Open Access Article
Open Access ArticleToward precision oncology: SERS microfluidic systems for multiplex biomarker analysis in liquid biopsy
Karthik Balaji
Shanmugasundaram†
 a,
Junrong
Li†
a,
Junrong
Li†
 a,
Abu Ibn
Sina
a,
Abu Ibn
Sina
 *a,
Alain
Wuethrich
*a,
Alain
Wuethrich
 *a and
Matt
Trau
*a and
Matt
Trau
 *ab
*ab
aCenter for Personalized Nanomedicine, Australian Institute for Bioengineering and Nanotechnology (AIBN), Corner College and Cooper Roads (Bldg 75), The University of Queensland, Brisbane, QLD 4072, Australia. E-mail: a.wuethrich@uq.edu.au; a.sina@uq.edu.au; Tel: +61-7-33464178
bSchool of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD 4072, Australia. E-mail: m.trau@uq.edu.au
First published on 6th January 2022
Abstract
Liquid biopsy-based diagnosis in precision oncology exhibits significant advantages over the traditional tissue biopsies by offering dynamic assessment of tumour heterogeneity, minimally invasive procedures for frequent sampling, and cost-effective tests. Implementation of liquid biopsy-based diagnosis for precision oncology could be the key to provide a confident cancer screening with tailored risk assessment, patient stratification, and real-time monitoring of cancer therapies. To achieve precision oncology with liquid biopsy, the simultaneous analysis of multiple circulating tumour biomarkers is a powerful strategy to establish an accurate signature for each individual patient. Among various developed approaches for tumour biomarker detection, microfluidic devices integrated with surface enhanced Raman scattering (SERS) are emerging as one of the powerful techniques to support precision oncology based on its potential to provide multiplexing and high sensitivity. Particularly, the microfluidic devices provide miniaturised channels for parallel reactions and SERS has the extremely narrow spectra for intrinsic multiplexing. This mini review will focus on recently reported SERS microfluidic techniques, which are capable of simultaneous detection of multiple cancer biomarkers in liquid biopsy and have the promise to be integrated into precision oncology.
1. Introduction
The emergence of precision oncology in the recent era has radically rejuvenated the field of oncology by introducing the concept of personalized treatment, which analyzes an individual's genetic, epigenetic, or phenotypic characteristics to maximize treatment efficiency and minimize side effects. Precision oncology is therefore classified as the cornerstone of next-generation cancer management.1 Although showing promises in providing personalized treatment, the diagnosis and routine monitoring in precision oncology remain mostly invasive and costly by relying on tissue biopsy to date.2,3 In this context, liquid biopsy has emerged as a promising alternative that involves analysis of tumour-derived components in body fluids (e.g., blood, urine, saliva). Due to the advantages of minimally invasive procedures and precise monitoring of tumour biomarkers in real-time, liquid biopsy has shown growing potential to become the next gold standard for cancer diagnostics and therapy monitoring.4,5 The recent surge in research for developing liquid biopsy technologies has enabled the discovery of many liquid biopsy-based cancer biomarkers, such as circulating tumour cells (CTCs), circulating tumour DNA and RNA (ctDNA and ctRNA), soluble proteins, and extracellular vesicles (EVs).6 These liquid biopsy cancer biomarkers are found to carry real-time cancer signatures that are displayed on the actual tumour sites.7 The analysis of these biomarkers in liquid biopsy could therefore revolutionize the early diagnosis and routine monitoring of cancer resulting in a significant improvement in patient outcome for precision oncology. Despite their immense potential, liquid biopsy technologies have not been established in the clinic yet mostly due to their low sensitivity in accurately identifying the cancer signatures and limited high throughput screening capabilities.8 However, these challenges can be addressed with the development of highly sensitive and multiplexed platforms that can specifically identify the biomarkers in trace concentrations in liquid biopsy.9Microfluidic devices have been gaining an increasing attention in the field of biosensing because of their advantageous features, flexible scale up, and mass production at relative low cost. Microfluidic devices are usually derived from micro or nanoscale materials ranging from a few micrometres to nanometres.10 The integration of 3D nanostructures into microfluidic devices can provide a larger surface area in a miniaturized system and ultra-sensitivity to detect trace concentration of biomolecules.11,12 The highly controllable micro/nano environment of such devices is further beneficial as it enables rapid fluid processing and efficient strategies that are required for high throughput biosensing. For instance, it can operate with a low volume of samples, incorporate more than one function in the same device, and enable the detection of multiple targets simultaneously.13,14 However, microfluidic devices rely on suitable readouts to detect biomarkers that are captured on the device. Fluorescence-based readouts have been widely used with microfluidic devices for molecular diagnostics. Nevertheless, fluorescence-based methods are limited by the overlapping spectra of the target molecules and photobleaching which significantly limits their sensitivity and multiplexing capability.15
Recent years have seen an upsurge in developing surface-enhanced Raman scattering (SERS) based biosensing techniques due to their advantages over fluorescence-based methods. SERS utilizes plasmonic substrates to substantially increase the light scattering resulting in structural characteristics of the molecule bound to or near the plasmonic surfaces.16 In SERS, there are two main types of surface enhancement mechanisms, i.e., long-range electromagnetic, and short-range chemical enhancement. The electromagnetic enhancement technique is generated by localized surface plasmon resonance which intensifies the scattering intensity of analytes using hot spots, whereas the chemical technique involves bonding interaction of absorbate and substrate to enhance the cross-section scattering of the analytes.17 SERS-based detection and characterization approaches have been recognized for their high sensitivity, efficient molecular fingerprint properties, and rapid sample preparation protocols.18 Significantly, SERS provides narrow spectral width with higher multiplexing capabilities, which makes it a superior readout candidate for biosensing.19
A multiplex system is particularly important for precision oncology because of the generation of a patient-specific multimolecular profile that can significantly facilitate accurate diagnosis and tailoring of therapy.20 The combination of microfluidics systems with SERS could be an ideal platform that can allow accurate identification of the multiple biomolecules on-chip and significantly enhance the detection capabilities in complex samples with ultra-sensitivity.21 As such, the combination of microfluidics and SERS has the potential to address limitations associated with sensitivity and multiplexity and could become the gold standard liquid biopsy technique for applications in precision oncology.
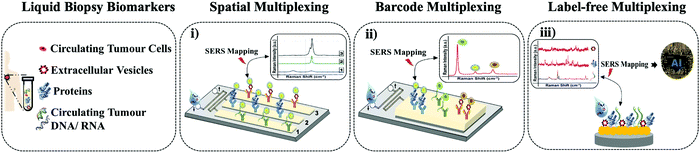
In this mini review, we focus on SERS microfluidic platforms that were evaluated to detect multiple biomarkers in liquid biopsy and could be potentially adapted in precision oncology. As shown in (Fig. 1), we grouped the techniques in different classes based on their three multiplexing types: (i) spatial multiplexing, (ii) barcode-enabled multiplexing, and (iii) label-free multiplexing, and discussed the methods to evaluate their potential for clinical applications.
2. Spatial multiplexing
Spatial multiplexing is a simple form of multiplexing and achieved by creating spatially separated zones for parallel reactions on the microfluidic system. By mapping the microfluidic system, the spatial information together with the Raman signal enable the biomarker identification. Typically, micro/nanofabrication methods (e.g., lithography) are used to prepare separated compartments (e.g., well structures) or parallel microfluidic channels. The micro/nanofabrication methods allow to integrate micro/nanofabricated structures such as patterning with metallic nanoparticles, which can be used to enhance the Raman signal and support trace biomarker detection in liquid biopsy.10 Furthermore, droplet microfluidics has also been explored for encapsulation of biomarkers and Raman reporter into individual droplets. The design and fabrication of separate zones for multiplexing and sample delivery to each zone need to be considered carefully. Furthermore, multiple separate zones increase the sample consumption and can also cause additional sample handling errors.Separated compartments were used by Wang and co-workers for highly sensitive 9-plex detection of miRNA using a microfluidic device with 9 spatially separated zones (Fig. 2a).11 After isothermal amplification of the miRNA, the amplified products were flown through the device. The device had 9 zones that consisted of silver nanoparticles (AgNPs) that were patterned on a silicon substrate. The AgNPs were functionalized with complementary DNA/Raman reporter hairpin probes. After binding of the amplified miRNA to the hairpin, enzymatic hydrolysation resulted in an opening of the hairpin structure that moved the Raman reporter away from the signal enhancing AgNPs, leading to a strong signal decrease. This “signal-on”/“signal-off” detection scheme was highly sensitive and enabled trace miRNA detection of as low as 1 aM in 9 min. This strategy was evaluated for molecular typing of breast cancer, where an 85% concordance with RT-qPCR was found.
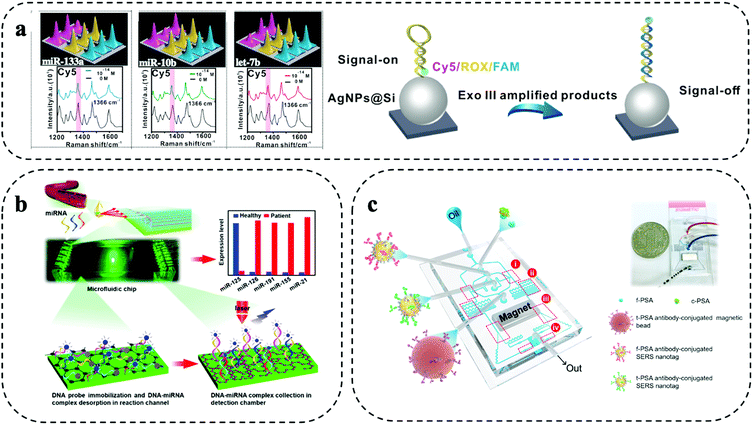 | ||
| Fig. 2 Spatial multiplexing on SERS microfluidic systems. (a) Separated compartments for 9-plex detection of miRNAs using isothermal amplification. Modified from ref. 11 with permission. Copyright 2021, Elsevier (b) parallel microfluidic channels for 5-plex miRNA detection in a device modified with graphene oxide and poly-l-lysine. Adapted from ref. 20 with permission. Copyright 2021, American Chemical Society (c) droplet microfluidic encapsulation to detect parallelly prostate-specific antigen (f-PSA) and total PSA (t-PSA) in prostate cancer patients. Reproduced with permission from ref. 22. Copyright 2018, Elsevier. | ||
Using parallel microfluidic channels, micro RNA (miRNA)20 and soluble proteins22,23 were detected. 4 breast cancer-associated miRNAs (i.e., miR-126, miR-191, miR-155, and miR-21) and tumour suppressing miR-125, were detected by Han and colleagues. Each channel comprised of a reaction and detection zone (Fig. 2b).20 After labelled DNA probes were immobilized in the reaction zones, the sample was added for DNA–miRNA complex formation by complementary hybridization. Subsequently, the complexes were desorbed and passed to the Raman detection chamber. The small features of the microfluidic device and dense immobilization of the DNA probes enabled highly efficient miRNA capture from as little as 2 μL of serum and provided a detection limit of 0.146 aM without requiring RNA amplification. Evaluated on 7 breast cancer patient samples and 3 healthy controls, increased levels of miR-126, miR-191, miR-155, and miR-21 were found in patients, while miR-125 was downregulated compared to the healthy samples.
Similarly, three parallel microfluidic channels were used by Cui et al. for 3-plex detection of soluble proteins CA153, CA125, and CEA that are indicated in breast cancer.24 To achieve highly sensitive biomarker detection, the microfluidic channels were patterned with AgNPs that were conjugated to Raman reporters and biomarker-specific antibodies. The AgNPs enhanced the Raman signal by approx. two orders of magnitude compared to the same strategy without AgNPs. Controlling the process to achieve a uniform patterning of the microfluidic channels with metallic nanoparticles is important for Raman signal enhancement and was investigated in detail by Zhang and colleagues.25 They found that a uniform patterning could be easily achieved through a spiral microfluidic geometry. High Raman signal enhancement can also be achieved using bimetallic substrates such as Au–Ag alloys, providing SERS enhancement factor of 107. Such strong enhancement factors are ideally suited for trace biomarker analysis in parallel microfluidic channels. Waluk et al. exploited microfluidic channels coated with a bimetallic Ag–Au surface for trace detection of cytokines (i.e., IL-6, IL-8, and IL-18) that are implicated in breast cancer.21 The three cytokines were captured in three separated microfluidic channels. The strong and homogenous signal enhancement also reduced Raman signal variation and contributed to the good assay reproducibility and picogram level detection sensitivity.
Using biomarker encapsulation by droplet microfluidics, Choo et al. developed a screening approach for prostate cancer based on the simultaneous detection of free prostate-specific antigen and total prostate specific antigen.22 The microfluidic device integrated a fully automated sandwich immunoassay including droplet generation and immunoreaction, formation of magnetic immunocomplexes, magnetic isolation of immunocomplexes, and SERS detection (Fig. 2c). The simultaneous detection of free and total prostate-specific antigen was achieved by splitting the sample into two droplet streams for separate incubation with SERS nanoparticles conjugated with antibodies against either free or total prostate-specific antigen. The immune-complex was then completed by incubation with antibody-functionalized magnetic beads to pull-down the antigen-SERS nanotag complex for read out. The automated process was completed within 10 min and enabled sensitive biomarker detection in the range of 0.05 to 100 ng mL−1. SERS droplet microfluidics has also been applied to the study prostate cancer cells.26
3. Barcode-enabled multiplexing
Barcode-enabled multiplexing refers to in situ detection of multiple biomarkers in non-confined space within the microfluidic system. Typically, the biomarkers are captured by antibodies immobilized on the microfluidic system, which is followed by a labelling step with a cocktail of target antibodies and Raman reporters co-conjugated SERS nanotags. By varying Raman reporters and target antibody, simultaneous capture and quantitative detection of multiple biomarkers can be achieved.10,19 Two main considerations are needed in the selection of Raman reporters. Firstly, the Raman reporters are expected to have separate characteristic peaks without significant spectral overall lap. For example, Raman reporter MPY and TFMBA have unique peaks at 1005 cm−1 and 1375 cm−1, respectively. As these two peaks are far away from each other, no mutual interferences can be observed due to spectra overlap. Secondly, the Raman reporters are required to have strong signals to indicate the low abundance of biomarkers. Due to the diverse enhancement capability, the Raman reporters having higher signals have the benefit of sensitively sensing targets for early detection. A novel method was reported by Li et al. for effective biomarker monitoring during nasopharyngeal carcinoma (NPC) using SERS sensing. They developed unique SERS reporters using metal carbonyls for enhancing the sensitivity and provided highly multiplex information during the sample testing.27 SERS nanotags with the use of different plasmonic nanostructures including gold nanorod (AuNRs), gold@silver (Au@Ag) nanorods, iron oxide–gold (IO–Au) core shell, gold nanoparticles (AuNPs), gold–silver (Au–Ag) alloy nanoboxes were reported for high-throughput assays with highly selective and sensitive detection. The barcode-enabled multiplexing offers a comprehensive target detection capability, which makes it well suited for wide range of clinical applications from early cancer diagnosis to profiling cancer heterogeneity at a single cell level to monitoring targeted drug therapies for precision oncology. Limitations of barcode-enabled multiplexing are the higher spectral complexity and potential Raman peak overlap. Furthermore, despite the claimed high multiplexing capability of Raman detection, the demonstration of simultaneous measurement of more than 6 biomarkers is rare in liquid biopsy. To deploy the rich Raman spectra for higher multiplexing detection, spectral decoding or deconvolution can be used to reduce signal overlap for identification. In addition, artificial intelligence (AI) based analysis is also increasingly used for Raman data processing.28,29 Another strategy to allow higher multiplexing is the use of novel Raman reporters that have lesser Raman peaks to avoid mutual interferences. For instance, research efforts are geared toward the synthesis of novel Raman reporters with few peaks (e.g., one) located beyond 2000 cm−1. For instance, Luo and colleagues, and Liang et al. explored the silent region with of the Raman spectrum ranging from ∼2050–2150 cm−1 as an extension strategy of the available Raman reporters, which has the promise of improving multi-biomarker detection.30,31Xiong and colleagues reported a SERS integrated microfluidic chip (MiChip) based on bio-conjugated magnetic nanochains (Magchains) for simultaneous multiplexing of three clinical serum protein biomarkers (i.e., CEA, AFP, PSA) expressed in prostate, colorectal and liver cancer (Fig. 3a).13 The integrated MiChip featured various compartments such as fluid ports for sample inlet and waste outlet, mixing chamber, detection chamber, and two pneumatic microvalves for controlling fluid flow in/out of mixing chamber. For the MiChip assay, the Magchains are conjugated with capture antibodies and SERS nanoprobes, the nanoprobes were prepared using AuNRs synthesized with Raman reporters and specific detection antibodies. The MiChip assay involved the collection of unprocessed liquid specimen, antibody conjugated Magchains and SERS nanoprobes in the mixing chamber followed by mixing driven by Magchains (i.e., synchronised rotation of Magchains in a spinning magnetic field generated by four orthogonal electromagnetic coils). Such a design resulted in the recognition of antigen–antibody and led to the formation of sandwich immune complex of MagChains, target biomarkers and SERS probes. The immune complex was collected in the detection chamber for Raman spectroscopic analysis. For the multiplexed biomarker detection, the specific peaks at 1341 cm−1, 1071 cm−1 and 885 cm−1 of three SERS nanoprobes allowed simultaneous detection of their encoded biomarkers. By measuring the same panel of biomarkers in serum samples, MiChip was able to analyze complex clinical specimens for profiling cancer biomarkers.
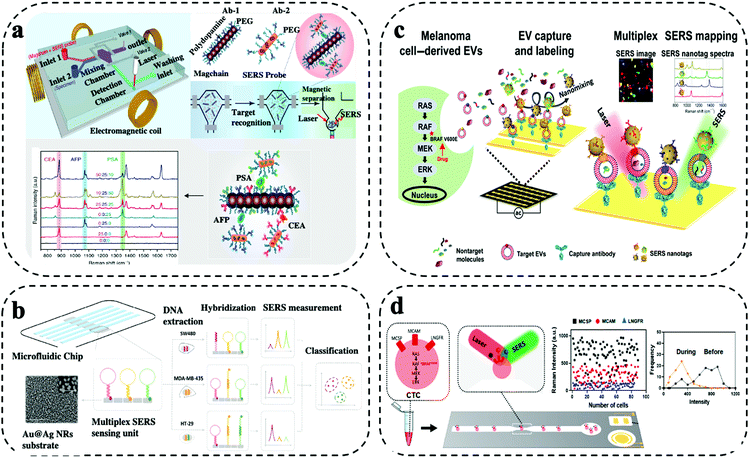 | ||
| Fig. 3 Barcode-enabled multiplexing on SERS microfluidic systems. (a) MiChip based on bio-conjugated magnetic nanochains for triplex detection of cancer specific protein biomarker. Modified from ref. 13 with permission. Copyright 2018, Springer Nature (b) Microfluidic biochip integrated with Au@Ag nanorods SERS substrate for amplification-free detection of three DNA mutation. Reproduced with permission from ref. 32. Copyright 2020, Elsevier (c) EPAC microchip using AuNPs for monitoring EV heterogeneity and profiling phenotypic variations. Adapted from ref. 33 with permission. Copyright 2020, Science (d) AuNPs based microfluidic chip for single CTC detection using multiple surface protein biomarkers. Adapted from ref. 15 with permission. Copyright 2021, American Chemical Society. | ||
Amplification-free SERS biochip integrated with microfluidic channels was demonstrated by Wu et al. for simultaneous identification and profiling of multiple point DNA mutations (i.e., KRAS, BRAF, PI3KCA genes) in melanoma and colorectal cancer cell lines (Fig. 3b).32 This barcoding-based in situ multiplexing approach was demonstrated on an Au@Ag nanorods SERS substrate assembled in individual microfluidic channel. Three kinds of hair-pin structured nucleic acid – molecular beacon probes were designed with three specific Raman reporter and were covalently linked to the Au@Ag nanorods substrate. The hybridization of target DNA caused a structural change of the molecular beacon probe, and the Raman reporter labelled at the free end were detached leading to low SERS intensity. The intensities of different Raman reporters varied corresponding to the target DNA hybridization. With SERS spectral encoding, signal changes were monitored for simultaneous detection of three DNA mutations in cancer patients. Supervised training algorithm based on classical least square (CLS)-linear discriminant analysis (LDA) was imposed to decode SERS spectra clustering of mutation assay for various cancer cells. SERS biochip demonstrated more than 90% accuracy from analyzing tumour DNAs from cancer patient samples showing its potential for clinical applications.
SERS assay based extracellular vesicles (EVs) phenotype analyser chip (EPAC) was reported by Wang et al. for monitoring treatment response in melanoma patients receiving targeted therapy (i.e., BRAF V600E inhibitors) (Fig. 3c).33 The EPAC based microfluidic system utilized alternating current electrodynamics (ac-EHD) for stimulating the fluid flow within the electrode channels that produced nanomixing strategy for enhancing the target-sensor interaction and to minimize the non-specific bindings. The multiplex assay targets four biomarkers in EVs such as MCSP, MCAM, LNGFR and ErbB3 using uniquely labelled SERS nanotags (i.e., AuNPs conjugated with different Raman reporters and target specific antibodies). EPAC enabled the detection of cancer-specific EV phenotypes in melanoma patient plasma and EVs phenotypic changes in eight melanoma patients receiving targeted therapy. The patient-specific EV profiles were promising for early indicating the development of drug resistance, an issue commonly found in targeted cancer treatment.
Single-cell analysis is of critical importance in gaining deep understanding of intercellular heterogeneity, identifying minority sub-populations of interest, as well as discovering for solutions to abnormal cell conditions and various diseases, thereby contributing to the development of diagnostic platforms and therapeutics decisions.34 Reza et al. developed a SERS immunoassay on a simple microfluidic channel integrated with SERS detection system for simultaneous in situ identification of single CTCs to unravel intercellular heterogeneity in their protein expression levels during the progression of disease and response to the treatment (Fig. 3d).15 This immunoassay approach was demonstrated using the SK-MEL-28 melanoma cell line for monitoring the expression of a panel of three cancer-specific surface protein biomarker (i.e., MCSP, MCAM, LNGFR). The SERS detection system was prepared by synthesizing spherical AuNPs conjugated with target antibodies and Raman reporters. The SERS characteristic peak intensities were obtained concurrently via Raman spectroscopy upon successful formation of antigen–antibody immune complexes by binding of different antibodies conjugated SERS nanotags for the different target proteins. Drug treatment studies were processed using BRAF inhibitors. The average SERS intensities showed the treatment response over the period. Day 0 was considered as un-treated and showed a high protein expression level, whereas after treatment from day 10 – day 60 suggested a decrease in the protein expression levels. The frequency vs intensity showed the transition from heterogeneous to homogeneous expression of the protein markers in single CTCs. However, certain cells showed higher expression even after treatment showing the necessity for change in drug treatment. Patient serum analysis using SERS immunoassay showed a promising potential for the simple microfluidic platform to be used in a clinical setting for monitoring of cancer and to design targeted drug therapies.
Several barcode-based in situ multiplexing using integrated microfluidic chip with enhanced SERS detection capabilities for identifying cancer-specific protein biomarkers, monitoring the immune activation levels of cytokine interleukins biomarkers and for profiling CTCs phenotypes were reported. Reza and co-workers have combined the utility of alternating current electrohydrodynamic (ac-EHD) to a SERS barcode integrated microfluidic platform consisting of an array of asymmetric electrode pairs for simultaneous in situ detection of four cancer-specific protein biomarkers that are clinically relevant for breast, lung, and ovarian cancer.35 This ac-EHD based approach provided an enhanced target-sensor interaction and minimized the non-specific binding by using four different SERS barcodes for spectral encoding of multiple cancer protein biomarkers.
Zhang et al. demonstrated a SERS immunoassay approach for monitoring epithelial–mesenchymal transition (EMT), a mechanism involved in cancer metastasis, through simultaneous detection of two soluble cadherins biomarkers in breast cancer.36 To enable the parallel cadherin detection, the sample was added on a microchip functionalized with anti-E-cadherin and anti-N-cadherin antibodies to specifically capture sE-cadherin and sN-cadherin. The captured targets were labelled with duplex SERS barcodes for detection of the targets. Specifically, the duplex SERS barcodes used MBA reporter and DTNB reporter that have characteristic Raman peaks at 1080 cm−1 and 1332 cm−1 to indicate E-cadherin and N-cadherin, respectively. This approach was developed on a microfluidic chip integrated with ac-EHD induced nanomixing for increasing the binding kinetics of the target and different SERS barcode nanotags. Kamińska et al. developed a sandwich type triplex SERS immunoassay on a microfluidic device embedded with Ag–Au bimetallic SERS-active substrate for simultaneous detection and monitoring the levels of immune activation of three cytokine interleukins biomarkers majorly responsible for breast cancer initiation and progression.21 The use of individual SERS barcoding for each target provided distinct signatures for detection and quantification of multiple biomarkers. SERS enabled microfluidic chip functionalized with modified graphene oxide (GO) platform was reported by Reza et al. for the simultaneous detection of three soluble immune checkpoint proteins biomarkers.37 Recombinant nanoyeast single chain variable fragments (NY-scFv) based affinity was introduced as an alternative to monoclonal antibody for capture and different SERS nanotags for multiple target protein detection.
Zhang and colleagues reported an in situ approach for CTC isolation combined with spectrally orthogonal SERS aptamer nanovectors for profiling multiple cell membrane protein biomarkers from breast cancer cells.38 The isolated CTCs from multiple breast cancer cells were simultaneously detected by three different DNA aptamers conjugated with the SERS nanovectors by the different target specific SERS intensities. An integrated Raman-Active Nanoprobe (RAN) system combined with microfluidic chip was developed by Cho et al., for simultaneous multiplex detection, isolation, and analysis of circulating cancer stem cells (CCSC) and various subtypes of CTCs in breast cancer cells.39 The simultaneous detection of targets (CD133, EpCAM, EGFR, HER2 and MUC1) was achieved by five different combinations of RANs, prepared by conjugating AuNPs with different Raman reporters and different detection antibodies along with biotinylated double stranded DNA (dsDNA). The isolation was performed using the microfluidic chip system functionalized with avidin–biotin and the restriction enzyme digestion of the dsDNA linker from the RANs. SERS immunoassay approach using a microtube based on magnetic beads and SERS tags was developed by Cheng and co-workers for determination of the ratio of free to total (f/t) prostate specific antigen for accurate diagnosis of prostate cancer.40 The initial part of the assay involved the fabrication of detection probes or SERS nanotags. Briefly, two Raman reporters (i.e., malachite green isothiocyanate (MGITC) and X-rhodamine-5-(and-6)-isothiocyanate (XRITC)) were immobilized on AuNPs, followed by conjugation of two antibodies (f-PSA and c-PSA) using SH-PEG-COOH on the surface of AuNPs. The capture substrate was prepared using carboxylic acid-functionalized magnetic beads to allow EDC/NHS assisted antibody immobilization. Ultimately, this in situ approach utilized a microtube for immune complex formation that were captured using a magnetic stand and later transferred capillary tube for SERS detection.16
As shown in Fig. 4a, Wilson Jr and co-workers demonstrated a combined microfluidic platform-based immunomagnetic enrichment with SERS detection system using magnetic multicolour SERS nanotags for simultaneous and quantitative detection of CTC using four surface protein biomarkers (i.e., EpCAM, HER2, CD44, IGF1R) on a single breast cancer cell for analyzing CTC subpopulation in clinical sample and monitoring cancer metastasis.41 For this combined assay, SERS nanotags developed using IO–Au core- shell nanoparticles served as dual function purpose, (i) isolation of CTC bound to the magnetic nanoparticle with the permanent external magnets under the device (i.e., immunomagnetic separation), (ii) subsequent detection of CTCs via absorbed Raman reporter under SERS spectroscopy. The on-chip multiplexing assay was performed by synthesizing IO–Au core- shell nanoparticles SERS nanotags with four organic dyes, which included, QSY21, BHQ3, QXL680 and IR740 conjugated with different Raman reporters and target antibody for targeting four different biomarkers in each individual cell. Although Raman reporter displayed different peaks, identification of one distinct peak for each tag when three or more colours needed simultaneously made it a difficult task. Signal deconvolution based on CLS regression method was performed for analysing the levels of each target surface protein biomarkers.
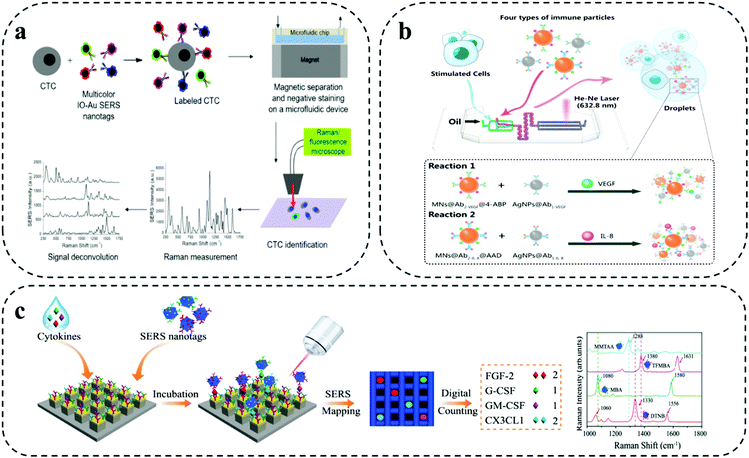 | ||
| Fig. 4 Barcode-enabled multiplexing on SERS microfluidic systems. (a) IO–Au core–shell nanoparticles based magnetic microfluidic chip for detection and quantification of four surface protein biomarker in individual CTC. Reproduced with permission from ref. 41. Copyright 2020, American Chemical Society. (b) SERS-Droplet microfluidic based dual signal enhancement for identification of two cytokines from a single cell. Adapted from ref. 42 with permission. Copyright 2019, American Chemical Society. (c) Single molecule capture nanopillar platform using SERS nanoboxes for detection of four cytokine biomarkers. Adapted from ref. 43 with permission. Copyright 2021, Springer Nature. | ||
A dual signal enhancement approach using SERS-based droplet microfluidics was reported by Sun et al. for simultaneous in situ detection of two cytokine biomarkers (i.e., VEGF, IL-8) secreted by a single-cell for studying cell–cell heterogeneity (Fig. 4b).42 This assay was built on a droplet based- cross-typed microfluidic SERS platform. The device involved the formation of water-in-oil microdroplets by cutting aqueous phase with oil phase for encapsulation of a single cell. The detection of cytokine biomarkers in the single cell was based on magnetic-field amplified SERS sandwich immunoassay. The immune particles complexes for the detection were prepared by conjugating monoclonal antibodies to silver nanoparticles and polyclonal antibodies to magnetic beads along with two different synthesized Raman active reporters for individual target spectral encoding of two cytokine biomarkers. The dual signal enhancement was obtained with the higher plasmon effect produced by adjacent AgNPs and magnetic field-induced aggregate amplification effect by the magnetic beads for analysing multiple cytokines from single cell. This provided the assay with 75 times enhanced SERS signal with a limit of detection at 1.0 fg mL−1 in one droplet.
As shown in Fig. 4c, Li and colleagues developed a digital nanopillar platform integrated with SERS barcode detection system for multiplex quantification of single cytokine molecule (i.e., FGF-2, G-CSF, GM-CSF, CX3CL1) to track immune toxicities in melanoma patients receiving immune checkpoint blockade therapy.43 Single cytokine molecule counting of four targets on nanopillar platform was based on three key concepts: (i) Rayleigh criterion separation for fabricating discrete single cytokine nanopillar chip with discretely separated compartments; (ii) probability-driven Poisson distribution for control of target concentration; and (iii) single particle active SERS nanotags for confocal SERS mapping. The gold topped nanopillars were functionalized with target specific capture antibodies for single cytokine capture and SERS nanotags were used for target recognition via SERS spectroscopy. The preparation of SERS nanotags involved the conjugation of different Raman reporters and four target antibodies on the Au–Ag alloy nanoboxes. The highly sensitive detection of four cytokines enabled the successful detection of immune toxicities in treated melanoma patients before the onset of symptoms. Furthermore, statistical analysis was performed with the inclusion of four cytokine biomarkers in LDA for wider and accurate patient sample analysis. The outcome from the LDA analysis showed a clear discrimination for the patients with severe cytokine level and was able to recognize minor differences in grade 1 immune toxicities patients.
4. Label-free multiplexing
Different from the previously discussed barcode-enabled multiplexing strategy, label-free multiplexing approach utilizes the distinct biomolecule-specific Raman signals for establishing the patient signature for personalized identification.44 Due to the diverse components and molecular structures of biomolecules, their vibrational and rotational information constitute the unique Raman fingerprints for differentiation.44 The direct utilization of the intrinsic Raman spectra from biomolecules for multiplexed analysis features streamlined operation. However, the relatively weak Raman peaks from biomolecules necessitates the employment of high performance plasmonic nanostructures to provide remarkable Raman enhancement.10 Typically, by integrating highly efficient plasmonic nanostructures with microfluidic chips, the label-free multiplexing analysis can be conducted by two approaches: (1) generating specific biomolecule Raman spectra with the assistance of capture probes to purify the targets; and (2) producing the ensemble spectra as a signature from a mixture, followed by performing multivariate statistical analysis for classification. Due to complex Raman spectra and interpretations, identifying biomarkers based on their intrinsic Raman fingerprint becomes increasingly difficult. Furthermore, the typical low Raman active cross-section of biomolecules (e.g., protein) results in the weak identifiable signals provides lower sensitivity. With the advantage of revealing subtle distinctions, deep learning and artificial neural networks are seeing a growing impact in the effective processing of big sensing data for complex spectra allowing classification and identification of specific targets.28,29 Particularly, with artificial intelligence (AI) for data analysis, the collection and construction of a SERS fingerprint database can have an increased opportunity for future clinical application.45 For instance, Shi and co-workers set up a label free SERS database of DNA and developed AI-based discrimination of tumor suppressor genes. On the AgNP-deposited silicon wafer SERS chip, they first collected three kinds of tumor suppressor genes with different base lengths (i.e., p16, p21, and p53 fragments). The collected SERS spectra were then processed to extract independent parameters (i.e., peak number and peak height) as the training data. A deep neural network (DNN) model was constructed with an input layer, multiple hidden layers, and an output layer to train the input data. Particularly, the DNN model involved a back-propagation algorithm and internal adjustable parameters to compute the data in each layer and in in turn trained the machine. Verifying the DNN model on new data, both the single DNA targets and DNA mixtures showed recognition with an accuracy rate of 90.28%.45In the first strategy of generating certain biomarker Raman signals for cancer detection, Sim et al. developed a head-flocked gold nanopillar chip to profile three miRNA biomarkers (i.e., miR-10b, miR-21, and miR-373) (Fig. 5a).46 Specifically, a gold nanopillar chip was fabricated by depositing gold via an electron beam evaporation onto a mask less etched Si nanopillar structure. After functionalized with capture DNA probes, the gold nanopillar chip was able to selectively capture the target miRNAs. Importantly, the gold nanopillars can spontaneously form the “flock” structures with the liquid flowing through the chip to induce the elastocapillary force, which was defined as the “head-flocking” effect. Sufficient hot spots generated by the head-flocking effect greatly contributed to the successful enhancement of three miRNA Raman signals. This label-free microfluidic SERS assay achieved ultra-high sensitivity down to fM level for three miRNA targets and quantified miRNAs directly from simulated patient serum, indicating the potential for future cancer diagnosis.
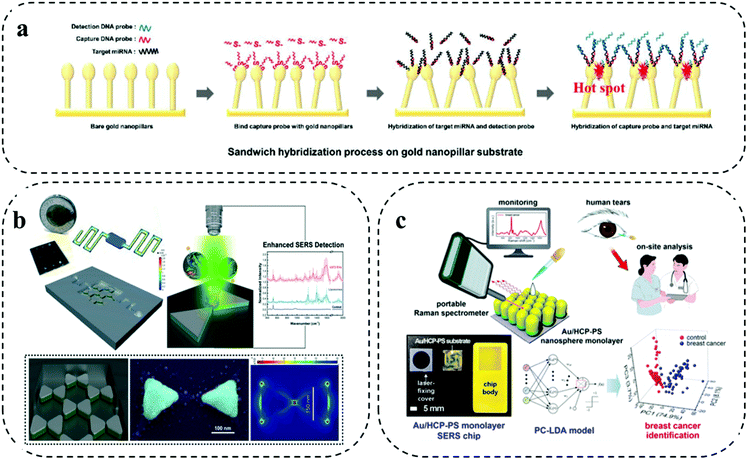 | ||
| Fig. 5 Label-free multiplexing on SERS microfluidic systems. (a) Gold nanopillar chip for specifically profiling three miRNA biomarkers. Reproduced with permission from ref. 46. Copyright 2019, Royal Society of Chemistry. (b) Nanobowtie structure integrated microfluidic device to enhance EV Raman signals. Adapted from ref. 47 with permission. Copyright 2021, Royal Society of Chemistry (c) Au/HPC-PS SERS chip for generating protein Raman signatures from human tears. Reproduced with permission from ref. 48. Copyright 2020, American Chemical Society. | ||
As for the second strategy of directly interrogating the sample, the ensemble Raman spectra reflects a panel of targets, which is difficult to clearly assign the peaks to specific biomarkers. Nevertheless, with the help of chemometric analysis, the Raman signature from the entire biomolecules is able to distinguish cancer samples from health ones. For instance, Jalali and co-workers reported the label-free Raman measurement of glioma extracellular vesicles (EVs) through integrating the plasmonic nanobowtie structures with a microfluidic device (Fig. 5b).47 By leveraging the locally enhanced electromagnetic field provided via the nanobowtie-shaped antennas and the controllable delivery of EVs via the microfluidic flow manipulation, the device can obtain uniform and reproducible Raman spectra. Significantly, the subsequent principle component analysis (PCA) of the Raman spectra unambiguously differentiated the clusters of cancerous cell lines (i.e., U373 and U87) from non-cancerous cell line (i.e., NHA), suggesting the potential of this label-free SERS microfluidic platform in future cancer diagnosis.
Recently, Kim et al. demonstrated a label-free SERS microchip for on-site breast cancer detection using human tears (Fig. 5c).48 The SERS microchip was fabricated by depositing a gold layer on a free-standing hexagonal-close-packed polystyrene monolayer (i.e., Au/HPC-PS SERS chip). Using human tears as a non-invasive source, the Au/HPC-PS SERS microchip showed the feasibility of generating fingerprint Raman signals from breast cancer patients and healthy controls, which was identified by the principle component linear discriminant analysis (PC-LDA). Furthermore, the leave-one-out cross-validation (LOOCV)-assisted PC-LDA analysis showed that the Au/HPC-PS SERS microchip label-free tear detection can achieve a clinical sensitivity and specificity of 92% and 100%, respectively. Having demonstrated the possibility of this technique in a small cohort of patient samples, a further verification of the performance in a larger patient cohort can help the assay transit into clinical use.
Using highly active silver nanoparticles and a droplet-based device, Hassoun and colleagues designed a microfluidic chip to diagnose different types of leukemia.49 In this system, tiny nanoliter droplets were firstly generated by a mixture of silver nanoparticles, KCl, and leukemia cell lysate, in which KCl was used as aggregation agent to create hot spots to enhance Raman signals. Within the confined microfluidic channel, the samples in the droplets were well mixed and allowed the acquisition of SERS spectra using a short integration time (i.e., 1 s). The generated reproducible spectra were mainly associated to the protein and lipids adsorbed to silver nanoparticle, which enabled a robust classification (i.e., accuracy above 99%) of three leukemia cell lines after applying to PCA. This work provided a good example of generating reproducible SERS spectra from nanoparticle colloids by combining with microfluidic devices.
5. Outlook
SERS microfluidic platforms have shown their potential to create multimolecular profiles in liquid biopsy. Table 1 provides an overview of three different multiplexing approaches. Current state of the arts such as polymerase chain reaction (PCR), enzyme linked immunosorbent assay (ELISA), immunohistochemistry, flow cytometry, cytology, fluorescence in situ hybridisation (FISH), or fluorescence microscopy have limitations for biomarker analysis.50,51 (For instance, technologies such as ELISA can have long analysis time and low sensitivity, fluorescence signals can suffer from photobleaching etc.). Microfluidic based SERS multiplexing platforms may overcome these drawbacks when applied with appropriate SERS strategies and microfluidic substrates.18 Yet, to translate these promising platforms from proof-of-concept to the clinical environment, future efforts should gear around strategies that make these platforms amendable for routine diagnosis and monitoring. This will require technology innovation from both the ‘fluidic’ side and SERS side to suit the clinical needs. From the ‘fluidic’ side, there were only a few demonstrations of fully automated sample-to-answer platforms. Sample preparation remained to be one of the major hurdles for integration into SERS microfluidic platforms. The automation of the platforms would further be advantageous as is it improves assay performance (e.g., reproducibility), which is a key factor for clinical implementation. Microfluidic platforms, and to a lesser degree nanofluidic system, can be produced at relative low cost and in large quantities. Thus, the development of disposable sample-to-answer chips seems feasible.| Multiplexing system | Biomarkers | Type of cancer | Detection method – Raman reporters (cm−1) | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|---|
| Spatial multiplexing | miRNA | Breast | FAM – 1332 | High sensitivity | High sample volume | 11 |
| Cy5 – 1366 | ||||||
| ROX – 1503 | ||||||
| Proteins | Breast | DTNB – 1333 | Design complications | 24 | ||
| 4MBA – 1080 | ||||||
| 2NAT – 1378 | ||||||
| Proteins | Prostate | MGITC – 1612 | Handling errors | 22 | ||
| Cytokines | Breast, Lung | FC – 1178 | 21 | |||
| p-MBA – 1078 | ||||||
| DTNB – 1326 | ||||||
| Barcode-enabled multiplexing | Proteins | Prostate, | NTP – 1341 | High sensitivity | Spectral overlapping | 13 |
| Colorectal, | BTP – 1071 | |||||
| Hepatocellular | TFTP – 885 | |||||
| EVs | Melanoma | MBA – 1075 | 33 | |||
| TFMBA – 1375 | ||||||
| DTNB – 1335 | ||||||
| MPY – 1000 | ||||||
| DNA | Melanoma | Cy5 – 1589 | 32 | |||
| Colorectal | FAM – 1325 | |||||
| ROX – 1493 | ||||||
| CTCs | Melanoma | DTNB – 1337 | High multiplexing | 15 | ||
| Breast | MBA – 1075 | |||||
| TFMBA – 1175 | ||||||
| CCSC & CTCs | Breast | NBA – 469 | 39 | |||
| QNT – 985 | ||||||
| NPT – 1051 | ||||||
| MDP – 1096 | ||||||
| TP – 1142 | ||||||
| Label-free multiplexing | miRNA | Breast | Nil | Label-free | Lower sensitivity | 41 |
| EVs | Glioblastoma | Nil | Intrinsic spectrum of samples | Spectral complexity | 42 | |
| Human tears | Breast | Nil | 43 | |||
As for the SERS side, several improvements can be made to go together with the microfluidic systems for the advanced and integrated chips. (1) Handheld Raman spectrometers that implement rapid and in situ signal readout should be designed to apply on microfluidic devices. The benefit of using handheld Raman spectrometer in chemical analysis has been emerging in the past decades. A further extension of the portable Raman spectrometer into microfluidic systems should open a new area for oncology analysis by allowing routine and point-of-care biomarker profiling. (2) The generation of reproducible and robust SERS signals is necessary to enhance the inter/intra-laboratory comparison and essential for the broad clinical applications. Particularly, the inconsistent signal is an issue for the Raman intensity-based biomarker quantification, which can witness large signal fluctuations due to the uncontrollable hot spots generated from nanoparticle aggregation. To solve this conundrum, the synthesis of highly uniform and stable plasmonic nanostructures as well as the use of an internal standard Raman reporter as a reference can be two promising strategies. (3) A larger panel of biomarkers is expected to achieve simultaneous analysis as the clinical need of a unique patient signature through screening a constellation of biomolecules. Though SERS has the advantage of high multiplexing capability (e.g., 31 plex), the current Raman barcoded detection confines in a relatively low target number (e.g., 4 or 5). To expand the multi-biomarker detection, the utilization of novel Raman reporters (e.g., alkyne molecules) that have minimal spectral overlap should be explored. As another option, the employment of advanced AI technique to learn and deconvolve the overlapped Raman spectra into identifiable codes can be a prospective approach.
Together, future research toward disposable micro/nanofluidics and combined with handheld, reproducible, and higher multiplexing SERS will be critical for the development of fully automated microfluidic SERS platform, which enables sample-to-answer analysis of multiple biomarkers. Such a next generation of microfluidic SERS is envisaged to deliver the detailed molecular profile to support accurate decision making in precision oncology.
6. Conclusion
Precision oncology is challenging the idea of “one solution for all” in terms of cancer treatment. Similarly, one marker-based analysis has also been proven not to be enough for diagnosis and monitoring of a complex disease like cancer. An ultrasensitive multiplexed platform could therefore be the central point of precision oncology for accurate diagnosis and monitoring of cancer at the very early stage. In this mini review, we evaluated SERS enabled microfluidic devices in terms of their sensitivity and multiplexing capability. We found that spatial multiplexing is unable to provide a true multiplexed platform. Label free multiplexing started attracting attention and showing promises supported by artificial intelligence which could overcome the limitations associated with the labelling. However, it is still a long way for label free multiplexing to be able to have a place in clinical settings. We conclude that the integration of SERS barcoded systems with spatial multiplexing could be highly powerful to provide a true multiplexed platform at this stage. Moreover, the barcode enabled multiplexed platforms with controllable and rapid fluid flow, 3D nanostructures as the sensor surface as well as the super plasmonic SERS nanoparticles can achieve the required sensitivity for detecting biomarkers at trace concentrations. Finally, we believe that SERS-microfluidic platforms with the support from AI could deliver the ultimate platform for precision oncology to significantly increase the patient survival rate.Abbreviations
| 3D | 3 Dimensional |
| ac-EHD | Alternating current electrohydrodynamics |
| AFP | Alpha-fetoprotein |
| AgNPs | Silver nanoparticles |
| aM | Attomolar |
| Au–Ag | Gold-silver |
| AuNPs | Gold nanoparticles |
| AuNRs | Gold nanorods |
| BTP | 4-Bromothiophenol |
| CCSC | Circulating cancer stem cells |
| CEA | Carcinoembryonic antigen |
| CLS | Classical least square |
| CTCs | Circulating tumour cells |
| ctDNA | Circulating tumour DNA |
| ctRNA | Circulating tumour RNA |
| CX3CL1 | Chemokine (C-X3-C motif) ligand |
| Cy5 | Cyanine 5 |
| DNN | Deep neural network |
| DTNB | 5,5′-Dithiobis (2-nitrobenzoic acid) |
| EDC/NHS | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial–mesenchymal transition |
| EPAC | Extracellular vesicles (EVs) phenotype analyser chip |
| EPCAM | Epithelial cellular adhesion molecule |
| ELISA | Enzyme linked immunosorbent assay |
| EVs | Extracellular vesicles |
| FAM | 5(6)-Carboxyfluorescein |
| fM | Femtomolar |
| FC | Fuchsin |
| FGF-2 | Fibroblast growth factor receptor |
| FISH | Fluorescence in situ hybridisation |
| G-CSF | Granulocyte colony stimulating factor |
| GM-CSF | Granulocyte macrophage colony stimulating factor |
| GO | Graphene oxide |
| HER2 | Human epidermal growth factor receptor 2 |
| IGF1R | Insulin-like growth factor 1 receptor |
| IL | Interleukin |
| IO–Au | Iron oxide-gold |
| LDA | Linear discriminant analysis |
| LNGFR | Low affinity nerve growth factor receptor |
| 4MBA | 4-Mercaptobenzoic acid |
| p-MBA | p-Mercaptobenzoic acid |
| MCAM | Melanoma cell adhesion molecule |
| MCSP | Melanoma-associated chondroitin sulphate proteoglycan |
| MGITC | Malachite green isothiocyanate |
| miRNA | MicroRNA |
| MPD | 4-Mercaptopyridine |
| MPY | 4-Mercaptopyridine |
| MUC1 | Mucin 1 |
| 2NAT | 2-Naphthalenethiol |
| NBA | Nile blue A |
| NPC | Nasopharyngeal carcinoma |
| NPT | 1-Naphthalenethiol |
| NTP | 4-Nitrothiophenol |
| NY-scFv | Nanoyeast single chain variable fragments |
| PCA | Principle component analysis |
| PC-LDA | Principle component linear discriminant analysis |
| PCR | Polymerase chain reaction |
| PSA | Prostate-specific antigen |
| QNT | 2-Quinolinethiol |
| RAN | Raman-active nanoprobe |
| ROX | X-Rhodamine |
| SERS | Surface enhanced Raman spectroscopy |
| SH-PEG-COOH | Thiol-polyethylene glycol-carboxyl |
| TFMBA | 2,3,5,6-Tetrafluoro-MBA |
| TFTP | 2,3,5,6-Tetrafluorothiophenol |
| TP | Thiophenol |
| XRITC | X-Rhodamine-5-(and-6)-isothiocyanate |
| VEGF | Vascular endothelial growth factor |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Australian Research Council for the support (DP160102836 and DP210103151). J. L. acknowledges support from The Commonwealth Scientific and Industrial Research Organisation (CSIRO). A. W. and A. A. I. S. thank the National Health and Medical Research Council for funding (APP1173669 and APP1175047). The support obtained through these funding agencies has significantly contributed to the current research.References
- R. Mirnezami, J. Nicholson and A. Darzi, N. Engl. J. Med., 2012, 366, 489–491 CrossRef PubMed.
- V. Prasad, T. Fojo and M. Brada, Lancet Oncol., 2016, 17, e81–e86 CrossRef PubMed.
- F. Di Nicolantonio, P. P. Vitiello, S. Marsoni, S. Siena, J. Tabernero, L. Trusolino, R. Bernards and A. Bardelli, Nat. Rev. Clin. Oncol., 2021, 1–20 Search PubMed.
- J. Kaiser, Science, 2018, 359, 259 CrossRef CAS PubMed.
- G. De Rubis, S. R. Krishnan and M. Bebawy, Trends Pharmacol. Sci., 2019, 40, 172–186 CrossRef CAS PubMed.
- N. S. Yee, J. Clin. Med., 2020, 9, 2556 CrossRef PubMed.
- M. R. Speicher and K. Pantel, Nat. Biotechnol., 2014, 32, 441–443 CrossRef CAS PubMed.
- C. Alix-Panabières and K. Pantel, Nat. Biomed. Eng., 2017, 1, 1–3 CrossRef.
- F. Tian, C. Liu, L. Lin, Q. Chen and J. Sun, TrAC, Trends Anal. Chem., 2019, 117, 128–145 CrossRef CAS.
- J. Li, A. Wuethrich, S. Dey, R. E. Lane, A. A. Sina, J. Wang, Y. Wang, S. Puttick, K. M. Koo and M. Trau, Adv. Funct. Mater., 2020, 30, 1909306 CrossRef CAS.
- S. Meng, R. Chen, J. Xie, J. Li, J. Cheng, Y. Xu, H. Cao, X. Wu, Q. Zhang and H. Wang, Biosens. Bioelectron., 2021, 190, 113470 CrossRef CAS PubMed.
- C. Wang, Y. Zhang, W. Tang, C. Wang, Y. Han, L. Qiang, J. Gao, H. Liu and L. Han, Anal. Chim. Acta, 2021, 338791 CrossRef CAS PubMed.
- Q. Xiong, C. Y. Lim, J. Ren, J. Zhou, K. Pu, M. B. Chan-Park, H. Mao, Y. C. Lam and H. Duan, Nat. Commun., 2018, 9, 1–11 CrossRef CAS PubMed.
- C. Liu, Q. Feng and J. Sun, Adv. Mater., 2019, 31, 1804788 CrossRef CAS PubMed.
- K. K. Reza, S. Dey, A. Wuethrich, J. Wang, A. Behren, F. Antaw, Y. Wang, A. A. I. Sina and M. Trau, ACS Nano, 2021, 15, 11231–11243 CrossRef CAS PubMed.
- Z. Wang, S. Zong, Y. Wang, N. Li, L. Li, J. Lu, Z. Wang, B. Chen and Y. Cui, Nanoscale, 2018, 10, 9053–9062 RSC.
- A. I. Pérez-Jiménez, D. Lyu, Z. Lu, G. Liu and B. Ren, Chem. Sci., 2020, 11, 4563–4577 RSC.
- J. Wu, X. Zhou, P. Li, X. Lin, J. Wang, Z. Hu, P. Zhang, D. Chen, H. Cai and R. Niessner, Anal. Chem., 2021, 93, 8799–8809 CrossRef CAS PubMed.
- J. Wang, K. M. Koo, Y. Wang and M. Trau, Adv. Sci., 2019, 6, 1900730 CrossRef CAS PubMed.
- Y. Chu, Y. Gao, W. Tang, L. Qiang, Y. Han, J. Gao, Y. Zhang, H. Liu and L. Han, Anal. Chem., 2021, 93, 5129–5136 CrossRef CAS PubMed.
- A. Kamińska, K. Winkler, A. Kowalska, E. Witkowska, T. Szymborski, A. Janeczek and J. Waluk, Sci. Rep., 2017, 7, 1–11 CrossRef PubMed.
- R. Gao, Z. Cheng, X. Wang, L. Yu, Z. Guo, G. Zhao and J. Choo, Biosens. Bioelectron., 2018, 119, 126–133 CrossRef CAS PubMed.
- L. Wu, Z. Wang, Y. Zhang, J. Fei, H. Chen, S. Zong and Y. Cui, Nano Res., 2017, 10, 584–594 CrossRef CAS.
- Z. Zheng, L. Wu, L. Li, S. Zong, Z. Wang and Y. Cui, Talanta, 2018, 188, 507–515 CrossRef CAS PubMed.
- Y. Nie, C. Jin and J. X. Zhang, ACS Sens., 2021, 6, 2584–2592 CrossRef CAS PubMed.
- M. R. Willner, K. S. McMillan, D. Graham, P. J. Vikesland and M. Zagnoni, Anal. Chem., 2018, 90, 12004–12010 CrossRef CAS PubMed.
- D. Lin, C.-L. Hsieh, K.-C. Hsu, P.-H. Liao, S. Qiu, T. Gong, K.-T. Yong, S. Feng and K. V. Kong, Nat. Commun., 2021, 12, 1–16 CrossRef PubMed.
- F. Cui, Y. Yue, Y. Zhang, Z. Zhang and H. S. Zhou, ACS Sens., 2020, 5, 3346–3364 CrossRef CAS PubMed.
- L. Guerrini, E. Garcia-Rico, A. O’Loghlen, V. Giannini and R. A. Alvarez-Puebla, Cancers, 2021, 13, 2179 CrossRef CAS PubMed.
- D. Liang, Q. Jin, N. Yan, J. Feng, J. Wang and X. Tang, Adv. Biosyst., 2018, 2, 1800100 CrossRef.
- S. Tian, H. Li, Z. Li, H. Tang, M. Yin, Y. Chen, S. Wang, Y. Gao, X. Yang and F. Meng, Nat. Commun., 2020, 11, 1–9 Search PubMed.
- L. Wu, A. Teixeira, A. Garrido-Maestu, L. Muinelo-Romay, L. Lima, L. L. Santos, M. Prado and L. Diéguez, Biosens. Bioelectron., 2020, 165, 112392 CrossRef CAS PubMed.
- J. Wang, A. Wuethrich, A. A. I. Sina, R. E. Lane, L. L. Lin, Y. Wang, J. Cebon, A. Behren and M. Trau, Sci. Adv., 2020, 6, eaax3223 CrossRef CAS PubMed.
- A. Rotem, O. Ram, N. Shoresh, R. A. Sperling, A. Goren, D. A. Weitz and B. E. Bernstein, Nat. Biotechnol., 2015, 33, 1165–1172 CrossRef CAS PubMed.
- K. Kamil Reza, J. Wang, R. Vaidyanathan, S. Dey, Y. Wang and M. Trau, Small, 2017, 13, 1602902 CrossRef PubMed.
- Z. Zhang, J. Wang, K. B. Shanmugasundaram, B. Yeo, A. Möller, A. Wuethrich, L. L. Lin and M. Trau, Small, 2020, 16, 1905614 CrossRef CAS PubMed.
- K. K. Reza, A. A. I. Sina, A. Wuethrich, Y. S. Grewal, C. B. Howard, D. Korbie and M. Trau, Biosens. Bioelectron., 2019, 126, 178–186 CrossRef CAS PubMed.
- Y. Zhang, Z. Wang, L. Wu, S. Zong, B. Yun and Y. Cui, Small, 2018, 14, 1704433 CrossRef PubMed.
- H.-Y. Cho, M. K. Hossain, J.-H. Lee, J. Han, H. J. Lee, K.-J. Kim, J.-H. Kim, K.-B. Lee and J.-W. Choi, Biosens. Bioelectron., 2018, 102, 372–382 CrossRef CAS PubMed.
- Z. Cheng, N. Choi, R. Wang, S. Lee, K. C. Moon, S.-Y. Yoon, L. Chen and J. Choo, ACS Nano, 2017, 11, 4926–4933 CrossRef CAS PubMed.
- R. E. Wilson Jr, R. O’Connor, C. E. Gallops, E. A. Kwizera, B. Noroozi, B. I. Morshed, Y. Wang and X. Huang, ACS Appl. Mater. Interfaces, 2020, 12, 47220–47232 CrossRef CAS PubMed.
- D. Sun, F. Cao, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 2551–2558 CrossRef CAS PubMed.
- J. Li, A. Wuethrich, A. A. Sina, H.-H. Cheng, Y. Wang, A. Behren, P. N. Mainwaring and M. Trau, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- L. A. Lane, X. Qian and S. Nie, Chem. Rev., 2015, 115, 10489–10529 CrossRef CAS PubMed.
- H. Shi, H. Wang, X. Meng, R. Chen, Y. Zhang, Y. Su and Y. He, Anal. Chem., 2018, 90, 14216–14221 CrossRef CAS PubMed.
- W. H. Kim, J. U. Lee, S. Song, S. Kim, Y. J. Choi and S. J. Sim, Analyst, 2019, 144, 1768–1776 RSC.
- M. Jalali, I. I. Hosseini, T. AbdelFatah, L. Montermini, S. W. Hogiu, J. Rak and S. Mahshid, Lab Chip, 2021, 21, 855–866 RSC.
- S. Kim, T. G. Kim, S. H. Lee, W. Kim, A. Bang, S. W. Moon, J. Song, J.-H. Shin, J. S. Yu and S. Choi, ACS Appl. Mater. Interfaces, 2020, 12, 7897–7904 CrossRef CAS PubMed.
- M. Hassoun, J. Rüger, T. Kirchberger-Tolstik, I. W. Schie, T. Henkel, K. Weber, D. Cialla-May, C. Krafft and J. Popp, Anal. Bioanal. Chem., 2018, 410, 999–1006 CrossRef CAS PubMed.
- J. Perumal, Y. Wang, A. B. E. Attia, U. Dinish and M. Olivo, Nanoscale, 2021, 13, 553–580 RSC.
- S. Abalde-Cela, L. Wu, A. Teixeira, K. Oliveira and L. Diéguez, Adv. Opt. Mater., 2021, 2001171 CrossRef CAS.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |