polyphenol forms inhibited cellular adhesion, however, the effect was more pronounced in the case of the oxidized form. The results were compared to the measurements performed on bare biosensor surfaces without fibronectin, and the roles of oxidation were investigated. It is suggested that the polyphenols can interact and block important cell adhesion protein motifs and affect the rigidity of the layers as well. Moreover, a novel molecular scale active mechanism involving the disulfide bridges of fibronectin was proposed to explain the recorded kinetic signals and highlight that these proteins can be active participants in the molecular scale transformations affecting adhesion.
1. Introduction
Fibronectin is a high-molecular-weight glycoprotein of the extracellular matrix that binds to integrin receptors.1–4 It is a protein dimer, consisting of two monomers linked by a pair of C-terminal disulfide bonds.5 It plays role in cell adhesion, growth, migration, differentiation, and migration. Furthermore, it is significant for processes like embryonic development and wound healing.1 Fibronectin has RGD (Arg-Gly-Asp) and PHSRN (Pro-His-Ser-Arg-Asn) integrin-binding sequences. The RGD sequence mediates the interaction of fibronectin with integrins, while the PHSRN sequence modulates the interaction.6 Integrin α5β1 was identified as a fibronectin-binding receptor in HeLa cells.7 Integrins are not always active. Activation of an integrin from the low to high ligand-binding affinity state requires conformational change due to cytoplasmic or extracellular interactions.8 For example, activation of α5β3, αIIbβ3, α11, and β1 integrin receptors can be triggered by modifications of extracellular disulfide bonds (thiol-disulfide exchange).8 Morphological alterations have been observed in tumors and cancer-derived cell lines; decreased fibronectin expression, increased fibronectin degradation, and/or decreased expression of integrin receptors, like the above-mentioned α5β1.9 According to the review article of Wang and Hielscher, it has been suggested that due to the interference with the processes of the immune system and engagement with several integrin receptors, fibronectin promotes cancer cell proliferation and survival.9 Furthermore, its isoforms participate in tumorigenesis as well, thus different variants of fibronectin not just interact with cell surface receptors, but their interactions also activate signaling pathways for tumor growth.9It has been shown that tumor cells produce an increased amount of ROS compared to normal tissue cells.10 Thus cancer cells have enhanced ROS production upon cell–matrix interaction as well, so oxygen and ROS affect the production of ECM proteins at both transcriptional and (post)translational levels. This increased extracellular redox potential increases the expression of cell adhesion molecules (P- and E-selectins, intercellular cell adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1)). This is important, because for example, in the case of leukocytes, this can generate inflammatory fibrosis. It has been suggested that the leg of the integrin α-subunit contains cysteine residues, which are sensitive to oxidizing agents. ROS and other oxidizing agents can establish disulfide bridges, furthermore, prevent their formation when two vicinal disulfide bridges are oxidized to cysteine sulfenic acid groups. The head domain (including the αA-domain) contains several cysteine residues, but it does not seem to be affected by reducing agents or ROS. Furthermore, the mutation of cysteines within the Aβ-domain of the β3 integrin subunit did not show any influence on the ligand-binding activity. Probably there are differences in the redox-regulation of EGF-domain-based cysteines between the two integrins α5β3 and αIIbβ3, although they share the same β3 integrin subunit. Thus it seems that the α-subunit influences the thiol-based redox regulation within the β integrin leg domains as well.10
The main compound of green tea, epigallocatechin-gallate, is probably the most studied polyphenol for decades.11–15 A lot of studies showed its beneficial effects on human health, for instance, its anticancer, anti-inflammatory, and anti-metastatic activities.12,13,16–21 These processes are in connection with cellular adhesion. Some experiments with cancer cell lines proved that this active substance effectively decreases adhesion to different extracellular matrix proteins like laminin,22 fibronectin,4 and collagen.23 These results highlight the potential anticancer effect of EGCG.2,12,23,24 Furthermore, EGCG has an impact on cancer cell viability as well. Our previous study showed that EGCG is cytostatic but not cytotoxic (IC50 > 500 μg mL−1) on HeLa cancer cells, as revealed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide end-point assay (MTT)) and flow cytometry.14 Of note, an anticancer compound that is rather cytostatic than cytotoxic has distinct advantages in mainly affecting cancer cells. Often the main goal is not the direct killing of all cells (cytotoxicity), but rather inhibiting the proliferation of the cancer cells selectively (cytostatic activity).14
The dimerization of EGCG even under mild oxidative conditions has been extensively reviewed.12,13,15,25–27 Tea polyphenols have antioxidant activities,26 and EGCG is the most effective in reacting with reactive oxygen species (ROS).12,15 These activities are due to the phenolic groups that are sensitive to oxidation and can generate quinone, an oxidized derivative of aromatic compounds.12,13 Oxidation is an irreversible reaction, and the oxidation species were found to correspond to Mw + 14 (where Mw is the molecular weight of EGCG), in which two hydrogen atoms are removed and one oxygen atom is added to the gallyl moiety in the B-ring of EGCG.12,25 When EGCG is oxidized in green tea, it forms two digallate dimers, theasinensin A (Mw 914) and P2 (Mw 884), finally theaflavin-3,3′-digallate (Mw 868.7) when oxidized in black tea.12,26–29
In this work, we aimed to reveal the interaction between fibronectin and EGCG and its oxidized form with the subsequent cellular adhesion by using a label-free optical sensor device. For this purpose, we applied fibronectin-coated and bare sensor surfaces. The adsorbed mass and the number of formed EGCG layers were also calculated from the recorded kinetic data. The presented sensor is sensitive to sub-nanometer scale changes in the cell membrane positioning or protein distribution while averaging the signals of thousands of adhering cells.30 Both small molecules and the larger cells could be easily examined in the same experiment with very high resolution.30 Furthermore, quantum-chemical modeling was used to reveal the molecular scale active mechanism and explain the recorded kinetic signals.
2. Materials and methods
All chemicals and reagents were obtained from Sigma-Aldrich Chemie GmbH, Munich, Germany unless stated otherwise.2.1. Preparation of EGCG and oxidized EGCG solutions
The stock solution of EGCG was prepared freshly in assay buffer (20 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) in Hank's balanced salt solution (HBSS), pH 7, hereafter HBSS-HEPES). The tested concentrations of the EGCG solution were 0.05, 0.5, 5, 20, 40, 50 and 500 μg ml−1. To get the oxidized EGCG forms, the freshly prepared solutions were subjected to heat treatment at 60–70 °C for 1.5 h in a water bath.302.2. Cell culture and cell adhesion assay buffer
HeLa cells (ECACC 93021013) were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum (Biowest SAS, France), 4 mM L-glutamine, 0.25 μg ml−1 amphotericin B, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin solution. Cells were cultured in a humidified incubator (37 °C, 5% CO2). On reaching 80% confluence, cells were detached every 3–4 days using 0.05% (w/v) trypsin and 0.02% (w/v) EDTA solution and were not used beyond passage 20.2.3. The resonant waveguide grating (RWG) biosensor and related protocols for in situ monitoring the EGCG binding and cell adhesion
The Epic BenchTop (BT) system (Corning Incorporated, NY, USA) employed in this study is an RWG base optical biosensor with high-throughput microplate-based label-free detection, the method is described in earlier studies.30–33 The experiments were run using 384-well uncoated biosensor microplates (5040, Corning) and 384-well fibronectin-coated plates (5042, Corning). During the experiments, the baseline with HBSS-HEPES buffer (30 μl) was recorded for approximately 40–60 min. After that, the buffer was removed from the wells. Then the EGCG solutions were pipetted (30 μl) into the wells and we measured them for approximately 80 min. Then washed 4 times, and the kinetic curves were recorded for 30 min again with HBSS-HEPES (30 μl). During this time, HeLa cells were brought into suspension by using trypsin–EDTA solution. trypsin–EDTA was removed before the complete detachment of HeLa cells and its activity was arrested by adding a complete culture medium. Harvested cells were centrifuged at 380 × g for 6 min and the cell pellet was resuspended in assay buffer. Cells were then counted in a hemocytometer, and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were added to each sensor well. The cells for buffer control received assay buffer instead of cell suspension. All measurements were carried out in triplicate using three different wells at room temperature. Cell spreading was monitored until saturation of the biosensor signals (2 h). Averaging every 5 subsequent data points, the effective sampling rate was 1/15 s−1.30–33
000 cells were added to each sensor well. The cells for buffer control received assay buffer instead of cell suspension. All measurements were carried out in triplicate using three different wells at room temperature. Cell spreading was monitored until saturation of the biosensor signals (2 h). Averaging every 5 subsequent data points, the effective sampling rate was 1/15 s−1.30–33
2.4. Quantumchemical modeling
A comparative modeling study on the binding of EGCG and its oxidized dimer form to fibronectin was performed using the Gaussian 09 program package.343. Results and discussion
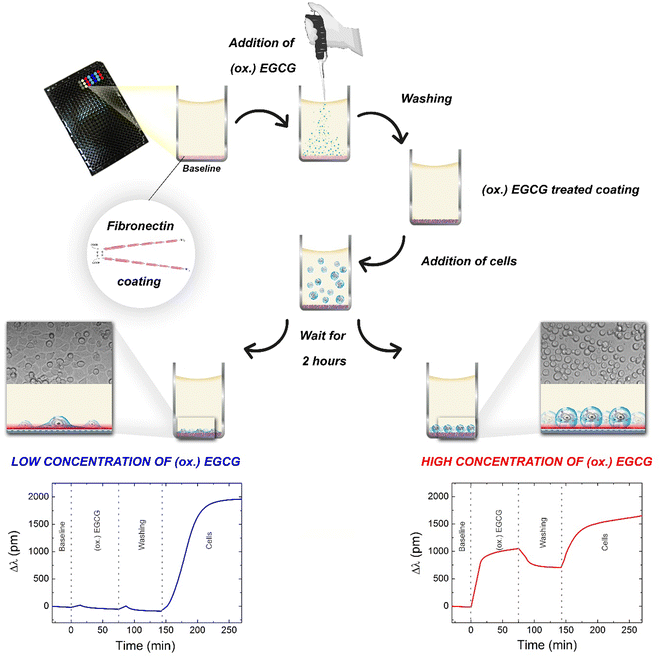
The adsorption kinetics of EGCG and its oxidized form at various concentrations and the subsequent cell adhesion process was monitored online using a high-throughput RWG biosensor. The measurement procedure and the typical kinetic curves obtained on fibronectin coating are shown in Fig. 1. The kinetic curves of the measurements with low (0.05 μg ml−1) and high concentrations (500 μg ml−1) of EGCG or oxidized EGCG exposure are shown. The biosensor measurements show that the (ox.) EGCG molecules irreversibly adsorbed on the fibronectin coating at higher concentrations (Fig. 1, bottom right).We measured the adhesion processes of the cells on EGCG and oxidized EGCG-treated fibronectin coatings for two hours because during this period these types of cells can reach the total spread morphology.32
3.1. Polyphenol adsorption and subsequent cellular adhesion on bare and fibronectin-coated surfaces
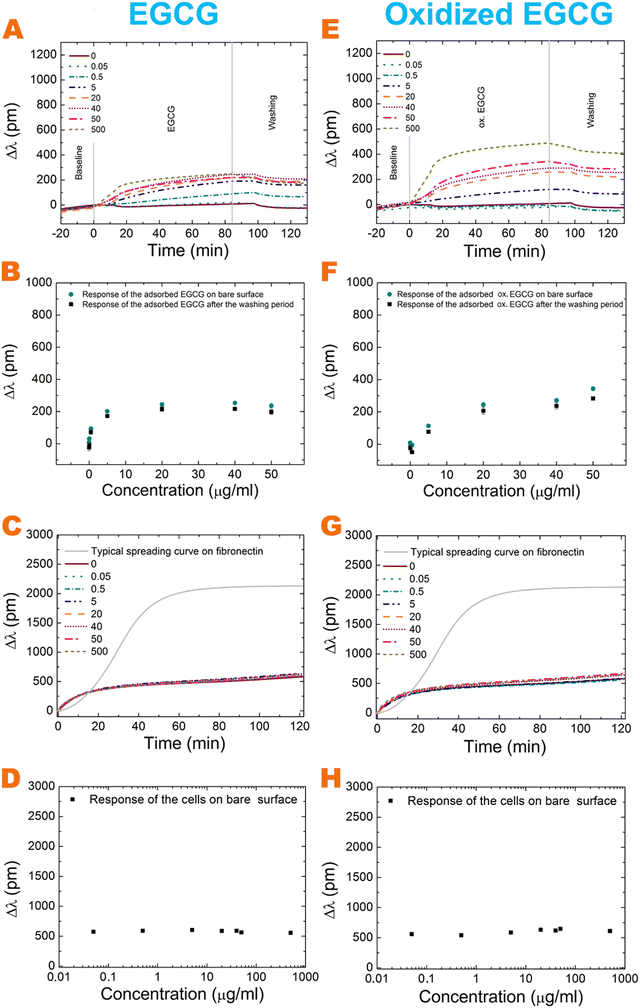
The effects of polyphenol exposure on fibronectin and the bare biosensor surface were investigated. The EGCG and its oxidized form adsorb on both surfaces in a concentration-dependent manner (Fig. 2 and 3A, E). A major amount adsorbed irreversibly and could not be removed by the washing procedure with buffer. The oxidized form adsorbs in a larger amount in all cases. On increasing the polyphenol concentration, the bound amount was also increased, but saturation was observed at a concentration of around 20 μg ml−1 for EGCG on the bare surface (Fig. 2B). However, for oxidized EGCG, we get increasing data points (Fig. 2F), just like in the case of fibronectin coating and EGCG and its oxidized form (Fig. 3B and F). The adsorbed amount is different on the bare surface and the fibronectin coating; at 500 μg ml−1 concentration the oxidized form adsorbed in almost two-fold amount to the fibronectin (Table 1). This proves that although (ox.) EGCG can adsorb on the bare surface irreversibly by physisorption, the polyphenol prefers binding to coatings, like fibronectin. Table 1 shows the exact adsorbed amount on bare and fibronectin surfaces.| Conc. (μg ml−1) | Bare surface | Fibronectin | ||||
|---|---|---|---|---|---|---|
| Δλ (pm) | ΔM (ng cm−2) | Number of layers | Δλ (pm) | ΔM (ng cm−2) | Number of layers | |
| 5 | 172.04 | 49.63 ± 3.31 | 1.31 ± 0.08 | 145.33 | 41.93 ± 0.66 | 1.11 ± 0.02 |
| 50 | 197.27 | 56.91 ± 4.52 | 1.51 ± 0.12 | 215.71 | 62.23 ± 0.64 | 1.65 ± 0.02 |
| 500 | 193.31 | 55.76 ± 1.59 | 1.47 ± 0.04 | 257.55 | 74.30 ± 1.10 | 1.97 ± 0.03 |
| ox. 5 | 76.68 | 22.17 ± 2.98 | 0.58 ± 0.08 | 138.01 | 39.82 ± 1.16 | 1.05 ± 0.03 |
| ox. 50 | 283.07 | 81.66 ± 3.32 | 2.16 ± 0.09 | 458.01 | 132.13 ± 2.07 | 3.49 ± 0.05 |
| ox. 500 | 409.05 | 118.01 ± 3.30 | 3.12 ± 0.09 | 723.76 | 208.80 ± 6.84 | 5.52 ± 0.18 |
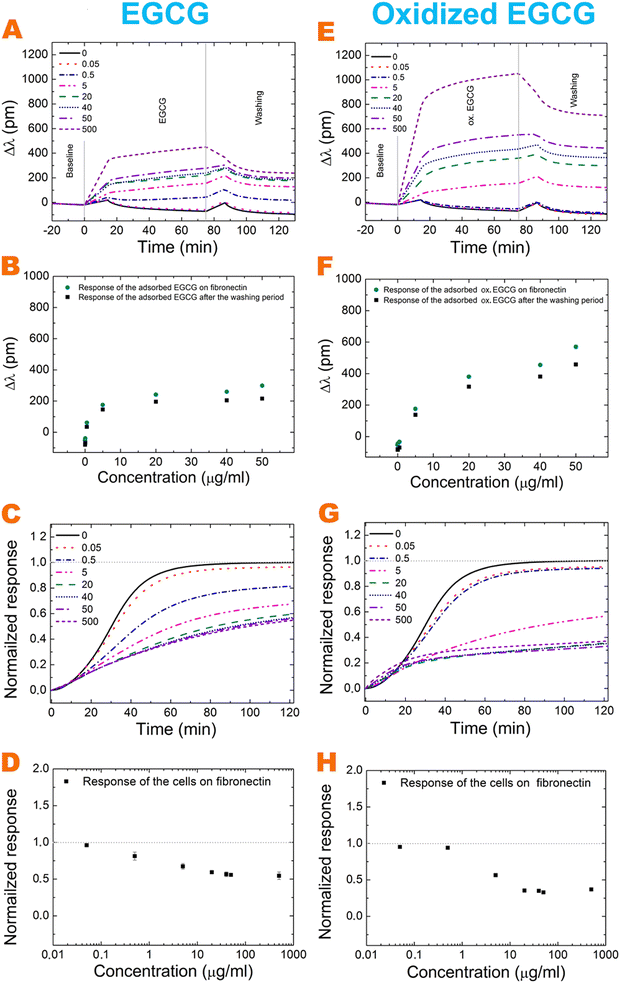
For active receptor-mediated cell adhesion and spreading a sigmoid-like kinetic curve is observed on a non-treated fibronectin coating (Fig. 2C, G grey curve and Fig. 3C, G black curve), while the non-specific cell adhesion results in an adsorption-like kinetic curve on the polyphenol-treated and untreated bare biosensor surface (Fig. 2C and G). EGCG and its oxidized form inhibited cellular adhesion onto fibronectin in a concentration-dependent manner, and the effect of the oxidized form is more pronounced (Fig. 3C, D, G and H). Interestingly, the oxidized 0.5 μg ml−1 EGCG may rather promote cellular adhesion compared to the non-oxidized form (Fig. 3C, D, G and H).
The highly hydrated poly(L-lysine)-graft-poly(ethylene glycol) (PLL-g-PEG) and its RGD (Arg-Gly-Asp) containing form, PLL-g-PEG-RGD (hereafter PP:PPR) employed in our previous work30 can be considered only as a simplified model system.
The two interactions revealed previously are (i) the binding of EGCG and oxidized EGCG with H-bonds to the polymer and (ii) the effective blocking of the RGD adhesion motifs by the bound polyphenols. The concentration-dependent effects of EGCG and oxidized EGCG in the case of fibronectin suggest that the above-mentioned interactions play important roles in fibronectin, too.
However, analyzing the results deeper, some marked differences are also observed. Namely, the adhesion strengthening effect of the bound polyphenols at low concentrations is present, but much less dominant in fibronectin (see Fig. 3). We attribute this to the differences in conformational flexibility between the two systems, and effects of the cross-coupling of EGCG on this. Clearly, the PEG chains have relatively large conformational flexibility.
Moreover, it is quite revealing that in contrast to the polymer system at high oxidized EGCG concentrations cell adhesion does not completely diminish in fibronectin. But, interestingly, the cell adhesion decreasing effect of high EGCG concentration is approximately the same in the two systems (50%). Moreover, analyzing the relative effects in Fig. 4 some differences in the measured concentration range are also observed.
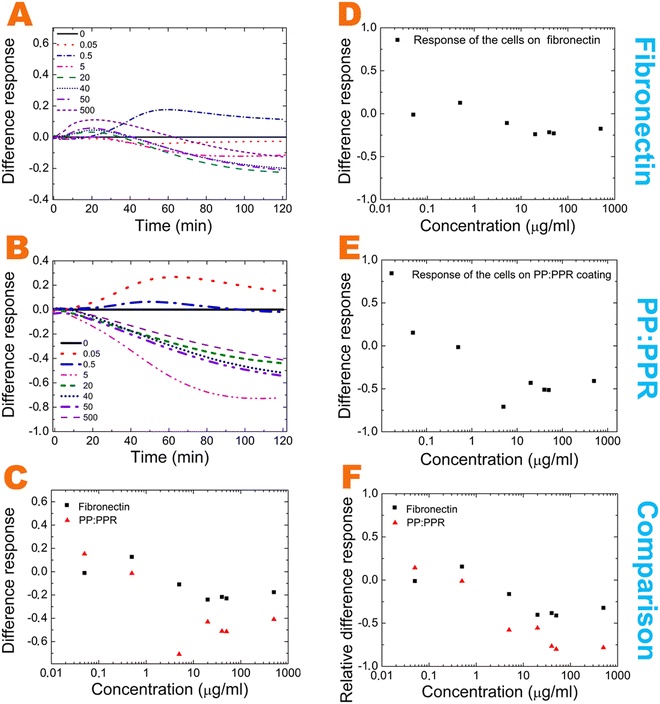
We analyzed the effect of EGCG oxidation by the subtraction of the EGCG normalized signal from the oxidized normalized signal. These differences between fibronectin and the PP:PPR systems and their comparisons are plotted in Fig. 4A–E. The comparison of the different cell responses is also plotted in Fig. 4C and F.
These differences suggest the existence of other dominant interactions in fibronectin. Such interactions are mainly affecting the fibronectin–oxidized EGCG interactions in the middle and high concentration range and the fibronectin–EGCG interactions in the middle concentration range.
3.2. Calculation of surface adsorbed mass density from the RWG biosensor data
The adsorbed mass can be calculated from the RWG data. The wavelength shift [Δλ (pm)] can be converted to a surface-adsorbed mass (ng cm−2) by using the calibration equation of Orgovan et al.35 This equation is valid for a polyelectrolyte solution with an RI increment of dn/dc = 0.1955 cm3 g−1.35 The dn/dc value of the EGCG and oxidized EGCG solution is 0.21 cm3 g−1 as determined earlier by measuring the RI of the EGCG solutions using a tabletop refractometer.30 Based on the previously developed methodology,33,35 this value leads to the following calibration equation:| ΔM = 0.2885 ng pm−1 cm−2 × Δλ | (1) |
3.3. Quantumchemical modeling and proposed molecular scale interactions
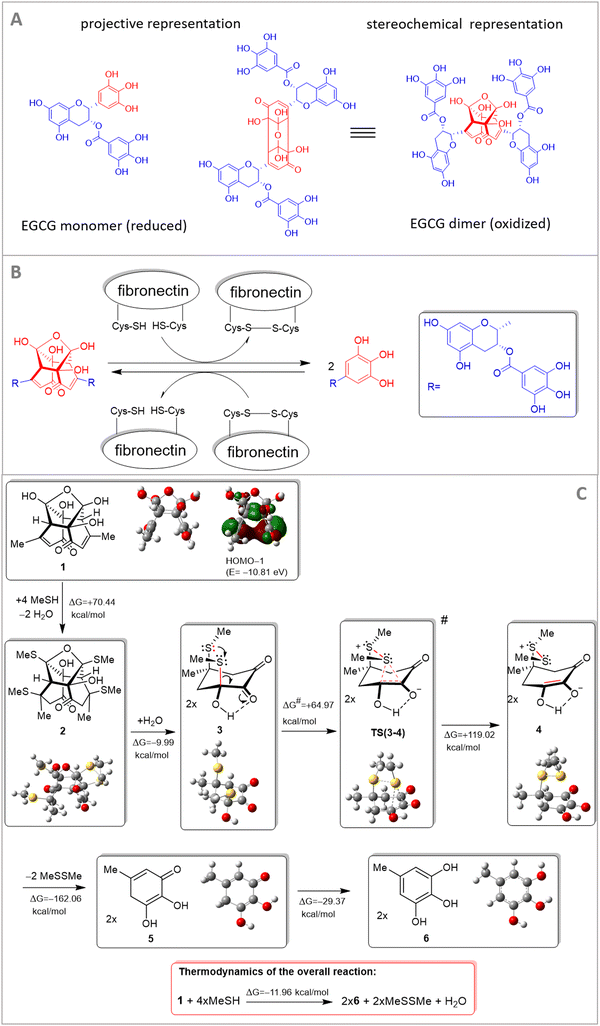
In our previous work,30 we used a semiempirical quantum-chemical method and showed that EGCG binds to PEG chains by hydrogen bonds and the binding is stronger for the oxidative products of EGCG.30 Regarding the dimer oxidative product of EGCG, we proposed a cross-coupling mechanism of polymer chains by the hydrogen-bonding network.30 This effect might be present in fibronectin, too. Besides this, a novel interaction mechanism is also proposed involving cysteine residues of fibronectin. Cysteine-based redox modifications within integrin heterodimers might be subject to redox-dependent conformational changes of integrin.10 This process can affect the binding activities and interactions of integrins and their exposed domains. Cell adhesion proteins, such as integrins, mediate the interaction of cells and other molecules and these interactions mainly depend on certain conformational status.Of note, the performed theoretical modeling provided a plausible interpretation of redox transformation mediated by fibronectin-linked thiol/disulfide residue. To better emphasize the main interaction partners, the central residue of the EGCG dimer exclusively implicated in the critical multistep redox process is presented in red color in Fig. 5. For the sake of simplicity, the residual molecular fragments with complex structures on EGCG and fibronectin, not involved in the redox transformation, are replaced with methyl groups. This molecular fragment (central residue) is only involved in the redox transformation and was only subjected to theoretical modeling.
Under experimental conditions, both EGCG and integrin are targets of redox-based modifications. In our study we have highlighted redox-relevant aspects of the EGCG monomer–dimer and integrin system, that can affect integrin-layer-based cell adhesion dynamics.
A theoretical study was directed at modeling a thiol-mediated reductive cleavage of the dimeric terminal of EGCG. The modeling process was strictly focused on the exact molecular fragments that are involved in the crucial elementary reaction steps. It is assumed that under the in vitro experimental conditions this redox process involves two triphenol fragments of EGCG, the dimeric fragment of oxidized EGCG, and a cysteine-containing fibronectin segment that can form disulfide bridges. Accordingly, the methyl group on the simplified EGCG models is a simplified representation of the pending molecular fragments including long polyethylene glycol chains (which are not involved in the redox transformations), while MeSH and MeSSMe are the simplified representations of a cysteine side chain and a disulfide bridge, respectively, in the cysteine-enriched segment of fibronectin. We proposed a mechanism for the cysteine-mediated reductive cleavage of dimer EGCG (Fig. 5) and supported our view about the assumed reaction sequence by quantum chemical modeling carried out at the HF/3-21G* level of theory36,37 complemented with the IEFPCM solvent model38 using the dielectric constant of water (ε = 80.1) to represent the biological environment. Focusing on the molecular regions involved in the actual reaction steps, in the course of calculation simplified structures (1 and MeSH representing an EGCG dimer and the pending cysteine side chains, respectively, along with 6 and MeSSMe representing monomeric EGCG and protein/peptide disulfide bridges, respectively) were analyzed and subjected to modeling studies (Fig. 5).
Although transformation 1 → 2 (its details are not presented) and the formation of the S–S bond leading to sulfonium-enolate (3 → 4) are accompanied by significant increases in Gibbs free energy, the multistep reaction also comprises the hydrolytic double retro aldol process (2 + H2O → 2 × 3) followed by disulfide-elimination (4 → 5 + MeSSMe) and sequential 1,5 hydrogen shift in the resulting monomeric cyclohexadienone (5 → 6), features favorable for overall thermodynamics (ΔG = −11.96 kcal mol−1). It must be noted here that in the course of thermodynamically unfavoured transformation 1 → 2, the feasible generation of the oxygen-bridged bis-thiosemiacetal moiety is accompanied by a double conjugate thiol-addition on the enone residues that breaks down the stabilizing π–π interaction in 1 as presented by HOMO−1 featuring marked electron density delocalized between the proximal C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) double bonds. On the other hand, the relatively large differences in the calculated energetics can at least partly be attributed to the use of a reasonably demanding calculation methodology optimizing the structures of 1–6 as separated species without any otherwise hardly predictable intermolecular interactions, e.g. the exact mode of solvation by definite numbers of water molecules. Utilizing intermolecular S+⋯O and O−⋯H interactions this solvent might significantly contribute to the stability of zwitterion 4 and transition state TS(3–4), markedly increasing both the thermodynamic- and the kinetic feasibility of the crucial elementary step 3 → 4 associated with the actual electron-transfer. Transition state TS(3–4) was localized as a saddle point on the potential energy surface by the QST2 method39 connecting the local minima representing intermediates 3 and 4. All calculations were performed using the Gaussian 09 software package.34
double bonds. On the other hand, the relatively large differences in the calculated energetics can at least partly be attributed to the use of a reasonably demanding calculation methodology optimizing the structures of 1–6 as separated species without any otherwise hardly predictable intermolecular interactions, e.g. the exact mode of solvation by definite numbers of water molecules. Utilizing intermolecular S+⋯O and O−⋯H interactions this solvent might significantly contribute to the stability of zwitterion 4 and transition state TS(3–4), markedly increasing both the thermodynamic- and the kinetic feasibility of the crucial elementary step 3 → 4 associated with the actual electron-transfer. Transition state TS(3–4) was localized as a saddle point on the potential energy surface by the QST2 method39 connecting the local minima representing intermediates 3 and 4. All calculations were performed using the Gaussian 09 software package.34
A simplified theoretical study was directed at modeling a thiol-mediated reductive cleavage of the dimeric terminal of EGCG. The modeling study was strictly focused on the molecular fragments that are involved in the crucial elementary steps. On the other hand, it is assumed that under the experimental conditions this redox process involves two triphenol fragments of EGCG, the dimeric fragment of oxidized EGCG and a cysteine-enriched fibronectin segment that is capable of forming disulfide bridges. Accordingly, the methyl group on the simplified EGCG models is the simplified representation of the pending molecular fragments including long polyethylene glycol chains, which are not involved in the redox transformation, while MeSH and MeSSMe are the simplified representations of a cysteine side chain and a disulfide bridge, respectively, in the cysteine-enriched segment of fibronectin.
Based on our quantum-chemical modeling, we can conclude that this redox process involves two triphenol fragments of EGCG, the dimeric fragment of oxidized EGCG and the cysteine-containing fibronectin segment that is capable of forming disulfide bridges. Furthermore, at lower EGCG concentrations, the oxidation process is slow, so there will likely be more dimers. At higher EGCG concentrations, the dimeric form immediately interacts with fibronectin (see Fig. 4A and B).
Overall, the presence of fibronectin shifts the balance to the monomer form. Therefore, the differences shown in Fig. 4C and F between the different cell adhesion signals of the oxidized and nonoxidized solutions might be explained. The difference between the effect of oxidized and nonoxidized solutions is less pronounced in fibronectin, especially at higher concentrations, perfectly in line with the described molecular scale mechanism.
It cannot be excluded that the disulfide bridges of fibronectin also play a role in the cell adhesive properties of the coatings. Of note, integrin activation by disulfide bond reduction was previously discovered.40 Therefore, ligand accessibility on fibronectin might be affected by the formation of the disulfide bonds. Future research is needed to more directly verify these interesting possibilities.
Cellular adhesion on fibronectin is an intensively researched field.2,3,9,41–43 The role of various peptide sequences and glycosylation states of fibronectin concerning cellular adhesion was studied before, but the interaction of fibronectin with polyphenols in this relation was never investigated. It has been published that EGCG bind to fibronectin,44,45 however, the adsorption kinetics of the oxidized solution and its effect on cellular adhesion remain uncovered. EGCG is unstable at high temperatures and under alkaline and neutral conditions (pH ≥ 7), and it dimerizes and oxidizes easily.12,26,46 In an aqueous solution, it changes from non-colored to yellow in higher pH regions.12,25 Although this is a relevant feature of this compound, the effect of the oxidized EGCG is poorly investigated.
4. Conclusions
In the present study, we examined the interaction between surface adsorbed fibronectin and EGCG and its oxidized form. Furthermore, the subsequent cellular adhesion on the coatings was real-time monitored using a high-throughput label-free optical biosensor. We employed both fibronectin-coated biosensor plates and bare sensor plates to quantify the differences between various circumstances.Based on the recorded label-free data, we identified EGCG and oxidized EGCG multilayer formation onto fibronectin and bare surface as well. The number of EGCG and oxidized EGCG layers were calculated using the geometrical parameters of the EGCG molecule and its molecular weight.30 As a result, at 500 μg ml−1 EGCG, approximately 2 layers, while in the case of 500 μg ml−1 oxidized EGCG, 5 layers were formed on the fibronectin coating. On the bare surface, 2 and 3 layers were adsorbed, respectively.
We proposed that the polyphenol molecules bound less between the fibronectin chains, and thus form fewer (approximately half) multilayers than in the case of PLL-g-PEG and PP:PPR coatings.30 We suggest that at high concentrations the formed multilayers can effectively block RGD or PHSRN (or both) cell adhesion motifs, decreasing cell adhesion and spreading on the polyphenol-exposed protein films.
Moreover, a novel molecular scale mechanism involving the disulfide bridges of fibronectin was proposed to possibly explain the recorded kinetic signals and highlight that these proteins can be active participants in molecular scale transformations affecting adhesion, too. The disulfide bonds are key cross-links in proteins and they are reactive and can undergo bimolecular nucleophilic substitution, a reaction with free thiol resulting in thiol-disulfide exchange.47,48 The function of some proteins is controlled by the cleavage of their disulfide bonds.49 The interaction of the EGCG dimer at the molecular level with thiol-containing fibronectin was modelled. This modeling provided a plausible interpretation of the redox transformation mediated by fibronectin-linked thiol/disulfide residues. Future research is needed to more directly verify the exact contribution of this effect.
The introduced methodology could be further continued with other extracellular matrix proteins and other small molecule active substances, the method is capable of illuminating the most important features of EGCG-adhesion matrix interactions, highlighting the importance of ligand oxidation during cellular interactions.
Author contributions
Beatrix Peter: investigation, data curation, writing – original draft, writing – review & editing, visualization, and funding acquisition. Nicolett Kanyo: investigation. Inna Szekacs: investigation and writing – review & editing. Antal Csampai: modeling, interpretation of the redox system and writing – original draft. Szilvia Bosze: interpretation of the redox system and writing – original draft. Robert Horvath: conceptualization, project administration, supervision, writing – review & editing, and funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The present work was supported by the Hungarian Academy of Sciences [Lendület (Momentum) Program], the National Research, Development, and Innovation Office (NKFIH) [ERC_HU, PD 131543 for B. P. and KKP_19 Programs], and the Eötvös Loránd Research Network (ELKH). Project no. TKP2021-EGA-04 has been implemented with the support provided from the Ministry of Innovation and Technology of Hungary from the National Research, Development, and Innovation Fund, financed under the TKP2021 funding scheme.References
- R. Pankov and M. Kenneth, J. Cell Sci., 2002, 115, 3861–3863 CrossRef CAS.
- M. Sazuka, T. Itoi, Y. Suzuki, S. Odani, T. Koide and M. Isemura, Biosci., Biotechnol., Biochem., 1996, 60, 1317–1319 CrossRef CAS.
- D. J. Iuliano, S. S. Saavedra and G. a Truskey, J. Biomed. Mater. Res., 1993, 27, 1103–1113 CrossRef CAS PubMed.
- Y. Suzuki and M. Isemura, Biomed. Res., 2013, 34, 301–308 CrossRef CAS PubMed.
- Y. Mao and J. E. Schwarzbauer, Matrix Biol., 2005, 24, 389–399 CrossRef CAS PubMed.
- K. Kimura, A. Hattori, Y. Usui, K. Kitazawa, M. Naganuma, K. Kawamoto, S. Teranishi, M. Nomizu and T. Nishida, Invest. Ophthalmol. Visual Sci., 2007, 48, 1110–1118 CrossRef.
- J. Friedrichs, J. Helenius and D. J. Müller, Proteomics, 2010, 10, 1455–1462 CrossRef CAS PubMed.
- M. Popielarski, H. Ponamarczuk, M. Stasiak, C. Watała and M. Świątkowska, Am. J. Cancer Res., 2019, 9, 1554–1582 CAS.
- J. P. Wang and A. Hielscher, J. Cancer, 2017, 8, 674–682 CrossRef CAS.
- J. A. Eble and F. F. De Rezende, Antioxid. Redox Signaling, 2014, 20, 1977–1993 CrossRef CAS PubMed.
- B. Peter, J. Nador, K. Juhasz, A. Dobos, L. Korosi, I. Székács, D. Patko and R. Horvath, J. Biomed. Opt., 2015, 20, 067002 CrossRef.
- B. Peter, S. Bosze and R. Horvath, Eur. Biophys. J., 2017, 46, 1–24 CrossRef CAS PubMed.
- B. N. Singh, S. Shankar and R. K. Srivastava, Biochem. Pharmacol., 2011, 82, 1807–1821 CrossRef CAS.
- B. Peter, R. Ungai-Salanki, B. Szabó, A. G. Nagy, I. Szekacs, S. Bösze and R. Horvath, ACS Omega, 2018, 3, 3882–3891 CrossRef CAS PubMed.
- H. Tachibana, Proc. Jpn. Acad., Ser. B, 2011, 87, 66–80 CrossRef CAS PubMed.
- R. C. C. de Pace, X. Liu, M. Sun, S. Nie, J. Zhang, Q. Cai, W. Gao, X. Pan, Z. Fan and S. Wang, J. Liposome Res., 2013, 23, 187–196 CrossRef.
- P. T. Devika and P. Stanely Mainzen Prince, Pharmacol. Res., 2008, 57, 351–357 CrossRef CAS.
- D. S. Hsieh, H. Wang, S. W. Tan, Y. H. Huang, C. Y. Tsai, M. K. Yeh and C. J. Wu, Biomaterials, 2011, 32, 7633–7640 CrossRef CAS.
- C.-F. Hung, T.-F. Huang, H.-S. Chiang and W.-B. Wu, J. Cell. Biochem., 2005, 96, 183–197 CrossRef CAS PubMed.
- T. Punathil, T. O. Tollefsbol and S. K. Katiyar, Biochem. Biophys. Res. Commun., 2008, 375, 162–167 CrossRef CAS.
- I. a Siddiqui, M. Asim, B. B. Hafeez, V. M. Adhami, R. S. Tarapore and H. Mukhtar, FASEB J., 2011, 25, 1198–1207 CrossRef CAS.
- Y. Suzuki and M. Isemura, Cancer Lett., 2001, 173, 15–20 CrossRef CAS.
- H. M. Lo, C. F. Hung, Y. Y. Huang and W. Bin Wu, J. Biomed. Sci., 2007, 14, 637–645 CrossRef CAS.
- D. Umeda, S. Yano, K. Yamada and H. Tachibana, J. Biol. Chem., 2008, 283, 3050–3058 CrossRef CAS PubMed.
- Y. Mizooku, M. Yoshikawa, T. Tsuneyoshi and R. Arakawa, Rapid Commun. Mass Spectrom., 2003, 17, 1915–1918 CrossRef CAS PubMed.
- J. Hong, H. Lu, X. Meng, H.-H. Colon, J. Ryu, Y. Hara and C. S. Yang, Cancer Res., 2002, 62, 7241–7246 CAS.
- Z. Hou, S. Sang, H. You, M. J. Lee, J. Hong, K. V. Chin and C. S. Yang, Cancer Res., 2005, 65, 8049–8056 CrossRef CAS PubMed.
- J. Xu, Z. Xu and W. Zheng, Molecules, 2017, 22, 1–18 Search PubMed.
- C. E. Isaacs, W. Xu, G. Merz, S. Hillier, L. Rohan and G. Y. Wen, Antimicrob. Agents Chemother., 2011, 55, 5646–5653 CrossRef CAS.
- B. Peter, E. Farkas, E. Forgacs, A. Saftics, B. Kovacs, S. Kurunczi, I. Szekacs, A. Csampai, S. Bosze and R. Horvath, Sci. Rep., 2017, 7, 42220 CrossRef CAS PubMed.
- I. Kurucz, B. Peter, A. Prosz, I. Szekacs, R. Horvath and A. Erdei, Sens. Actuators, B, 2017, 240, 528–535 CrossRef CAS.
- N. Orgovan, B. Peter, S. Bősze, J. J. Ramsden, B. Szabó and R. Horvath, Sci. Rep., 2014, 4, 4034 CrossRef PubMed.
- B. Peter, I. Lagzi, S. Teraji, H. Nakanishi, L. Cervenak, D. Zámbó, A. Deák, K. Molnár, M. Truszka and I. Szekacs, ACS Appl. Mater. Interfaces, 2018, 10, 26841–26850 CrossRef CAS PubMed.
- M. J. Frisch, et al., Gaussian 09, Revision E.01, Gaussian Inc., Wallingford CT, 2009 Search PubMed.
- N. Orgovan, B. Kovacs, E. Farkas, B. Szabó, N. Zaytseva, Y. Fang and R. Horvath, Appl. Phys. Lett., 2014, 104, 1–5 CrossRef.
- C. Froese Fischer, Comput. Phys. Commun., 1987, 43, 355–365 CrossRef CAS.
- K. D. Dobbs and W. J. Hehre, J. Comput. Chem., 1986, 7, 359–378 CrossRef CAS.
- J. Tomasi, B. Mennucci and E. Cancès, J. Mol. Struct.: THEOCHEM, 1999, 464, 211–226 CrossRef CAS.
- P. Chunyang and H. B. Schlegel, Isr. J. Chem., 1993, 33, 449–454 CrossRef.
- B. Yan and J. W. Smith, Biochemistry, 2001, 40, 8861–8867 CrossRef CAS PubMed.
- P. Speziale and G. Pietrocola, Front. Microbiol., 2020, 11, 1–13 CrossRef.
- D. J. Vaca, A. Thibau, M. Schütz, P. Kraiczy, L. Happonen, J. Malmström and V. A. J. Kempf, Med. Microbiol. Immunol., 2020, 209, 277–299 CrossRef CAS PubMed.
- S. Y. Boateng, S. S. Lateef, W. Mosley, T. J. Hartman, L. Hanley and B. Russell, Am. J. Physiol.: Cell Physiol., 2005, 288, C30–C38 CrossRef CAS.
- C.-M. Chan, J.-H. Huang, H.-S. Chiang, W.-B. Wu, H.-H. Lin, J.-Y. Hong and C.-F. Hung, Mol. Vis., 2010, 16, 586–595 CAS.
- E. Melgarejo, M. Á. Medina, F. Sánchez-Jiménez and J. L. Urdiales, Br. J. Pharmacol., 2009, 158, 1705–1712 CrossRef CAS PubMed.
- S. Hirun and P. D. Roach, Int. Food Res. J., 2011, 18, 1261–1264 CAS.
- P. Carl, C. H. Kwok, G. Manderson, D. W. Speicher and D. E. Discher, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 1565–1570 CrossRef CAS PubMed.
- K. Kolšek, C. Aponte-Santamaría and F. Gräter, Sci. Rep., 2017, 7, 1–10 CrossRef PubMed.
- P. J. Hogg, Trends Biochem. Sci., 2003, 28, 210–214 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |