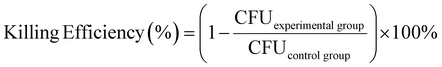Open Access Article
Open Access ArticleThe chemodynamic antibacterial effect of MnOX nanosheet decorated silicon nanowire arrays†
Mingkang
Li‡
,
Guize
Li‡
,
Hongwei
Wang
* and
Lin
Yuan
 *
*
State and Local Joint Engineering Laboratory for Novel Functional Polymeric Materials, College of Chemistry, Chemical Engineering and Materials Science, Soochow University, 199 Ren’ai Road, Suzhou 215123, P. R. China. E-mail: wanghw@suda.edu.cn; yuanl@suda.edu.cn
First published on 6th November 2021
Abstract
Bacteria exist everywhere and pose a threat to human health. Reactive oxygen species (ROS), as a highly reactive chemical substance, can effectively oxidize and break lipids, DNA, protein and other bioactive molecules, leading to the death of bacteria. Modification of manganese oxide on the surface of silicon nanowire arrays deposited with polydopamine (SN@PDA@MnOX) can significantly increase the yield of ROS in a short period of time, which provides SN@PDA@MnOX with a strong chemodynamic antibacterial effect. Both manganese ions and manganese oxides in the reaction system promote the generation of ROS, and their mechanisms of action can be synergistic with each other. The results show that SN@PDA@MnOX can achieve a 99.99% antibacterial effect by using the ROS generated by chemodynamic reactions within 1 hour. SN@PDA@MnOX has great potential as a novel antibacterial material.
1. Introduction
Bacteria are one of the most abundant types of organisms, widely distributed in soil and water, or in symbiosis with other organisms. Bacteria are the pathogens of many diseases. They can spread diseases between humans in various ways, seriously threatening human life and health, and hindering the development of society.1 In the long struggle against bacteria, humans already possess a variety of weapons, such as the application of physical methods (temperature,2,3 radiation,4,5 and electrical stimulation6), chemical methods (antibiotics,7,8 metals, and metal oxides9–11), and biological methods (bacteriophage,12 and biological control bacteria13). However, physical methods are often limited by the lack of exogenous stimuli. Chemical methods may cause bacterial resistance and inevitable biological toxicity. Biological methods are difficult to be developed due to their low antibacterial efficiencies. Therefore, it is particularly important to develop new antibacterial materials as soon as possible to protect human health.Reactive oxygen species (ROS) are one of the main causes for the bactericidal effect of many natural substances.11,14 ROS are a single-electron reduction product of oxygen. They have high chemical activity due to their structure containing unpaired electrons and can oxidize and break lipids, DNA, protein and other bioactive molecules. ROS are widely used in antibacterial applications,15,16 tumor treatment,17,18 immune regulation19 and other biomedical fields. ROS can be produced through the catalysis of metals and metal oxides,20,21 electron transfer in redox reactions, normal mitochondrial metabolism,22etc. The excitation conditions include chemodynamics,23,24 photodynamics25,26 and acoustic dynamics.27,28 The generation of ROS by metal oxide nanosheets often requires external energy stimulation.29,30 Thomas et al. used the ROS generated on the AuPd catalyst during the synthesis of hydrogen peroxide from hydrogen and air to achieve efficient sterilization and disinfection, providing a new technique for water disinfection.20 Liu et al. developed a catechol–chitosan film that could continuously produce ROS. This film can inhibit the growth of bacteria at the wound site in the presence of ascorbic acid, reduce inflammation and promote wound healing.31 Although these materials can produce ROS effectively, they must rely on a large amount of a reducing agent to achieve excellent antibacterial performance, which limits their wide application.
Manganese is a transition metal element abundant in nature. Manganese oxides (oxides, hydroxides and oxyhydroxides) produced by spontaneous oxidation have the advantages of low cost, simple preparation, and low biological toxicity.32 Because of its multiple valence states and multiple structural morphologies,33 and the rapid transfer of electrons and oxygen,34 manganese oxide is considered to be a potential catalyst. By interacting with various peroxides, manganese oxide can catalyze the production of ROS.35,36 Jia et al. designed a low-cost, highly active MnOX/g-C3N4 (MCN) nanocomposite material, which could stimulate a variety of active substances, such as ˙O2−, ˙OH and h+, under the synergistic effect of photoelectricity.37 Forooshani et al. functionalized the microgel with catechol and heme. The material uses heme to react with H2O2 generated in the process of oxidizing catechol to produce hydroxyl radicals with obvious antibacterial effects.38 However, due to the limited autooxidation of dopamine, the generation of H2O2 is still at a low level. Therefore, the overall production efficiency of ROS is unsatisfactory, which restricts the sterilization effect of related materials.
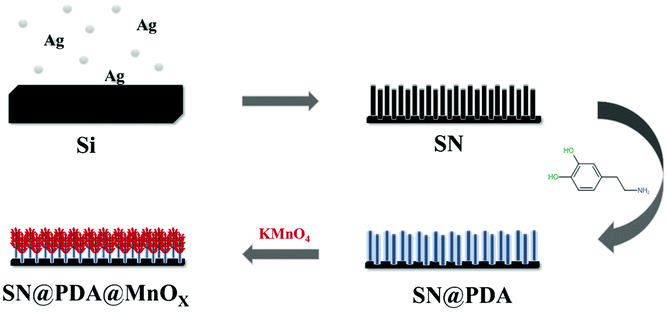
In order to solve these problems, based on the efficient promotion of manganese oxide on the generation of ROS, we modified the silicon nanowire arrays deposited with polydopamine in situ reduced manganese oxide nanosheets (SN@PDA@MnOX). Silicon nanowires were good candidates for energy conversion and were selected as substrates because of their good biocompatibility. The silicon nanowire arrays strongly support the adhesion of dopamine and dispersion of manganese oxide nanomaterials, which is beneficial to increase the specific surface area. Polydopamine was widely used as a material with excellent biocompatibility and degradability. The exceptional adhesive capability of polydopamine allows the nanomaterials to adhere closely to the substrate, ensuring the stability of the material.39 At the same time, the reducibility of polydopamine due to its own structure also provided convenience for the production of manganese oxide.40,41 SN@PDA@MnOX uses a chemodynamic, only in the presence of water and oxygen without addition of external stimuli, pathway to significantly generate the ROS in a short period of time. It can release a high concentration of ROS equivalent to more than 115 mM H2O2 within 2 h, which can achieve a 99.99% antibacterial effect, greatly improving the antibacterial efficiency.
2. Experimental
2.1. Materials
Silicon wafers were obtained from Guangzhou Institute of Semiconductor Materials (Guangzhou, China), and the specific model was (1,0,0)-oriented, with 0.56 mm thickness, 100 mm diameter, n-doped, and one side polished. Tris(hydroxymethyl)–aminomethane hydrochloride (Tris–HCl) was bought from Beijing Solarbio Science & Technology (China). Dopamine hydrochloride (DA), hydrofluoric acid (HF), nitric acid (HNO3), hydrochloric acid (HCl), silver nitrate (AgNO3), acetone, potassium permanganate (KMnO4), hydrogen peroxide (H2O2), manganese dichloride tetrahydrate (MnCl2·4H2O), and manganese dioxide (MnO2) were purchased from Sinopharm Chemical Reagent (China). Gram-negative E. coli MG1655 was provided by the China General Microbiological Culture Collection Center (Beijing, China). Bacterial culture media (tryptone, yeast extract and agar) were obtained from Oxoid (England). Ultrapure water was prepared using a Millipore water purification system to a minimum resistivity of 18.2 MΩ cm.2.2. Fabrication of silicon nanowire arrays (SN)
Pre-cut silicon wafers (0.5 cm × 0.5 cm) were ultrasonically cleaned alternately with ultrapure water and acetone 3 times, each for 2 min. The reaction solution (5 M HF, 0.05 M AgNO3) was prepared in a polytetrafluoroethylene container soaked in aqua regia (HCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3 = 3
HNO3 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for more than 4 h, and it was preheated in an oven at 50 °C for 5 min. The silicon wafer was placed evenly in the polytetrafluoroethylene reaction dish, and the preheated reaction solution was slowly poured into the reaction dish to completely immerse the silicon wafer. After reacting at 50 °C for 15 min, the reaction solution was sucked out, and 20% HNO3 was added to terminate the reaction. When there were no more bubbles on the surface of the silicon wafer, the solution was sucked out, and another 20% HNO3 was added to clean it. After being washed with ultrapure water, the wafer was put in an oven at 50 °C for drying.
1) for more than 4 h, and it was preheated in an oven at 50 °C for 5 min. The silicon wafer was placed evenly in the polytetrafluoroethylene reaction dish, and the preheated reaction solution was slowly poured into the reaction dish to completely immerse the silicon wafer. After reacting at 50 °C for 15 min, the reaction solution was sucked out, and 20% HNO3 was added to terminate the reaction. When there were no more bubbles on the surface of the silicon wafer, the solution was sucked out, and another 20% HNO3 was added to clean it. After being washed with ultrapure water, the wafer was put in an oven at 50 °C for drying.
2.3. Depositing polydopamine on the surface of silicon nanowire arrays (SN@PDA)
20 mg of dopamine hydrochloride was weighed and dissolved in 10 ml Tris–HCl solution (10 mM, pH = 8.5), and the silicon nanowire arrays were quickly immersed in the reaction solution for 3 h. After the reaction was finished, SN@PDA was washed three times with ultrapure water, then dried naturally at room temperature.2.4. Surface modification of polydopamine-deposited silicon nanowire arrays with MnOX (SN@PDA@MnOX)
The polydopamine-deposited silicon nanowire arrays were put into the prepared KMnO4 solution (10 μg ml−1) and reacted for 12 h. Then SN@PDA@MnOX was rinsed with ultrapure water and dried at room temperature.2.5. Modified MnOX on the surface of silicon nanowire array after reduction treatment (SN@MnOX)
After immersing the etched silicon nanowires in HF solution (5%, V/V) for 5 min, the reduced silicon nanowire arrays were cleaned with ultrapure water. The processed SN was put into the prepared KMnO4 solution (10 μg ml−1) and reacted for 12 h. Then SN@MnOX was taken out and rinsed with ultrapure water. Finally, it was dried at room temperature.2.6. Characterization of material structure
SEM (FESEM, S4700, Hitachi, Japan) is used to characterize the morphology of the material surface, and XPS (EscaLab 250Xi, Thermo Scientific, USA) is used to qualitatively, quantitatively and chemically analyze the chemical composition of the material surface.2.7. The release of reactive oxygen species (ROS) of SN, SN@PDA, SN@PDA@MnOX (converted to H2O2 amount)
Non-fluorescent DCFH can be oxidized by ROS to produce fluorescent DCF, and the concentration of the generated ROS can be analyzed by detecting the intensity of the fluorescence signal.42 As a kind of ROS, hydrogen peroxide is often used to quantify the concentration of ROS produced by materials due to its relatively stable chemical properties.2.8. Release of manganese ions from SN@PDA@MnOX
SN@PDA@MnOX was soaked in 500 μl PBS for 0.5 h, 1 h, and 2 h respectively, and the solution was diluted to 5 ml with a volume fraction of 2% hydrochloric acid solution. Manganese ions were measured by atomic absorption spectroscopy (AA240FS + GTA120, Varian, USA).2.9. Antibacterial test
E. coli strain (E. coli MG1655) was inoculated into the liquid medium with a pipette tip and cultivated overnight in a shaking incubator at constant temperature (37 °C). After centrifugation, the supernatant was discarded, and PBS was added into it. The centrifugation was repeated twice. Bacteria were diluted to an OD600 value of 0.01 and dropped on the surface of the material. Then the material was put into an incubator and incubated at 37 °C for 0.5 h, 1 h, and 2 h, respectively. After the incubation, the bacteria were diluted, and applied onto LB agar plates. The killing efficiency or the antibacterial effect of the material was calculated via the colony forming unit (CFU) using the equation2.10. Observation of the morphology of bacteria
100 μl of bacterial solution (OD600 = 0.01) was dropped on SN@PDA@MnOX and incubated at 37 °C for 2 h. 400 μl of 2.5% glutaraldehyde solution was added and allowed to stand for 1.5 h. Then all the liquid was sucked out, and ethanol at different concentrations (30–100%) was added step by step from low to high concentration. The liquid was aspirated at every 10 min of reaction and the next concentration of ethanol was added. The morphological changes of bacteria after being treated were observed by SN@PDA@MnOX with SEM (FESEM, S4700, Hitachi, Japan).3. Results and discussion
3.1. Characterization of SN@PDA@MnOX
The preparation process of SN@PDA@MnOX is shown in Fig. 1. First, we fabricated silicon nanowire arrays (SN) by chemical etching.43 The cross-sectional analysis of SEM shows that the silicon nanowires are uniformly and vertically distributed (Fig. 2a). Dopamine uses its excellent adhesion to form polydopamine to be fixed and attached to the surface of silicon nanowires. At the same time, it can also use its own reducibility to reduce KMnO4 to MnOX and bond to silicon nanowires. According to Fig. 2b, a large number of MnOX nanosheet structures appear on the surface of the silicon nanowires.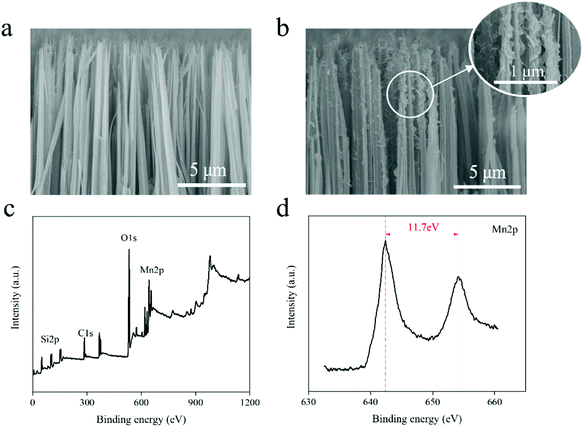 | ||
| Fig. 2 (a) SEM image of SN (cross section). (b) SEM image of SN@PDA@MnOX (cross section). (c) XPS full spectrum of SN@PDA@MnOX. (d) XPS narrow scan of SN@PDA@MnOX for Mn. | ||
In order to know the chemical properties of the achieved material, SN@PDA@MnOx was analyzed by XPS (Fig. 2c and d). The result showed that SN@PDA@MnOx has two specific peaks at 642.4 eV and 654.1 eV, which correspond to Mn2p3/2 and Mn2p1/2, respectively. The energy difference between the two peaks is 11.7 eV, indicating that Mn4+ in MnOX is the main oxidation state of Mn.41,44 MnO2 can induce the production of ROS, increase the activity of ATPase and cause the leakage of electrolyte and protein content, leading to the death of bacteria.45
3.2. Antibacterial effect of SN@PDA@MnOX
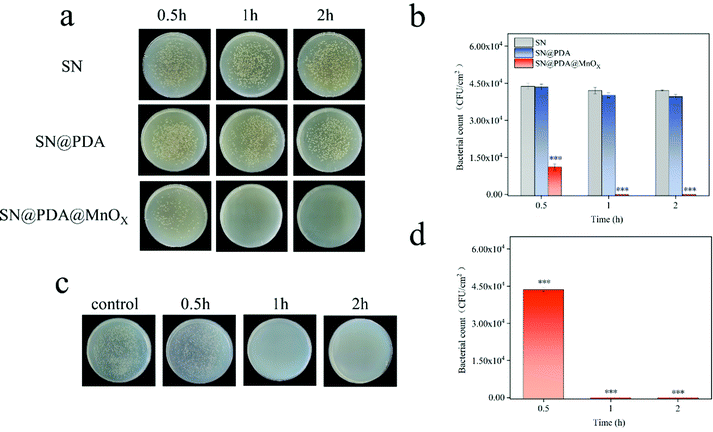
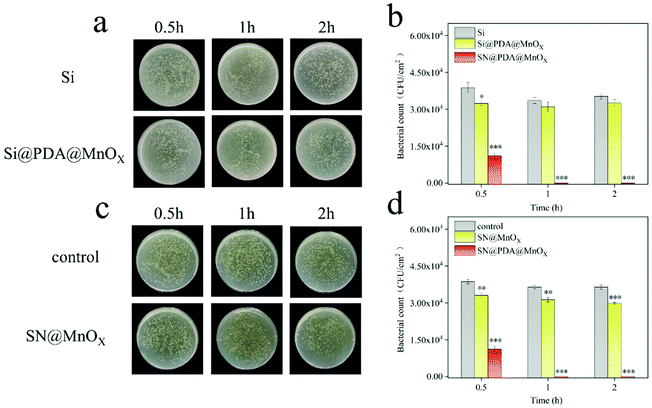
The antibacterial properties of SN, SN@PDA, and SN@PDA@MnOX were tested by the colony formation assay (Fig. 3a and b). After the bacteria were incubated on the SN for 0.5 h, 1 h and 2 h, the number of bacteria did not decrease significantly, indicating that SN has no inhibitory effect on the growth of bacteria. When SN is modified by PDA, it could be found that the number of bacteria on SN@PDA decreased to a certain extent, with an antibacterial rate of less than 10%. However, SN@PDA@MnOX showed a very strong antibacterial effect. After 0.5 h of incubation, the number of bacteria can be reduced by 70%. And almost 99.99% of the antibacterial effect could be achieved when the incubation time was prolonged to at least 1 h. In addition, Staphylococcus aureus was also used to test the antibacterial properties of SN@PDA@MnOX (Fig. 3c and d). After 0.5 h of incubation, the number of bacteria was reduced by 20%. After more than 1 h of incubation, the antibacterial effect of 99.99% can be achieved. These results show that SN@PDA@MnOX itself can achieve excellent antibacterial effects against Gram-negative bacteria and Gram-positive bacteria, independent of the addition of any reducing agent.It can be seen from the above results that the presence of MnOX may be the main reason for the enhanced antibacterial effect of the material. In order to verify the role of the SN and PDA components in SN@PDA@MnOX in the antibacterial process, we compared their antibacterial performance differences with Si@PDA@MnOX and SN@MnOX. According to Fig. 4a and b, there was no significant difference between the number of bacteria after 2 h incubation on the silicon chip and the number of bacteria after 0.5 h incubation, only a reduction of less than 10%. But after the bacteria were incubated on Si@PDA@MnOX for 0.5 h, 1 h and 2 h, the number of bacteria decreased only slightly, and the antibacterial rate was about 10%. The huge difference in antibacterial effect with SN@PDA@MnOX highlights the advantages of SN in the system. The silicon nanowire array can deposit more dopamine due to its huge specific surface area, so that it can load more MnOX nanosheets and promote the antibacterial effect (Fig. S1, ESI†). According to Fig. 4c and d, after the bacteria were incubated on SN@MnOX for a period of time, the number of bacteria decreased to a certain extent, and the antibacterial rate was calculated to be about 15%. Compared with the 99.99% antibacterial effect of SN@PDA@MnOX, the antibacterial performance of SN@MnOX is greatly reduced due to the lack of PDA. The adhesion of polydopamine makes it form a thin film on the surface of the silicon nanowire array, which is conducive to the dispersion of MnOX nanosheets. At the same time, the H2O2 produced by polydopamine in the self-circulating redox process might also be one of the key reasons that SN@PDA@MnOX can produce ROS.
3.3. The generation of reactive oxygen species (ROS) by SN@PDA@MnOX
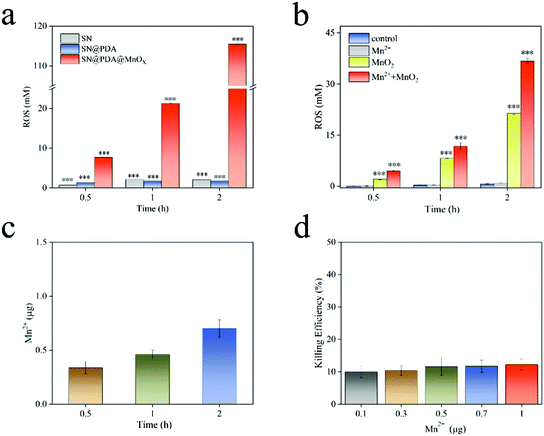
ROS is a one-electron reduction product of oxygen, including one-electron reduction product of oxygen superoxide anion (O2−), two-electron reduction product hydrogen peroxide (H2O2), three-electron reduction product hydroxyl radical (˙OH), etc. ROS has a high reactivity and a short lifespan, which can cause damage to biological molecules such as bacterial cell walls, protein peptide chains and nucleic acids, and ultimately produce an antibacterial effect. After SN and SN@PDA were incubated in PBS for a period of time, the concentration of ROS generated was about 2 mM (Fig. 5a). There is little difference between SN and SN@PDA in the amount of ROS released. However, SN@PDA@MnOX had a strong activity for generating ROS through its specific chemodynamic properties. It was found that ROS produced by SN@PDA@MnOX was 7 mM after just 0.5 h of incubation in PBS (Fig. 5a), which was higher than that produced by the catechol reaction for 50 h.38 And surprisingly, the concentrations of ROS generated by SN@PDA@MnOX increased significantly with time, reaching 115 mM within 2 h. This result corresponds to the enhancement of the antibacterial effect with the prolonged reaction time with the bacteria, indicating that the antibacterial performance of SN@PDA@MnOX might be directly related to the continuous production of ROS through chemical reactions catalyzed by MnOX sheets.Since MnO2 was the important component in MnOX sheets on SN@PDA@MnOX and Mn2+ could be released from MnOX materials,46,47 Mn2+ and MnO2 might be mainly responsible for the production of ROS. In order to explore the influence of different components in the system on the generation of ROS, we added Mn2+ and MnO2 to the PBS solution soaked with SN@PDA. It can be seen from Fig. 5b that the ROS released by both SN@PDA and Mn2+ treated SN@PDA were less than 1 mM even after 2 h. However, the addition of MnO2 could effectively increase the production of ROS so that the production of reactive oxygen species could reach 21 mM within 2 h. When SN@PDA was treated by both Mn2+ and MnO2, it could generate the concentration of active oxygen of as high as 37 mM. Generally, manganese oxides such as manganese dioxide achieve catalysis through the mechanism of adsorption of oxygen and the mechanism of lattice oxygen.48,49 Manganese ions achieve catalysis through the formation of complexes and other intermediates.50,51 Therefore, Mn2+ and MnO2 might be synergistic with each other for promoting ROS generation on SN@PDA, which can effectively increase the catalytic efficiency of manganese oxide.
Transition metal ions are important for the formation of ROS. The ability of these ions to remove electrons is the basis for the formation and expansion of many highly toxic ROS reactions. Because of the presence of polydopamine, manganese oxide can be reduced and dissolved in the solution, thereby leaching trace manganese ions to catalyze the production of ROS. Actually, the release of manganese ions was observed from SN@PDA@MnOX (Fig. 5c). The result showed that there was about 0.3 μg of manganese ions released from a chip of SN@PDA@MnOX (0.5 cm × 0.5 cm) in 0.5 h. With the prolongation of the immersion time in PBS, more manganese ions released from the material. However, even after 2 h, the amount of manganese ions released from SN@PDA@MnOX was as low as around 0.7 μg. It may be due to both the oxidation reaction of polydopamine and the reduction process of manganese oxide in the solution were slow, leading to a low amount of manganese ions being released.
Considering that the antibacterial effects of metal ions (such as Ag+) might come from the disturbance of the bacterial metabolism, the effect of Mn2+ in inhibiting bacterial growth was investigated. It was observed that the killing efficiency of Mn2+ in an amount of 0.1–1 μg was only about 10%. This result further proved that manganese ions were not the main reason of the antibacterial effect of SN@PDA@MnOX. However, the addition of manganese ions will inhibit the activity of enzymes in the microorganisms,52 resulting in a decrease in the activity of microorganisms, which may be favorable for killing bacteria.
3.4. Research on the mechanism of bacterial death
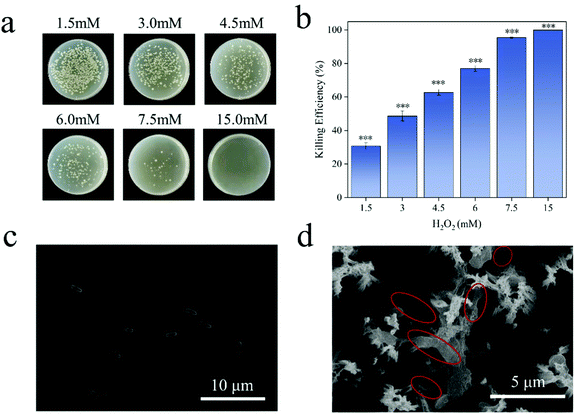
E. coli is a Gram-negative bacterium. Under normal conditions, both ends of E. coli are blunt, round and full in shape (Fig. 6c). ROS can enter the bacteria and react with biological macromolecules, such as protein, lipid and DNA, etc. They will destroy the antioxidant capacity of the intracellular protective enzymes, and cause changes in the permeability of the bacterial cell membrane, leading to the death of the bacteria.53 It can be seen from Fig. 6a and b that the bacteria have a very low tolerance to hydrogen peroxide, and 15 mM hydrogen peroxide can achieve a 99.99% killing efficiency of E. coli. The concentration of ROS released by SN@PDA@MnOX catalyzed by chemodynamic action is calculated by the equivalent of hydrogen peroxide, which was 115 mM, far exceeds this value. Therefore, the generation of ROS might the main reason for the excellent sterilization performance of SN@PDA@MnOX. Such high concentrations of ROS released from SN@PDA@MnOX had a significant effect on changing the morphology of the bacteria. From the SEM image (Fig. 6d), it can be found that E. coli had obviously shrunk, indicating that the permeability of the bacterial cell membrane had changed, which also confirms that SN@PDA@MnOX achieves the antibacterial effect through ROS.4. Conclusions
In the present study, the specific adhesion property and weak reducibility of polydopamine (PDA) were used to modify manganese oxide (MnOX) on the surface of the silicon nanowire arrays (SN) to prepare SN@PDA@MnOX. SN@PDA@MnOX has excellent antibacterial effects, which reaches 99.99% killing efficiency in 1 h treatment. Mn2+ and MnO2 from SN@PDA@MnOX have a synergistic effect for generating ROS through a chemodynamic reaction. The concentration of ROS increased to as high as 115 mM, leading to bacterial death. These results demonstrate that SN@PDA@MnOX has great potential in antibacterial applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21774088), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Jiangsu Clinical Research Center for Cardiovascular Surgery.References
- A. P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hindler, G. Kahlmeter, B. Olsson-Liljequist, D. L. Paterson, L. B. Rice, J. Stelling, M. J. Struelens, A. Vatopoulos, J. T. Weber and D. L. Monnet, Clin. Microbiol. Infect., 2012, 18, 268–281 CrossRef CAS PubMed.
- J. Huo, Q. Jia, H. Huang, J. Zhang, P. Li, X. Dong and W. Huang, Chem. Soc. Rev., 2021, 50, 8762–8789 RSC.
- Y. Xie, Y. Qian, Z. Li, Z. Liang, W. Liu, D. Yang and X. Qiu, ACS Sustainable Chem. Eng., 2021, 9, 6479–6488 CrossRef CAS.
- V. K. Sharma, S. T. Tan, Z. Haiyang, S. Shendre, A. Baum, F. Chalvet, J. Tirén and H. V. Demir, Adv. Opt. Mater., 2021, 9, 2100072 CrossRef CAS.
- J. Wadsworth and C. S. Cockell, Sci. Rep., 2017, 7, 4662 CrossRef.
- P. Liu, K. Jin, W. Wong, Y. Wang, T. Liang, M. He, H. Li, C. Lu, X. Tang, Y. Zong and C. Li, Chem. Eng. J., 2021, 415, 129025 CrossRef CAS.
- I. Keren, Y. Wu, J. Inocencio, L. R. Mulcahy and K. Lewis, Science, 2013, 339, 1213 CrossRef CAS.
- F. Baquero and B. R. Levin, Nat. Rev. Microbiol., 2021, 19, 123–132 CrossRef CAS PubMed.
- A. Bhargava, V. Pareek, S. Roy Choudhury, J. Panwar and S. Karmakar, ACS Appl. Mater. Interfaces, 2018, 10, 29325–29337 CrossRef CAS.
- S. Rtimi and J. Kiwi, Appl. Catal., B, 2017, 213, 62–73 CrossRef CAS.
- R. Pachaiappan, S. Rajendran, P. L. Show, K. Manavalan and M. Naushad, Chemosphere, 2021, 272, 128607 CrossRef CAS.
- K. A. McCarthy, M. A. Kelly, K. Li, S. Cambray, A. S. Hosseini, T. van Opijnen and J. Gao, J. Am. Chem. Soc., 2018, 140, 6137–6145 CrossRef CAS.
- A. Soenens and J. Imperial, Phytochem. Rev., 2020, 19, 577–587 CrossRef CAS.
- P. L. Lam, R. S. M. Wong, K. H. Lam, L. K. Hung, M. M. Wong, L. H. Yung, Y. W. Ho, W. Y. Wong, D. K. P. Hau, R. Gambari and C. H. Chui, Chem.-Biol. Interact., 2020, 320, 109023 CrossRef CAS.
- G. Wang, W. Jin, A. M. Qasim, A. Gao, X. Peng, W. Li, H. Feng and P. K. Chu, Biomaterials, 2017, 124, 25–34 CrossRef CAS PubMed.
- Y. Wu, L. Zhang, Y. Zhou, L. Zhang, Y. Li, Q. Liu, J. Hu and J. Yang, Chin. J. Catal., 2019, 40, 691–702 CrossRef CAS.
- S. Wang, G. Yu, Z. Wang, O. Jacobson, L.-S. Lin, W. Yang, H. Deng, Z. He, Y. Liu, Z.-Y. Chen and X. Chen, Angew. Chem., Int. Ed., 2019, 58, 14758–14763 CrossRef CAS PubMed.
- S. Wang, Z. Wang, G. Yu, Z. Zhou, O. Jacobson, Y. Liu, Y. Ma, F. Zhang, Z.-Y. Chen and X. Chen, Adv. Sci., 2019, 6, 1801986 CrossRef.
- J. Lloberas, J. P. Muñoz, M. I. Hernández-Álvarez, P.-J. Cardona, A. Zorzano and A. Celada, Autophagy, 2020, 16, 2307–2309 CrossRef CAS PubMed.
- T. Richards, J. H. Harrhy, R. J. Lewis, A. G. R. Howe, G. M. Suldecki, A. Folli, D. J. Morgan, T. E. Davies, E. J. Loveridge, D. A. Crole, J. K. Edwards, P. Gaskin, C. J. Kiely, Q. He, D. M. Murphy, J.-Y. Maillard, S. J. Freakley and G. J. Hutchings, Nat. Catal., 2021, 4, 575–585 CrossRef CAS.
- M. Misawa and J. Takahashi, Nanomedicine, 2011, 7, 604–614 CrossRef CAS.
- P. Hernansanz-Agustín and J. A. Enríquez, Antioxidants, 2021, 10 Search PubMed.
- Q. Tian, F. Xue, Y. Wang, Y. Cheng, L. An, S. Yang, X. Chen and G. Huang, Nano Today, 2021, 39, 101162 CrossRef CAS.
- Z. Ren, S. Sun, R. Sun, G. Cui, L. Hong, B. Rao, A. Li, Z. Yu, Q. Kan and Z. Mao, Adv. Mater., 2020, 32, 1906024 CrossRef CAS.
- C. Mao, Y. Xiang, X. Liu, Y. Zheng, K. W. K. Yeung, Z. Cui, X. Yang, Z. Li, Y. Liang, S. Zhu and S. Wu, ACS Appl. Mater. Interfaces, 2019, 11, 17902–17914 CrossRef CAS PubMed.
- Q. Jiang, F. E. J. Tian, J. Yang, J. Zhang and Y. Cheng, ACS Appl. Mater. Interfaces, 2020, 12, 16150–16158 CrossRef CAS.
- Y. He, S. Hua Liu, J. Yin and J. Yoon, Coord. Chem. Rev., 2021, 429, 213610 CrossRef CAS.
- S. Son, J. H. Kim, X. Wang, C. Zhang, S. A. Yoon, J. Shin, A. Sharma, M. H. Lee, L. Cheng, J. Wu and J. S. Kim, Chem. Soc. Rev., 2020, 49, 3244–3261 RSC.
- Y. Cao, T. Wu, W. Dai, H. Dong and X. Zhang, Chem. Mater., 2019, 31, 9105–9114 CrossRef CAS.
- X. Li, H. Yang, K. Lv, L. Wen and Y. Liu, Appl. Surf. Sci., 2020, 503, 144080 CrossRef CAS.
- H. Liu, X. Qu, E. Kim, M. Lei, K. Dai, X. Tan, M. Xu, J. Li, Y. Liu, X. Shi, P. Li, G. F. Payne and C. Liu, Biomaterials, 2018, 162, 109–122 CrossRef CAS.
- S. K. Ghosh, ACS Omega, 2020, 5, 25493–25504 CrossRef CAS PubMed.
- S. Zhu, S.-H. Ho, C. Jin, X. Duan and S. Wang, Environ. Sci.: Nano, 2020, 7, 368–396 RSC.
- H. Fang, C. Wang, D. Li, S. Zhou, Y. Du, H. Zhang, C. Hang, Y. Tian and T. Suga, J. Mater. Sci. Technol., 2021, 91, 5–16 CrossRef.
- S. Zhu, X. Li, J. Kang, X. Duan and S. Wang, Environ. Sci. Technol., 2019, 53, 307–315 CrossRef CAS.
- A. Wang, H. Wang, H. Deng, S. Wang, W. Shi, Z. Yi, R. Qiu and K. Yan, Appl. Catal., B, 2019, 248, 298–308 CrossRef CAS.
- X. Jia, L. Xie, Z. Li, Y. Li, R. Ming, Q. Zhang, X. Mi and S. Zhan, Sci. Total Environ., 2021, 768, 144368 CrossRef CAS PubMed.
- P. Kord Forooshani, R. Pinnaratip, E. Polega, A. G. Tyo, E. Pearson, B. Liu, T.-O. Folayan, L. Pan, R. M. Rajachar, C. L. Heldt and B. P. Lee, Chem. Mater., 2020, 32, 8182–8194 CrossRef CAS PubMed.
- I. Singh, G. Dhawan, S. Gupta and P. Kumar, Front. Microbiol., 2021, 11, 3326 Search PubMed.
- S.-C. Chou, C. Lin, B.-Y. Sun, K.-C. Tso, T.-S. Chan and P.-W. Wu, J. Taiwan Inst. Chem. Eng., 2021, 119, 196–203 CrossRef CAS.
- Z. Wang, H. Yu, Y. Xiao, L. Guo, L. Zhang and X. Dong, J. Hazard. Mater., 2021, 407, 124795 CrossRef CAS.
- S. Liu, D. Lu, X. Wang, D. Ding, D. Kong, Z. Wang and Y. Zhao, J. Mater. Chem. B, 2017, 5, 4918–4925 RSC.
- H. Wang, W. Jiang, L. Yuan, L. Wang and H. Chen, ACS Appl. Mater. Interfaces, 2013, 5, 1800–1805 CrossRef CAS.
- Z. Wang, H. Yu, Y. Xiao, L. Zhang, L. Guo, L. Zhang and X. Dong, Chem. Eng. J., 2020, 394, 125014 CrossRef CAS.
- T. Du, S. Chen, J. Zhang, T. Li, P. Li, J. Liu, X. Du and S. Wang, Nanomaterials, 2020, 10 Search PubMed.
- T. He, C. Jiang, J. He, Y. Zhang, G. He, J. Wu, J. Lin, X. Zhou and P. Huang, Adv. Mater., 2021, 33, 2008540 CrossRef CAS.
- Y. Cai, J. He, J. Li, J. Zhang and Y. Luo, Biochem. Eng. J., 2020, 159, 107600 CrossRef CAS.
- H. Wang, H. Chen, Y. Wang and Y.-K. Lyu, Chem. Eng. J., 2019, 361, 1161–1172 CrossRef CAS.
- J. Jia, P. Zhang and L. Chen, Appl. Catal., B, 2016, 189, 210–218 CrossRef CAS.
- M. Xiao, Y. Qi, Q. Feng, K. Li, K. Fan, T. Huang, P. Qu, H. Gai and H. Song, Chemosphere, 2021, 269, 129436 CrossRef CAS.
- X. Li, P. Gao, J. Tan, K. Xiong, M. F. Maitz, C. Pan, H. Wu, Y. Chen, Z. Yang and N. Huang, ACS Appl. Mater. Interfaces, 2018, 10, 40844–40853 CrossRef CAS PubMed.
- H. Chaudhry, N. Fatima and I. Z. Ahmad, BioMed Res. Int., 2015, 2015, 708691 Search PubMed.
- D. Wang, Q. Li, J. Qiu, X. Zhang, N. Ge and X. Liu, Adv. Mater. Interfaces, 2019, 6, 1900514 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ma00794g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |