Open Access Article
Open Access ArticleWater solubilization of paclitaxel using polypeptides for cancer therapy†
Riku
Kawasaki‡
 *a,
Shogo
Kawamura‡
a,
Shodai
Hino‡
*a,
Shogo
Kawamura‡
a,
Shodai
Hino‡
 ab,
Keita
Yamana
a and
Atsushi
Ikeda
ab,
Keita
Yamana
a and
Atsushi
Ikeda
 *a
*a
aProgram of Applied Chemistry, Graduate School of Advanced Science and Engineering, Hiroshima University, 1-4-1 Kagamiyama, Higashi Hiroshima, 739-8527, Japan. E-mail: riku0528@hiroshima-u.ac.jp; aikeda@hiroshima-u.ac.jp
bBiomedical Research Institute, National Institute of Advanced Industrial Science and Technology, AIST; 1-8-31 Midorigaoka, Ikeda, 563-8577, Japan
First published on 12th November 2021
Abstract
To address problems in drug discovery, we developed approaches to solubilize hydrophobic compounds using polypeptides including poly-L-lysine (PLL) via high-speed vibration milling. The current method can be used to prepare highly concentrated dispersions of the anticancer drug paclitaxel. Moreover, the complex of paclitaxel with PLL efficiently induced cancer cell death via apoptosis without harmful side effects on healthy cells. Finally, we demonstrated that actively growing cancer cell spheroid growth was efficiently suppressed after treatment.
Introduction
In recent decades, pharmaceutical companies have developed numerous technologies for drug discovery, such as structure-based drug design,1 combinatorial chemistry,2 and high-throughput screening.3 Although these technologies accelerated drug discovery, the pharmaceutical industry has faced a major problem in finding and developing novel therapeutics.4 Despite enormous increases in pharmaceutical R&D spending, the productivity of the industry has gradually decreased for several reasons including insufficient therapeutic efficacy, acute toxicity, and poor water solubility of compounds.5–7 More than 40% of candidate compounds have not advanced beyond the initial screening phase with no evaluation of biological activity.5From these viewpoints, the development of water-solubilizing techniques is urgently required for hydrophobic compounds to accelerate drug discovery by expanding and opening the bioavailability of lead compounds.8 To improve the water solubility of hydrophobic compounds, several approaches have been employed such as conjugation with hydrophilic moieties,9 pH adjustment,10,11 cosolvency,10,12 the use of surfactant,13,14 and encapsulation.9,15–17 Among these approaches, complexation with biocompatible molecules such as phospholipids18 and polysaccharides9,16,17 is a robust strategy because this approach does not require conjugation techniques, which can cause unfavorable side effects in patients.17
In this aim, we have developed a water-solubilizing technique based on mechanochemical ball milling known as high-speed vibration milling (HSVM) using cyclodextrins and polysaccharides.19–26 The HSVM technique represents a powerful means of obtaining highly concentrated dispersions of hydrophobic compounds such as fullerene derivatives,19 porphyrin derivatives,20–23 and phthalocyanine derivatives24 in water without any using harmful organic solvents.27 Recently, we found that the technique can preserve information from crystals such as chirality25 and mechanochromism.26 These fascinating properties of HSVM encouraged us to develop polypeptide-based water solubilization methodologies because of their excellent biocompatibility, biodegradability, and functionality.28–31
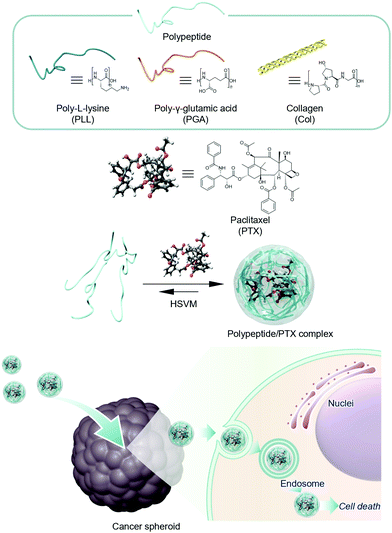
In this work, we report the water solubilization of the hydrophobic anticancer drug paclitaxel (PTX) using polypeptides, including poly-L-lysine (PLL), poly-L-γ-glutamic acid (PGA), and collagen (Col), for cancer treatment (Fig. 1). The developed systems improved the solubility and stability of PTX in water. We evaluated the anticancer effects of PTX complexes against murine colon cancer. Our system had enhanced cytotoxicity against cancer cell lines and 76-fold stronger antitumor effects than free PTX in DMSO. In addition, the results of a spheroid growth inhibition assay suggested that our system was more practicable for cancer treatment.
Results and discussion
Preparation and characterization of polypeptide/PTX complexes
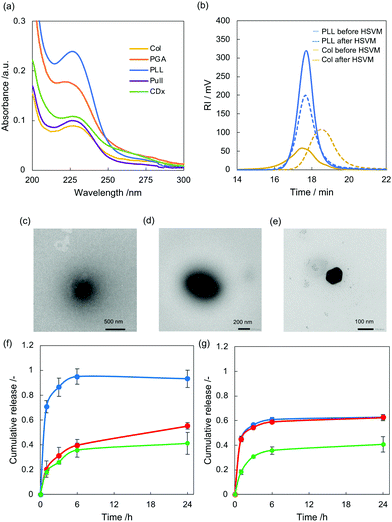
The complexes of PTX with polypeptides including PLL, PGA, and Col were prepared via HSVM as previously reported.19–26 The absorption spectra of the resulting dispersions were measured to confirm the water solubilization of PTX (Fig. 2a). PLL, PGA, and Col could dissolve PTX at concentrations of 80.8, 60.1, and 29 μM, respectively (Table 1). To prove the usefulness of HSVM techniques in water solubilization using polypeptides, we compared the solubility of PTX with conventional water solubilization techniques including heating32 and sonication methods (Fig. S1, ESI†).33 In addition, polypeptides could dissolve larger amount of hydrophobic PTX than conventional water solubilizers including β-cyclodextrin (CDx) and pullulan (Pull) (Fig. 2a). The HSVM technique generated higher concentrations of PTX than the other conventional methods. This result supports that the HSVM method using polypeptides is a powerful means to improve the solubility of hydrophobic compounds. We next examined changes in the molecular weights of the polypeptides following HSVM via GPC measurement (T = 40 °C; eluent, hexafluoroisopropanol; calibration standards, polymethylmethacrylate, 2.78 × 103–1.01 × 106 g mol−1).34 As presented in Fig. 2b, the elution time of PLL did not change after HSVM. By contrast, the elution time of Col was delayed after HSVM, suggesting a molecular weight decrease (Table S1, ESI†). These differences may be caused by differences in the persistent length of the polymer35–37 because stiffer polymers should be more easily degraded. Unfortunately, GPC measurement for PGA could not be performed because PGA was not dissolved in hexafluoroisopropanol.| PTX (μM) | D hy (nm) | PDI | ζ-potential (mv) | |
|---|---|---|---|---|
| All samples were measured in water (pH, 7.4; 25 °C). PDI was calculated using the cumulant method. Zeta potential was measured using capillary cells. | ||||
| PLL/PTX | 81 | 240 ± 10 | 0.26 | +21 ± 6 |
| PGA/PTX | 60 | 310 ± 20 | 0.21 | −47 ± 1 |
| Col/PTX | 29 | 140 ± 1 | 0.21 | +1.5 ± 0.3 |
Dynamic light scattering (DLS) measurement revealed that the hydrodynamic diameter (Dhy) of the complex of PTX with PLL (PLL/PTX complex), PGA (PGA/PTX complex), and Col (Col/PTX complex) were 235, 314, and 140 nm (Table 1), respectively. The sizes of the complexes are reasonable for enhanced permeable and retention effects.38,39 In addition, the electrical charges of the complex are highly dependent on the electrical character of the polymer backbone.
We additionally conducted morphological observation of these complexes via transmission electron microscopy (Fig. 2c–e). Spherical morphologies were found in each sample, and their size distribution corresponded to the DLS results. We next carried out long-term sustainability of the polypeptides/PTX complex was evaluated by measuring absorbance from PTX using UV-Vis absorption spectra (Fig. S2, ESI†). After 7 days incubation in water, Col/PTX exhibited the highest sustainability among these three polypeptides. In addition, we examined thermal stability of the polypeptides/PTX complex (Fig. S3, ESI†). After heating at 80 °C for 30 min, over 70% of PTX was still encapsulated within polypeptides in all the systems, suggesting polypeptides/PTX complex are thermally stable. PLL/PTX complex could retain 100% of hydrophobic PTX within their polymer matrix. We further evaluated colloidal stability of polypeptides/PTX complex in water and in cell culture medium (Fig. S4, ESI†). Hydrodynamic diameter of polypeptides/PTX complex did not significantly change in water with maintaining narrow dispersity. In contrast, hydrodynamic diameter of PLL/PTX complex and Col/PTX complex gradually increased in the presence of cell culture media, suggesting serum protein bind to the complexes. Anionic PGA/PTX was relatively stable in cell culture media.
To address the mechanism of complex formation between hydrophobic PTX and polypeptides, we investigated the hydrophobicity of the polymer matrix in polypeptides using pyrene.40,41 The ratio of fluorescence intensity at 375 (I375) and 386 nm (I386) is known as the polarity indicator of the microenvironment established by polymer chains. Pyrene encapsulated by polypeptides was prepared via HSVM, and the fluorescence spectrum of each complex was measured using a fluorometer (excitation wavelength, 336 nm; Fig. S5, ESI†). The I375/I386 ratios for PLL, PGA, and Col were 1.08, 0.68, and 0.98, respectively. This result suggests that polypeptides provide a hydrophobic nano-space to solubilize hydrophobic compounds. Judging from the solubilization efficiency of PTX, we performed the following experiments using PGA and PLL.
We next examined pH-triggered cargo release using Coumarin6 (C6) as a model drug for PLL and PGA have been used as pH responsive delivery platforms. The complex of C6 with polypeptides was prepared via HSVM. C6 was efficiently encapsulated similarly as PTX (Fig. S6, solid line and Table S2, ESI†). Although the concentration of C6 complexed with PLL was much higher than that of C6 complexed with PGA, the PGA complex produced stronger fluorescence emission than the PLL complex (Fig. S6, dashed line, ESI†). This is because aggregation induced quenching as previously reported.21–24 After incubation with varying pH value (5, 7, and 9), the release of C6 was quantified by measuring fluorescence at each time point. Release of C6 was found even at neutral pH in both C6 complexed with PLL (PLL/C6 complex) and C6 complexed with PGA (PGA/C6 complex) with time and the differences in release profiles of these two systems are not significant (Fig. 2f and g). In case of PLL/C6 complex, the release was enhanced only when the complex was incubated with basic pH. In addition, all the C6 was released in basic pH at 24 h. In contrast, release ratio of C6 increased by exposing both acidic pH and basic pH in case of PGA/C6 complex. These results suggested that current systems can work as pH-responsive payload release systems.
Anticancer effects in vitro
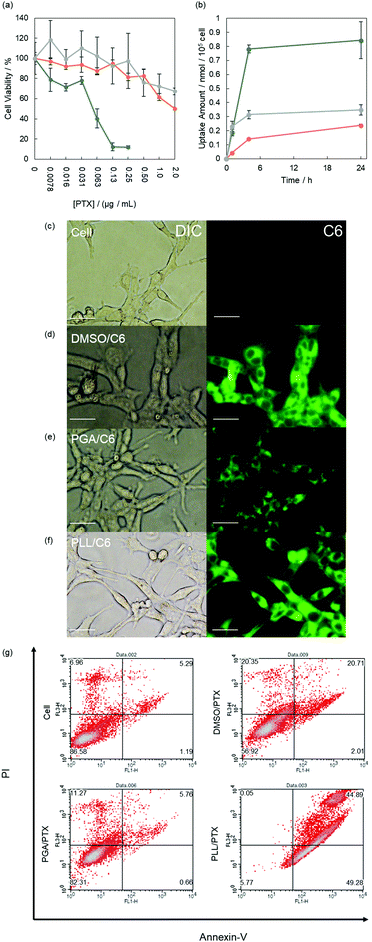
The anticancer effects of PTX complexed with polypeptides were investigated in murine colon carcinoma cells (Colon-26), and their viability was quantified using a modified MTT assay.42 In this study, we used free PTX prepared in DMSO as the control.43 After 24 h of incubation, the viability of the treated cells decreased in a PTX concentration-dependent manner (Fig. 3a), and the half-maximal inhibitory concentrations of PLL/PTX, PGA/PTX, and PTX in DMSO were 0.064, 2.2, and 4.9 nM, respectively. These results suggest that the PLL system strongly interacted with cancer cells because of its electrically cationic character. Moreover, none of the complexes caused apparent cytotoxicity in murine fibroblast-like cells (L929), suggesting our system can lessen the severe side effects of PTX (Fig. S7, ESI†).To clarify the deliverability of the polypeptide systems with that of free drugs prepared in DMSO in vitro,43–45 we quantified the cellular uptake of C6 as a model drug (Fig. 3b). The cellular uptake of C6 increased over time in all systems. After 24 h of incubation, the PLL system delivered cargo molecules with the highest efficiencies among the three systems. Furthermore, the cellular uptake at 4 °C using polypeptides was significantly lower than that at 37 °C, suggesting polypeptides systems are mainly accumulated via endocytosis (Fig. S8, ESI†).43,45,46 By contrast, changes in cellular uptake were not induced by free C6 prepared in DMSO, indicating that C6 is internalized by cells mainly via diffusion.46 We further examined the cellular uptake of C6 using fluorescence microscopy. Fluorescence signals from delivered C6 were detected within cells incubated with all systems (Fig. 3c–f). In addition, their fluorescence intensity corresponded to the results obtained in cellular uptake studies. In case of PTX delivery, subcellular distribution of delivered PTX is critical in therapeutic effects because PTX work as mitotic inhibitor within cytosol.47 To attain cytosolic delivery, endosomal escape of cargo molecules is one of the greatest keys. For the point of view, we carried out subcellular distribution by confocal laser scanning microscope (CLSM). Late endosomes and lysosomes were stained with commercially available dye (Lysotracker Red), and we quantified the overlap ratio of delivered C6 with Lysotracker Red at each time point (3, 6, and 24 h). In case of C6 dissolved in DMSO, large part (over 70%) was not distributed in lysosome because this system delivers cargo molecules mainly via diffusion (Fig. S9 and S10, ESI†) and the co-localization ratio did not change with time. In contrast, C6 delivered with PLL and C6 delivered with PGA were highly overlapped with lysosome at 3 h (over 50%). This fact is corresponding to endocytosis inhibition assay. The co-localization ratio decreased with time in both PLL system and PGA systems and the capability to escape from endosome of PLL system (80%) are higher than that of PGA system (70%). This may be caused by proton sponge effects from the nature of cationic PLL.48
We additionally conducted an apoptosis detection assay based on the Annexin V–PI method.46,49 The cells treated with PTX were analyzed by flow cytometry. For PTX complexed with PGA (PGA, 48.5 μg mL−1; PTX, 4.68 μM), the population of the treated cells were similar to that of nontreated cells, indicating apoptosis was not induced under the current condition (Fig. 3g). Contrarily, a population shift was found following incubation with the PLL/PTX complex and PTX dissolved in DMSO. Moreover, the PLL/PTX complex more strongly induced apoptosis (ratio of early apoptosis, 49%; ratio of late apoptosis, 44%) than free PTX (ratio of early apoptosis, 2%; ratio of late apoptosis, 20%; Fig. S11, ESI†). These results agree with the observed anticancer effects.
Anticancer effects against spheroids
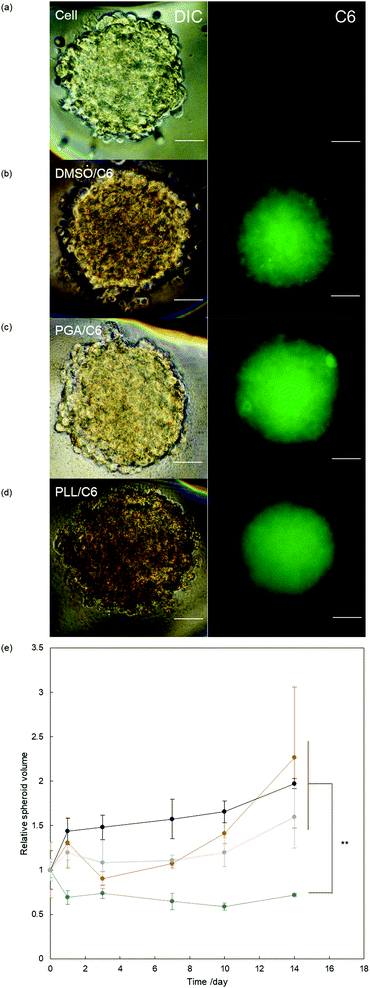
Finally, we examined the anticancer effects of the complexes against cancer spheroids, which represent an advanced tumor tissue model, to demonstrate the potential of this platform for cancer therapy. Initially, we confirmed the deliverability of our systems to deeper sites in cancer spheroids using fluorescence microscopy. All systems could deliver C6 to cancer spheroids (Fig. 4a–d). We further examined distribution of delivered C6 in spheroid by CLSM (Fig. S12, ESI†). As a result, PLL system could deliver cargo molecules deeper site from the surface of spheroids compared to other systems.We evaluated growth inhibition in the spheroids by applying the PGA/PTX complex, PLL/PTX complex, and free PTX (PTX, 2.34 μM). The PGA system and free PTX suppressed spheroid growth in the initial 10 days, but spheroid regrowth was found after 14 days in both groups (Fig. 4e). Conversely, the PLL/PTX complex exhibited the greatest effects on spheroid growth. Thus, the current system using PLL is effective as an anticancer agent in a complex biological setting.
Conclusion
In conclusion, we developed an anticancer agent delivery nanoplatform using polypeptides. Our system exhibited improved water solubility and stability of PTX. The PLL/PTX complex exerted 75-fold stronger antitumor effects than free PTX prepared in DMSO. Moreover, the PLL/PTX system displayed excellent anticancer effects even in 3D spheroids, which are used as a more complicated tumor model. We believe that supramolecular chemistry-based approaches such as that described in this stud have unique features that can be applied in cancer therapy.Experimental methods
Preparation of the complexes of PTX with polypeptides
A polypeptide (PLL, PGA, or Col; 10 mg) and PTX (1.7 mg, 2.0 μmol) were placed in a vial for processing in a high-speed vibrating mill. The mixture of a polypeptide and PTX was processed in the high-speed vibrating mill at 30 Hz for 20 min. The resulting solid mixture was then suspended in Milli-Q water (2.0 mL). The precipitates were removed by centrifugation (4500 rpm, 20 min, 25 °C). The size of each complex was controlled using a probe-type sonicator. Complexes containing pyrene (1.0 g, 5.0 μmol) or C6 (1.8 mg, 5 μmol) were prepared using the same method. Dissolution was also attempted by heating (24 h, 80 °C) or ultrasonic treatment (2 h). After lyophilization and redissolution in DMSO, the concentration of the complexed PTX or C6 was determined by measuring its absorbance using a UV–Vis spectrometer (3600 UV–Vis–NIR spectrometers, Shimadzu, Kyoto, Japan).Characterization of the complexes of PTX with polypeptides
D hy of the complexes of PTX with polypeptides was measured using a DLS instrument (Zeta-sizer Nano ZS; Malvern, Malvern, UK). PDI was calculated by cumulant fitting. The zeta potential of each complex was measured by a Zeta-sizer Nano ZS using capillary cells. Morphological observations were performed using a transmission electron microscope (JEM-1400, JEOL Ltd. Co., Tokyo, Japan). The samples were cast on a hydrophilized, ultrathin, carbon-deposited Cu grid and incubated for 30 min. Afterward, the samples were stained with ammonium molybdate (3 wt%). The samples were observed by transmission electron microscope (acceleration voltage, 100 keV).Cell viability assay
The viability of adherent Colon-26 or L929 cells were measured using the colorimetric MTT assay. Cells (5 × 103) were seeded in 96-well plates (100 μL of medium per well) and cultured for 24 h at 37 °C in an atmosphere of 5% CO2. Then, cells were incubated at 37 °C in a 5% CO2 atmosphere for 24 h in 100 μL of medium containing different concentrations of the polypeptide/PTX complexes. The cells were incubated with 100 μL of medium containing MTT at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution in an atmosphere of 5% CO2 at 37 °C for 3 h. The absorbance was measured using a microplate reader at 450 nm. Data are expressed as the percentage of the absorbance of treated cells relative to the untreated control cells and represented as the means of quadruplicate measurements ± SD.
20 dilution in an atmosphere of 5% CO2 at 37 °C for 3 h. The absorbance was measured using a microplate reader at 450 nm. Data are expressed as the percentage of the absorbance of treated cells relative to the untreated control cells and represented as the means of quadruplicate measurements ± SD.
Cellular uptake of the complex of C6 with polypeptides
Colon26 cells (1 × 105 cells) were seeded in 12-well plates (1000 μL of medium per well) and cultured for 24 h at 37 °C in an atmosphere of 5% CO2. Then, cells were incubated (at 37 °C for 1 h, 4 °C for 4 h, 37 °C for 4 h, or 37 °C for 24 h) in medium containing polypeptide/C6 complexes (4.7 μM). Cells were washed three times with PBS and lysed in 500 μL of RIPA lysis buffer. The precipitates were removed by centrifugation (3500 rpm, 5 min, 25 °C). C6 in the supernatant solution was then extracted using ethyl acetate (800 μL) and evaluated by measuring the fluorescence spectrum (excitation wavelength, 540 nm) in the extract.The deliverability of the complexes to Colon-26 cells
Colon-26 cells (1 × 105 cells) were seeded in 12-well plates (1000 μL of medium per well) and cultured for 24 h at 37 °C in an atmosphere of 5% CO2. Then, cells were incubated at 37 °C in an atmosphere of 5% CO2 for 24 h in medium containing polypeptide/C6 complexes (4.7 μM). Then, cells were washed with PBS and observed using fluorescence microscopy.Cell apoptosis assay
Colon-26 (1 × 105 cells) were seeded in 12-well plates (1000 μL of medium per well) and cultured for 24 h at 37 °C in an atmosphere of 5% CO2. Then, cells were incubated at 37 °C in an atmosphere of 5% for 24 h in medium containing the polypeptide/PTX complexes (4.7 μM). After 24 h of incubation, cells were washed twice with PBS and detached from the plate using trypsin–EDTA. The volume of medium was added, and cells were collected by centrifugation (1000 rpm, 3 min, 25 °C). Next, an Annexin V–FITC Apoptosis Detection Kit (Nacalai Tesque) was chosen to detect and quantify the apoptosis by flow cytometric.Spheroid preparation
Colon-26 cells (200 cell per well) were seeded in 96-well plates (100 μL of medium per well) and cultured for 4 days at 37 °C in an atmosphere of 5% CO2.The deliverability of the complexes to spheroids
Spheroids were incubated 37 °C in an atmosphere of 5% for 24 h in medium containing polypeptide/C6 complexes (2.3 μM). Then, spheroids were washed with PBS and were observed via fluorescence microscopy.Spheroid growth-inhibitory effects
Spheroids were exposed to the polypeptide/PTX complexes (2.3 μM). The spheroidal volume was measured after 0, 1, 3, 7, 10, and 14 days. Spheroid volumes were calculated using the following eqn (1).| V = 4π(long axis)2 × (Short axis)/3 | (1) |
Author contributions
R. K., S. K., and S. H. mainly carried out all the experiments. All the authors conducted and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science, KAKENHI (R. K., JP19K15401). Experiments using transmission electron microscopy were carried out in Natural Science Center for Basic Research and Development, (N-BARD). The author would like to thank Enago (http://www.enago.jp) for the English language review.Notes and references
- H. Sun and D. O. Scott, Chem. Biol. Drug Des., 2010, 75, 3–17 CrossRef CAS.
- R. Liu, X. Li and K. S. Lam, Curr. Opin. Chem. Biol., 2017, 38, 117–126 CrossRef CAS.
- V. Zoete, A. Grosdidier and O. Michielin, J. Cell. Mol. Med., 2009, 13, 238–248 CrossRef CAS PubMed.
- S. Venkatesh and R. A. Lipper, J. Pharm. Sci., 2000, 89, 145–154 CrossRef CAS.
- A. Santos, F. Veiga and A. Figueiras, Materials, 2019, 13, 65 CrossRef.
- R. M. Moshikur, M. R. Chowdhury, M. Moniruzzaman and M. Goto, Green Chem., 2020, 22, 8116–8139 RSC.
- S. Manju and K. Sreenivasan, J. Colloid Interface Sci., 2011, 359, 318–325 CrossRef CAS.
- P. Nkansah, A. Antipas, Y. Lu, M. Varma, C. Rotter, B. Rago, A. El-Kattan, G. Taylor, M. Rubio and J. Litchfield, J. Controlled Release, 2013, 169, 150–161 CrossRef CAS PubMed.
- X. Zhang, R. Zhang, J. Huang, M. Luo, X. Chen, Y. Kang and J. Wu, J. Mater. Chem. B, 2019, 7, 3537–3545 RSC.
- A. Jain, Y. Ran and S. H. Yalkowsky, AAPS PharmSciTech, 2004, 5, 65–67 CrossRef.
- S. Tommasini, M. L. Calabrò, D. Raneri, P. Ficarra and R. Ficarra, J. Pharm. Biomed. Anal., 2004, 36, 327–333 CrossRef CAS PubMed.
- N. Seedher and S. Bhatia, AAPS PharmSciTech, 2003, 4, E33 CrossRef CAS.
- K. T. Oh, T. K. Bronich and A. V. Kabanov, J. Controlled Release, 2004, 94, 411–422 CrossRef CAS PubMed.
- P. Zi, C. Zhang, C. Ju, Z. Su, Y. Bao, J. Gao, J. Sun, J. Lu and C. Zhang, Eur. J. Pharm. Sci., 2019, 134, 233–245 CrossRef CAS.
- G. Davidov-Pardo and D. J. McClements, Trends Food Sci. Technol., 2014, 38, 88–103 CrossRef CAS.
- J. Zhang and P. X. Ma, Adv. Drug Delivery Rev., 2013, 65, 1215–1233 CrossRef CAS.
- P. Pattekari, Z. Zheng, X. Zhang, T. Levchenko, V. Torchilin and Y. Lvov, Phys. Chem. Chem. Phys., 2011, 13, 9014–9019 RSC.
- S. K. Jena, C. Singh, C. P. Dora and S. Suresh, Int. J. Pharm., 2014, 473, 1–9 CrossRef CAS.
- A. Ikeda, T. Iizuka, N. Maekubo, K. Nobusawa, K. Sugikawa, K. Koumoto, T. Suzuki, T. Nagasaki and M. Akiyama, Chem. – Asian J., 2017, 12, 1069–1074 CrossRef CAS.
- T. Yumoto, S. Satake, S. Hino, K. Sugikawa, R. Kawasaki and A. Ikeda, Org. Biomol. Chem., 2020, 18, 6702–6709 RSC.
- S. Hino, R. Funada, K. Sugikawa, K. Koumoto, T. Suzuki, T. Nagasaki and A. Ikeda, Photochem. Photobiol. Sci., 2019, 18, 2854–2858 CrossRef CAS.
- K. Yamana, R. Kawasaki, K. Sugikawa and A. Ikeda, ACS Appl. Bio Mater., 2020, 3, 3217–3225 CrossRef CAS.
- R. Kawasaki, K. Yamana, R. Shimada, K. Sugikawa and A. Ikeda, ACS Omega, 2021, 6, 3209–3217 CrossRef CAS.
- Y. Goto, S. Hino, K. Sugikawa, R. Kawasaki and A. Ikeda, Asian J. Org. Chem., 2020, 9, 1589–1596 CrossRef CAS.
- S. Hino, K. Sugikawa, R. Kawasaki, H. Funabashi, A. Kuroda and A. Ikeda, ChemPhotoChem, 2020, 4, 577–581 CrossRef CAS.
- R. Omokawa, R. Kawasaki, K. Sugikawa, T. Nishimura, T. Nakaya and A. Ikeda, ACS Appl. Polym. Mater., 2021, 3, 3708–3713 CrossRef CAS.
- K. Kuroda, T. Komori, K. Ishibashi, T. Uto, I. Kobayashi, R. Kadokawa, Y. Kato, K. Ninomiya, K. Takahashi and E. Hirata, Commun. Chem., 2020, 3, 163 CrossRef CAS.
- K. Lin, D. Zhang, M. H. Macedo, W. Cui, B. Sarmento and G. Shen, Adv. Funct. Mater., 2019, 29, 1804943 CrossRef.
- L. L. Wang, J. T. Chen, L. F. Wang, S. Wu, G. Zhang, H. Q. Yu, X. Ye and Q. S. Shi, Sci. Rep., 2017, 7, 12787 CrossRef.
- Y. Shen, X. Fu, W. Fu and Z. Li, Chem. Soc. Rev., 2015, 44, 612–622 RSC.
- K. Matsumoto, A. Kawamura and T. Miyata, Chem. Lett., 2015, 44, 1284–1286 CrossRef CAS.
- T. Loftsson, D. Hreinsdóttir and M. Másson, Int. J. Pharm., 2005, 302, 18–28 CrossRef CAS.
- H. Chen, C. Khemtong, X. Yang, X. Chang and J. Gao, Drug Discovery Today, 2011, 16, 354–360 CrossRef CAS PubMed.
- D. F. Farrar and R. K. Gillson, Biomaterials, 2002, 23, 3905–3912 CrossRef CAS.
- A. M. Rosales, H. K. Murnen, S. R. Kline, R. N. Zuckermann and R. A. Segalman, Soft Matter, 2012, 8, 3673–3680 RSC.
- Y. L. Sun, Z. P. Luo, A. Fertala and K. N. An, Biochem. Biophys. Res. Commun., 2002, 295, 382–386 CrossRef CAS.
- L. Shi, F. Carn, F. Boué and E. Buhler, Phys. Rev. E, 2016, 94, 032504 CrossRef.
- Y. Herdiana, N. Wathoni, S. Shamsuddin, I. M. Joni and M. Muchtaridi, Polymers, 2021, 13, 1717 CrossRef CAS PubMed.
- M. Shang, X. Sun, L. Guo, D. Shi, P. Liang, D. Meng, X. Zhou, X. Liu, Y. Zhao and J. Li, Int. J. Nanomed., 2020, 15, 537–552 CrossRef CAS.
- A. Kessler, O. Menéndez-Aguirre, J. Hinrichs, C. Stubenrauch and J. Weiss, Faraday Discuss., 2013, 166, 399–416 RSC.
- S. Kancharla, N. A. Zoyhofski, L. Bufalini, B. F. Chatelais and P. Alexandridis, Polymers, 2020, 12, 1831 CrossRef CAS PubMed.
- I. J. Gomez, B. Arnaiz, M. Cacioppo, F. Arcudi and M. Prato, J. Mater. Chem. B, 2018, 6, 5540–5548 RSC.
- J. Wang, W. Liu, Q. Tu, J. Wang, N. Song, Y. Zhang, N. Nie and J. Wang, Biomacromolecules, 2011, 12, 228–234 CrossRef CAS.
- D. H. Yu, Q. Lu, J. Xie, C. Fang and H. Z. Chen, Biomaterials, 2010, 31, 2278–2292 CrossRef CAS PubMed.
- N. Song, W. Liu, Q. Tu, R. Liu, Y. Zhang and J. Wang, Colloids Surf., B, 2011, 87, 454–463 CrossRef CAS PubMed.
- R. Bawa, S. Y. Fung, A. Shiozaki, H. Yang, G. Zheng, S. Keshavjee and M. Liu, Nanomedicine, 2012, 8, 647–654 CrossRef CAS.
- M. V. Blagosklonny, Int. J. Cancer, 1999, 83, 151–156 CrossRef CAS.
- R. V. Benjaminsen, M. A. Mattebjerg, J. R. Henriksen, S. M. Moghimi and T. L. Andresen, Mol. Ther., 2013, 21, 149–157 CrossRef CAS.
- X. Zhang, R. Zhang, J. Huang, M. Luo, X. Chen, Y. Kang and J. Wu, J. Mater. Chem. B, 2019, 7, 3537–3545 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ma00800e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |