Open Access Article
Open Access ArticleSema4D–plexin-B1 signaling in recruiting dental stem cells for vascular stabilization on a microfluidic platform†
Lili
Zhang
,
Yuanyuan
Han
,
Qixin
Chen
and
Waruna Lakmal
Dissanayaka
 *
*
Applied Oral Sciences & Community Dental Care, Faculty of Dentistry, The University of Hong Kong, Hong Kong SAR. E-mail: warunad@hku.hk; Tel: +852 2859 0484
First published on 27th October 2022
Abstract
The recruitment of mural cells is critical for stabilization of nascent vessels. Stem cells from human exfoliated deciduous teeth (SHED) are considered to have mural cell-like properties. However, the signaling mechanisms that regulate the cross-talk between endothelial cells and SHED in recruiting them as mural cells is much less well understood. Herein, using a 3D biomimetic microfluidic device, for the first time, we unraveled the role of semaphorin 4D (Sema4D)–plexin-B1 signaling in the recruitment of SHED as mural cells during angiogenic sprouting and vasculature formation by endothelial cells (ECs) in a 3D fibrin matrix. The specific compartmentalized design of the microfluidic chip facilitated recreation of the multi-step dynamic process of angiogenesis in a time and space dependent manner. Enabled by the chip design, different morphogenic steps of angiogenesis including endothelial proliferation, migration & invasion, vascular sprout formation and recruitment of mural cells as well as functional aspects including perfusion and permeability were examined under various pharmacological and genetic manipulations. The results showed that Sema4D facilitates the interaction between endothelial cells and SHED and promotes the recruitment of SHED as mural cells in vascular stabilization. Our results further demonstrated that Sema4D exerts these effects by acting on endothelial–plexin-B1 by inducing expression of platelet-derived growth factor (PDGF)-BB, which is a major mural cell recruitment factor.
1. Introduction
Achievement of a stable and mature vascular network, which guarantees the survival and function of the target tissue, remains a fundamental challenge in tissue engineering.1,2 Angio-/vasculogenesis is a sequential process that involves endothelial cells and mural cells, where initiation of vascular morphogenesis by endothelial cells is followed by mural cell recruitment by nascent vessels. Endothelial cell-derived factors are critical for mural cell recruitment.3 Mural cells, including pericytes and vascular smooth muscle cells (vSMCs), play an essential role in vascular maturation and stability by direct contact with endothelial cells and through paracrine growth factor secretion.1,4 In the absence of support from mural cells, the nascent vascular tubes become unstable and prone to regress.5,6 Therefore, the recruitment of mural cells is vital for the longevity of tissue-engineered vessels.For tissue engineering applications, primary mural cells from human tissue are scarcely available, and the variability and limited proliferative capacity of tissue-specific phenotypes would hinder their clinical application.7 Stem cells from human exfoliated deciduous teeth (SHED) are considered a promising population of mesenchymal stem cells for regenerative applications and could be a feasible source of mural cells as well. It has been reported that SHED may have originated from a perivascular microenvironment,8 and possess pericyte-like characteristics, through which they facilitate vessel formation by guiding endothelial cells and stabilizing newly formed blood vessels.9 Despite this evidence, the signaling mechanisms that regulate the cross-talk between endothelial cells and SHED in recruiting SHED as mural cells is much less well understood.
Semaphorin 4D (Sema4D), also known as cluster of differentiation 100 (CD100), is a member of the class IV semaphorin family. Sema4D expression was found in many tissues including brain, kidney, and heart10,11 and has been identified for its roles in immune regulation,12 axon guidance,13 angiogenesis,14 and tumor progression.15 The angiogenic properties of Sema4D have been reported to be activated through high-affinity receptor plexin-B1 on endothelial cells both in vitro and in vivo.14,16 Further studies have shown that macrophage-derived Sema4D contributes to the maturation of tumor vessels by facilitating the recruitment of pericytes by activating plexin-B1 on endothelial cells.14 A more recent study reported that treatment of tumor tissue with anti-Sema4D antibodies resulted in decreased platelet-derived growth factor BB (PDGF-BB) levels and decreased number of α-SMA+ pericytes, which suggested the role of Sema4D in inducing PDGF expression and regulating pericyte recruitment and differentiation.17 Based on the above-mentioned evidence, we hypothesized that Sema4D–endothelial–plexin-B1 could be a candidate signaling pathway that recruits SHED as mural cells.
Animal models have been commonly used in examining angiogenesis and related signaling mechanisms. Although these models provide physiologically relevant microenvironments, differences in genetic, immunological and cellular factors could compromise the validity of the results. Furthermore, the difficulty to isolate individual parameters and reproduce parametric conditions as well as challenges in imaging live cells in high resolution make animal studies not so optimal to study angiogenic processes. Herein, organ-on-a-chip devices provide micro-scale systems that mimic the physiological in vivo environment closely while facilitating the spatiotemporal control over different parameters. Hus et al.18 had reported a similar microfluidic chip that can develop nearly identical human microtissues with interconnected vascular networks by controlling the chemical and mechanical microphysiological environment.
In the current study, using a three-dimensional (3D) biomimetic angiogenesis microfluidic chip, for the first time, we unraveled the role of Sema4D–plexin-B1 signaling in the recruitment of SHED as mural cells during angiogenic sprouting and vasculature formation by ECs in a 3D fibrin matrix. The specific compartmentalized design of the microfluidic chip facilitated to recreate the multi-step dynamic process of angiogenesis in a time and space dependent manner. Enabled by the chip design, different morphogenic steps of angiogenesis including endothelial proliferation, migration & invasion, vascular sprout formation and recruitment of mural cells as well as functional aspects including perfusion and permeability were examined under various pharmacological and genetic manipulations. Using exogenous recombinant Sema4D, we demonstrated its role in the recruitment of SHED as mural cells in vascular stabilization. Our results further showed that Sema4D exerts these effects by acting on endothelial–plexin-B1 by inducing expression of platelet-derived growth factor (PDGF)-BB, which is a major mural cell recruitment factor (Fig. 1).
 | ||
| Fig. 1 Sema4D promotes vascular stabilization by recruiting SHED through endothelial-derived PDGF-BB. | ||
2. Materials and methods
2.1 Cell culture
SHED were purchased from AllCells (Alameda, CA, USA) and cultured in α-minimum essential medium (α-MEM) supplemented with 10% FBS and 1% penicillin/streptomycin. Mesenchymal origin and multipotent differentiation capacity of the cells were evaluated and published in our previous study.19 Human umbilical vein endothelial cells (HUVECs) were purchased from ScienCell (Carlsbad, CA, USA) and cultured in endothelial cell medium (ECM, ScienCell) supplemented with 5% (v/v) fetal bovine serum, 1% (v/v) endothelial cell growth supplement and 1% (v/v) penicillin/streptomycin. Green fluorescent protein-expressing human umbilical vein endothelial cells (GFP-HUVECs) were purchased from Angio-Proteomie (Boston, MA, USA) and cultured in ECM. HUVECs were cultured on plates coated with 2 μg cm−2 bovine plasma fibronectin (ScienCell). All cell cultures were kept in a 37 °C and 5% CO2 incubator. Passage 4–7 of SHED and passage 3–6 of HUVECs and GFP-HUVECs were used in all the downstream experiments.2.2 Sema4D
Human recombinant Sema4D was purchased from ACRO Biosystems, Newark, DE, USA. Several concentrations of Sema4D (0, 0.5, 1, 2, 4 μg ml−1) were used in the preliminary experiments depending on the effective concentrations of previous studies that have investigated the angiogenic properties of Sema4D (0.4–1 μg mL−1).16,20,21 A concentration of 1 μg ml−1 Sema4D was identified as the optimum concentration for further studies based on the findings of our preliminary experiments (Fig. S1†).2.3 Plexin-B1 knockdown by small interfering RNA (siRNA)
siRNA targeting plexin-B1 was purchased from Invitrogen. A siRNA with no target human sequence was used as negative control. HUVECs were transfected with siRNA (final concentration 50 nM) in Opti-MEM (Invitrogen) using Lipofectamine™ 3000 reagent (Invitrogen) according to manufacturer's instructions. After 24 h, plexin-B1KD-HUVECs and negative-HUVECs were used for downstream experiments. SHED were transfected with siRNA (final concentration 50 nM) in Opti-MEM (Invitrogen) using Lipofectamine™ 3000 Reagent (Invitrogen), according to manufacturer's instructions. After 48 h, Plexin-B1KD–SHED and negative-SHED were used for downstream experiments. Plexin-B1 knockdown was confirmed by western blotting.2.4 Microfluidic assays
In order to investigate the interaction between HUVECs and SHED under Sema4D treatment, AIM Biotech 3D Cell Culture Chips (AIM Biotech, Ayer Rajah Crescent, Singapore) were used in different settings. Schematic diagram of the microfluidic assay setting used for examining the recruitment of SHED to support the endothelial vascular network was shown in Fig. 2A. HUVECs were re-suspended at a concentration of 12 × 106 cells per mL. The fibrinogen solution and the cell suspension were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to yield a final fibrinogen concentration of 2.5 mg mL−1 with 0.15 U mL−1 aprotinin (Sigma-Aldrich). The mixture was then mixed with thrombin (Sigma-Aldrich) and injected into the central channel immediately. The side channels were coated with bovine plasma fibronectin (30 μg mL−1, ScienCell) for 1 h followed by washing with PBS. Then, HUVECs in ECM were seeded in the upper channel at a concentration of 2 × 106 cells per mL. After HUVECs formed a vascular network within the gel, the SHED was seeded in the lower channel at a concentration of 2 × 106 cells per mL. At the same time, Sema4D (1 μg mL−1) was added to the upper channel. The culture medium of each channel was changed every day. On day 7, the samples were fixed and stained. The microfluidic assay was repeated using plexin-B1KD HUVECs (schematic diagram, Fig. S2†) and PDGFR-β inhibitor (CP-673451, 0.4 μM, MedChemExpress, USA) (schematic diagram, Fig. S3†) to investigate the potential mechanism.
1 to yield a final fibrinogen concentration of 2.5 mg mL−1 with 0.15 U mL−1 aprotinin (Sigma-Aldrich). The mixture was then mixed with thrombin (Sigma-Aldrich) and injected into the central channel immediately. The side channels were coated with bovine plasma fibronectin (30 μg mL−1, ScienCell) for 1 h followed by washing with PBS. Then, HUVECs in ECM were seeded in the upper channel at a concentration of 2 × 106 cells per mL. After HUVECs formed a vascular network within the gel, the SHED was seeded in the lower channel at a concentration of 2 × 106 cells per mL. At the same time, Sema4D (1 μg mL−1) was added to the upper channel. The culture medium of each channel was changed every day. On day 7, the samples were fixed and stained. The microfluidic assay was repeated using plexin-B1KD HUVECs (schematic diagram, Fig. S2†) and PDGFR-β inhibitor (CP-673451, 0.4 μM, MedChemExpress, USA) (schematic diagram, Fig. S3†) to investigate the potential mechanism.
2.5 Immunofluorescent staining
Samples were washed with PBS and fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 min. Then samples were blocked with 5% bovine serum albumin (BSA, Beyotime Institute of Biotechnology, China) containing 0.2% (v/v) Triton-X100 for 1 hour at room temperature. Mouse monoclonal anti-CD31 antibody (Cell Signaling Technology, Danvers, MA, USA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300), rabbit polyclonal anti-SM22α antibody (Abcam, Cambridge, MA, USA, 1
300), rabbit polyclonal anti-SM22α antibody (Abcam, Cambridge, MA, USA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300), mouse monoclonal anti-collagen IV-Alexa Fluor™ 647 (eBioscience™, 10 μg mL−1), rabbit monoclonal anti-PDGFR-β (Abcam, 1
300), mouse monoclonal anti-collagen IV-Alexa Fluor™ 647 (eBioscience™, 10 μg mL−1), rabbit monoclonal anti-PDGFR-β (Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) were used in immunofluorescence staining. Alexa Fluor® 488 conjugated goat anti-mouse IgG (Cell Signaling Technology) and Alexa Fluor® 555 conjugated goat anti-rabbit IgG (Invitrogen) were used as secondary antibodies. Cell nuclei were stained with DRAQ5 (1
100) were used in immunofluorescence staining. Alexa Fluor® 488 conjugated goat anti-mouse IgG (Cell Signaling Technology) and Alexa Fluor® 555 conjugated goat anti-rabbit IgG (Invitrogen) were used as secondary antibodies. Cell nuclei were stained with DRAQ5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Thermo Scientific) or hoechst (Invitrogen, 2 μg mL−1). The images were captured under confocal laser scanning microscope (Olympus IX81, Tokyo, Japan; Zeiss LSM 880 and Zeiss LSM 900 confocal microscope with Airyscan laser, Carl Zeiss, Germany). The SM22α positive SHED coverage was defined as the ratio of SM22α positive SHED covered vessel length to total vessel length. The length was quantified through ImageJ software (National Institutes of Health, Bethesda, MD).
1000, Thermo Scientific) or hoechst (Invitrogen, 2 μg mL−1). The images were captured under confocal laser scanning microscope (Olympus IX81, Tokyo, Japan; Zeiss LSM 880 and Zeiss LSM 900 confocal microscope with Airyscan laser, Carl Zeiss, Germany). The SM22α positive SHED coverage was defined as the ratio of SM22α positive SHED covered vessel length to total vessel length. The length was quantified through ImageJ software (National Institutes of Health, Bethesda, MD).
2.6 Dextran permeability assay
Alexa fluor 555 labelled dextran (Invitrogen) was introduced to visualize the perfusion of microvasculature and the permeability coefficient was calculated. Briefly, after removing the media in one of the reservoirs in upper channel, 70 μl of 10 kDa Alexa fluor 555 labelled dextran was added. Dextran flowed into the endothelial tubes and then across from the intravascular to extravascular region over time. Sequential images were captured using confocal microscope at every 60 seconds. Permeability coefficient was calculated based on the fluorescent intensity of the intravascular and extravascular regions as described previously.22,232.7 Migration assay using microfluidic chip
Schematic diagram of the microfluidic assay setting used for examining the effects of Sema4D on migration of SHED was shown in Fig. S4.† Fibrinogen (2.5 mg mL−1, Sigma-Aldrich) with 0.15 U mL−1 aprotinin (Sigma-Aldrich) was injected into the central channel and incubated for 30 min to allow for fibrin cross-linking. The side channels were coated with bovine plasma fibronectin (30 μg mL−1, ScienCell) for 1 h followed by washing with PBS. HUVECs were labeled with CellTracker™ Red CMTPX (Invitrogen) and SHED were labeled with CellTracker™ Green CMFDA (Invitrogen). HUVECs were then suspended in ECM at a concentration of 2 × 106 cells per mL and seeded in the upper channel with 10 μL in each reservoir. SHED was suspended in serum free α-MEM at a concentration of 1 × 106 cells per mL and seeded in the lower channel with 10 μL in each reservoir. The device was incubated at 37 °C overnight. Sema4D (1 μg mL−1) was added to the HUVECs for 48 h. The cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 min and permeabilized with 0.1% (v/v) Triton-X100 for 10 min. Cell nuclei were stained with DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for 10 min. Images were captured by fluorescence microscope (Nikon, Tokyo, Japan).
000) for 10 min. Images were captured by fluorescence microscope (Nikon, Tokyo, Japan).
2.8 Live imaging of microfluidic model
The methods of using microfluidic model were described in detail under 2.4. GFP-HUVECs and SHED (CellTracker™ Red CMTPX labeled) were cultured with Sema4D treatment in microfluidic device, which was incubated at 37 °C, 5% CO2, and 95% humidity. Time-sequential images were captured by Leica Microscope hourly for 24 hour durations up to day 7 at 200× magnification and produced time-lapse videos.2.9 Conditioned media
HUVECs were cultured until 80% confluence and treated with Sema4D (1 μg mL−1) in ECM. Conditioned medium (CM) was collected after 24 h and centrifuged at 1500 rpm for 5 min.2.10 Proliferation assay of SHED in conditioned media
SHED were seeded on a 96-well plate at a density of 3000 cells per well. After 24 h of incubation, the culture medium was replaced by CM. At time points of 24 h, 48 h, and 72 h, cell proliferation was quantified using Cell Counting Kit-8 (CCK-8) (Abcam, Cambridge, MA, USA) according to the manufacturer's protocol and OD values were measured at 460 nm by SpectraMax® M2 microplate reader (Molecular Devices, CA, US).2.11 Trans-well migration assay
Trans-well assay (8 μm; Thermo Scientific, Grand Island, NY, USA) was conducted to investigate the migration capacity of SHED in Sema4D-treated HUVEC CM. SHED were seeded on the insert at a density of 3 × 104/150 μL in serum free α-MEM. Then, 500 μL of ECM, ECM containing Sema4D (1 μg mL−1), HUVEC CM and Sema4D-treated HUVEC CM were added to the respective lower compartment. After 16 h, cells were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violet (Sigma-Aldrich) for 30 min. The cells on the upper compartment of the insert were gently removed with a cotton swab. The migrated cells were photographed under Nikon TE-2000 camera (Japan). Nine randomly chosen photos were used for statistics. CM from negative-HUVECs and plexin-B1KD-HUVECs treated with or without Sema4D were used to investigate the role of Sema4D–plexin-B1 signaling in recruitment of SHED. Furthermore, the trans-well assay was repeated after adding PDGFR-β inhibitor to Sema4D-treated HUVEC CM to investigate the role of PDGF-BB.2.12 Western blotting
To examine the mural cell marker expression, SHED were seeded on 6 well plates at a density of 3 × 105 cells per well. After 24 h, Sema4D-treated HUVEC CM was added to SHED and cultured for 24 h and 72 h. Total protein was collected using M-PER protein extraction buffer containing 1× protease inhibitor cocktail (Thermo Scientific). To examine the expression of endothelial cell derived factors, HUVECs were seeded in 6 well plates at a density of 3 × 105 cells per well until they reached 80% confluence followed by treated with Sema4D (1 μg mL−1) for 24 h. Total protein was collected. The knockdown efficiency of plexin-B1 was also confirmed by western blotting. After vortex and centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, the total protein concentrations were quantified by a BCA kit (Thermo Scientific). A total amount of 20 μg of protein from each sample was subjected to 7.5%, 10% or 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) followed by transfer onto the polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences, Little Chalfont, UK). The membranes were blocked for 1 h with 5% (w/v) nonfat dry milk in Tris-phosphate buffer containing 0.1% (v/v) Tween 20 (TBST). Subsequently, the membranes were incubated overnight at 4 °C with corresponding primary antibodies, including rabbit monoclonal anti-NG2 (neural/glial antigen 2) (1
000 rpm for 10 min, the total protein concentrations were quantified by a BCA kit (Thermo Scientific). A total amount of 20 μg of protein from each sample was subjected to 7.5%, 10% or 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) followed by transfer onto the polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences, Little Chalfont, UK). The membranes were blocked for 1 h with 5% (w/v) nonfat dry milk in Tris-phosphate buffer containing 0.1% (v/v) Tween 20 (TBST). Subsequently, the membranes were incubated overnight at 4 °C with corresponding primary antibodies, including rabbit monoclonal anti-NG2 (neural/glial antigen 2) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Abcam), rabbit monoclonal anti-PDGFR-β (platelet derived growth factor receptor beta) (1
1000; Abcam), rabbit monoclonal anti-PDGFR-β (platelet derived growth factor receptor beta) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Abcam), rabbit monoclonal anti-α-SMA (α-smooth muscle actin) (1
1000; Abcam), rabbit monoclonal anti-α-SMA (α-smooth muscle actin) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Cell Signaling Technology), rabbit polyclonal anti-SM22α (smooth muscle protein 22-alpha) (1
1000; Cell Signaling Technology), rabbit polyclonal anti-SM22α (smooth muscle protein 22-alpha) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Abcam), rabbit polyclonal anti-PDGF-BB (1
1000; Abcam), rabbit polyclonal anti-PDGF-BB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800; Abcam), mouse monoclonal anti-plexin-B1 (1
800; Abcam), mouse monoclonal anti-plexin-B1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Santa Cruz Biotechnology, Dallas, TX, USA), mouse monoclonal anti-GAPDH (1
200; Santa Cruz Biotechnology, Dallas, TX, USA), mouse monoclonal anti-GAPDH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Cell Signaling Technology), and mouse monoclonal anti-β-actin (1
1000; Cell Signaling Technology), and mouse monoclonal anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Santa Cruz Biotechnology); after three washes with TBST for 5 min each, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. After repeating the washing step, the target protein signal was measured by enhanced chemiluminescence (Thermo Scientific). Then, the membranes were visualized using a gel imaging system (Bio-Rad, Hercules, CA). The expression of protein was quantified by ImageJ software.
1000; Santa Cruz Biotechnology); after three washes with TBST for 5 min each, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. After repeating the washing step, the target protein signal was measured by enhanced chemiluminescence (Thermo Scientific). Then, the membranes were visualized using a gel imaging system (Bio-Rad, Hercules, CA). The expression of protein was quantified by ImageJ software.
2.13 ELISA
The secretory protein levels of PDGF-BB, b-FGF, ANGPTL4, VEGF, HB-EGF and TGF-β1 were examined using respective ELISA kits (R&D Systems, Minnesota, US). Briefly, HUVECs or SHED were seeded on 6 well plates at a density of 3 × 105 cells per well until they reached 80% confluence and treated with Sema4D (1 μg mL−1). Supernatants were collected after culturing for 24 h and centrifuged to remove any particulates. The 96-well microplate was coated with capture antibody overnight, followed by blocking with reagent diluent for 1 h at room temperature. Then, 100 μL of standards diluted in reagent diluent and supernatants were added to the microplate, incubating for 2 h at room temperature. Subsequently, the microplate was incubated with detection antibody for 2 h, followed by incubating with HRP-conjugated streptavidin for 20 min in the dark. Finally, each well was incubated with 100 μL of substrate solution for 20 min. 50 μL of stop solution was added to each well and absorbance readings were measured immediately under 450 nm and 540 nm by SpectraMax® M2 microplate reader (Molecular Devices).2.14 Quantitative real-time polymerase chain reaction (RT-PCR)
RT-PCR was performed to detect the expression of PDGF-BB and plexin-B1 in Sema4D-treated HUVECs as described in our previous study.24 Briefly, HUVECs were seeded on 6 well plates at a density of 3 × 105 cells per well. Sema4D (1 μg mL−1) was added at 80% confluence. After culturing for 24 h, total RNA was extracted using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and then mixed with SuperScript® VILO™ Master Mix (Invitrogen) to reverse transcription of RNA into cDNA. RT-PCR was performed with a StepOne Real-Time PCR system (Thermo Scientific) using SYBR™ Select Master Mix (Thermo Scientific). The delta–delta Ct method was used to calculate gene expression levels normalized to GAPDH and control group values. Primers (Sigma Aldrich) were as follows: GAPDH: 5′-GTCTCCTCTGACTTCAACAGCG-3′ (forward); 5′-ACCACCCTGTTGCTGTAGCCAA-3′ (reverse); plexin-B1: 5′-CCTTCACGGGCACGCCCTGGGCCT-3′ (forward); 5′-AAGAACCCCAAGCTGATGCTGCGCAGG-3′ (reverse); PDGF-BB: 5′-GAGATGCTGAGTGACCACTCGA-3′ (forward); 5′-GTCATGTTCAGGTCCAACTCGG-3′ (reverse).2.15 Statistical analysis
The experiments were performed in triplicate with three independent experiments. All data are shown as mean ± standard deviation (SD). Statistical analyses were carried out using Student's t test or one-way analysis of variance with a Tukey's post hoc test to determine significant differences between groups. All analyses were performed using the GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA). P < 0.05 was considered statistically significant.3. Results and discussion
3.1 Sema4D enhances vessel formation and percentage of SHED-covered vascular structures
To investigate the effects of Sema4D on SHED supported vessel formation and stabilization, in vitro 3D biomimetic microfluidic device was used. We encapsulated HUVECs within the fibrin gel and laid in the central channel, followed by seeding HUVECs in the upper channel (Fig. 2A). After 24 h, once the nascent endothelial tubes were formed, SHED were seeded in the lower channel and Sema4D was added into the HUVEC-seeded upper channel. SHED were migrated into the fibrin gel and co-localized with HUVEC formed vessel network in the central channel (Fig. 2B). A significant increase in total length of the vessel structures was observed in Sema4D-treated group (p < 0.01) compared with that of the non-treated group (Fig. 2C). We believe that the total vessel length in initial stages could be the same among different groups. When SHED were recruited to the abluminal surface of vessels formed by HUVECs, the vessels become stabilized and last longer. Therefore, after culturing for 7 days, the Sema4D-treated groups had more vessels remained compared to control group giving rise to higher total vessel length. Additionally, Sema4D-treated group showed significantly higher number of SHED co-localizing with HUVEC formed vascular tube network, as indicated by the higher percentage of SM22α+SHED covered vascular network (p < 0.05) (Fig. 2C). At high magnification (Fig. 2D), clear vessel structures with CD31+ endothelial lumens supported by SM22α+SHED in the perivascular area were observed. These findings indicated that Sema4D could facilitate the direct interaction between HUVECs and SHED in enhancing the vessel formation and stabilization. To further confirm the functional effects of Sema4D on vessel stabilization, dextran permeability assay was conducted. After introducing 10 kDa Alexa fluor 555 labelled dextran into upper channel, dextran flowed into the vascular tubes and leaked out to the perivascular region. Interestingly, Sema4D-treated group showed lower vascular permeability as shown by significantly less amount of dextran in perivascular region compared to the non-treated group, which indicates a more mature and stable vessel structures (Fig. 2E). Sema4D-treated group showed lower permeability coefficient (0.86 × 10−6 cm s−1, Fig. 2F), which is comparable to those measured in rat cerebral microvessels (0.31 × 10−6 cm s−1).25In order to further confirm the incorporation of SHED onto the vessel wall, we performed immunofluorescent staining for vascular basement membrane marker type IV collagen. Triple staining for CD31, SM22α, and collagen IV demonstrated that SM22α+SHED truly get incorporated to the endothelial wall of the vessels formed by CD31+HUVECs (Fig. 3). Furthermore, both SM22α+SHED and CD31+HUVECs were localized within the collagen IV positive basement membrane, which is another important criterion for mural cells (Fig. 3). The video containing different layers of Z-stacks further confirmed this feature (Video S1†).
We also conducted the live imaging to visualize the behavior of SHED migration and incorporation on to the vessel wall. The videos showed that during early time points (on day 3), SHED (CellTracker™ Red CMTPX labeled) migrated from the side channel to central channel where HUVECs (GFP labeled) formed the vascular tubes (Video S2†). At later time points (on day 6), the SHED incorporated onto the abluminal surface of endothelial vessels (GFP-HUVEC) were stable without moving along the vessel walls (Video S3†), indicating incorporation onto the vessel wall to act as mural cells.
Mural cells have been known to play a role in the regulation of vessel diameter, vascular permeability, and blood flow.26 Kim et al. reported that mural cells could reduce the vessel diameter,22 while Huang et al. showed that there is no difference in vessel diameter when the NG2 positive mural cells are deficient.27 In our experiments, vessels did not show a significant difference in diameter between control and Sema4D groups (data not shown).
3.2 Sema4D indirectly increases the migration of SHED through endothelial derived factors
As mural cells play a critical role in vascular stabilization and homeostasis, the recruitment of mural cells is a prerequisite for achieving mature vessels. Once we observed that under Sema4D treatment, SHED co-localized with endothelial lumens, we asked the question how does Sema4D–HUVEC interaction recruit SHED to stabilize endothelial lumens. To answer this question, we examined the differentiation, proliferation and migration of SHED under Sema4D or Sema4D-treated HUVEC CM.We found that SHED express relatively high levels of mural cell markers NG2, PDGFR-β, α-SMA, and SM22α (Fig. 4A) under normal culture conditions, which indicates their inherent potential to act as mural cells. This is in accordance with previous studies that reported mural cell like characteristics of SHED.8,9 Mural cells, including vSMCs and pericytes, are a heterogenous population of cells28,29 which share some common markers, such as α-SMA, vimentin, and desmin.6,30,31 Besides these common markers, SM22α, calponin, and smooth muscle myosin heavy chain (SM-MHC) are widely used as markers for vSMCs,19,28 while NG2, PDGFR-β, and RGS5 are often considered as markers for pericytes.32 Up to date, there is no consensus on markers specific for either vSMCs or pericytes. Due to these reasons, most studies have adopted an approach to assess several markers to characterize the mural cells of concern. In our study, we performed immunofluorescence for PDGFR-β additional to SM22α to demonstrate the mural cell phenotype of SHED (Fig. S5†).
Furthermore, we examined the expression of mural cell markers after culturing SHED for 24 h and 72 h in different CM (Fig. S6A and B†). At 24 h, the expression of NG2 was highest in SHED cultured in Sema4D-treated HUVEC CM, among all the groups (p < 0.05). There was no significant difference in the expression of PDGFR-β, α-SMA and SM22α among all the groups at 24 h. Furthermore, there was a significantly higher (p < 0.05) expression of NG2, α-SMA and SM22α in SHED cultured in Sema4D-treated HUVEC CM than that in ECM control group at 72 h (Fig. S6A and B†). Interestingly, a significant downregulation of PDGFR-β was observed in Sema4D-treated HUVEC CM than that in ECM control group at 72 h compared to the similarly high levels of expression among all groups at 24 h. These results indicated that Sema4D induced endothelial factors drive SHED differentiation towards a more mature mural cell phenotype. It has been reported that NG2 is expressed during the early stages of angiogenesis and α-SMA is considered as a late stage marker.33,34
When we examined the proliferation, compared with the ECM group, SHED cultured in ECM containing Sema4D or HUVEC CM with or without Sema4D had a significantly higher (p < 0.01) proliferation rate at 24 h (Fig. 4B). At 48 and 72 h, SHED cultured in HUVEC CM or Sema4D-treated HUVEC CM demonstrated a significantly higher (p < 0.01) proliferation than that in ECM with or without Sema4D. However, no statistically significant difference in proliferation of SHED cultured in HUVEC CM and Sema4D-treated HUVEC CM was observed at 48 h and 72 h. These results indicated that although endothelial derived factors significantly enhance SHED proliferation, Sema4D does not have a direct or indirect effect on that.
Migration capacity of SHED in Sema4D-treated HUVEC CM was examined using microfluidic chip and trans-well assay. In microfluidic assay, (Fig. S4†), fibrin gel was laid in the central channel of microfluidic device followed by seeding of HUVECs in the upper channel and SHED in the lower channel. Sema4D was added to the upper channel after 24 hours of cell seeding. SHED were migrated from the lower channel towards HUVECs. There was no difference in the number of migrated SHED between Sema4D-treated and non-treated groups (Fig. 4C). However, the mean migration distance of SHED in Sema4D-treated group was significantly higher than the control group (p < 0.05) (Fig. 4C).
To further investigate whether Sema4D drives the migration of SHED, trans-well assay was conducted under different CM. The migratory capacity of SHED, when Sema4D was directly added to ECM, was similar to that of the control group (Fig. 4D). In contrast, in Sema4D-treated HUVEC CM, SHED showed a markedly increased migration compared to all other groups (p < 0.01). This finding demonstrated that Sema4D does not have a direct effect on the migration of SHED, but induces the migration indirectly through acting on endothelial cells to secrete paracrine factors.
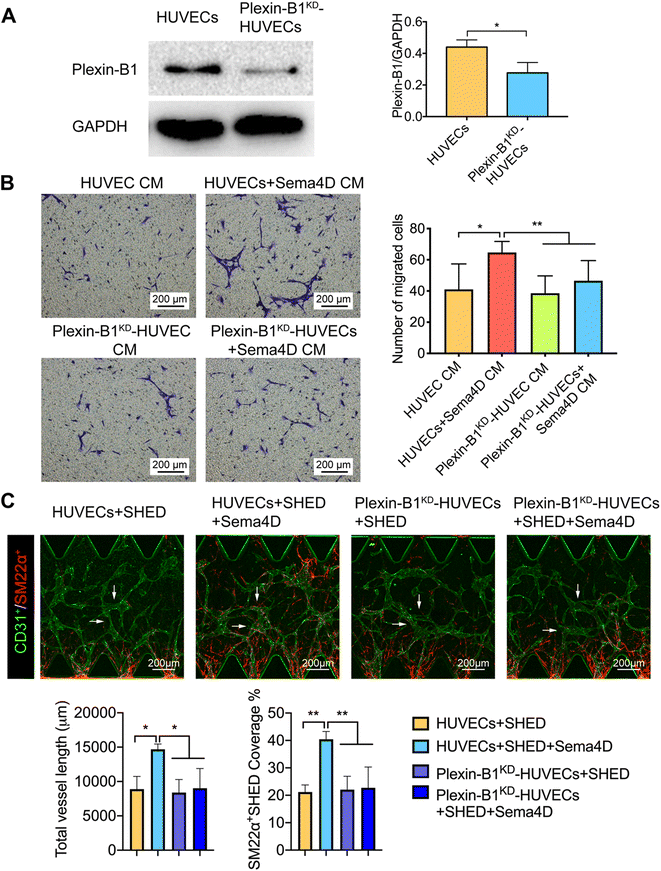
3.3 Knockdown of plexin-B1 on HUVECs (plexin-B1KD-HUVECs) impairs the recruitment of SHED
In order to confirm that the above-mentioned effects are Sema4D meditated, the expression levels of Sema4D receptor plexin-B1 were examined in SHED and HUVECs by western blotting. The results revealed that HUVECs express significantly higher (p < 0.01) level of plexin-B1 than SHED (Fig. S7†). In order to further clarify the role of endothelial plexin-B1 in Sema4D induced mural cell recruitment, loss of function experiment was done. Plexin-B1 was knocked down in HUVECs via siRNA and confirmed by western blotting (Fig. 5A). Migration of SHED in Sema4D-treated plexin-B1KD-HUVEC CM was significantly decreased (p < 0.01) compared with that in Sema4D-treated HUVEC CM (Fig. 5B). In contrast, there was no effect on migration of SHED when plexin-B1 was knocked down in SHED and cultured in HUVEC CM or Sema4D-treated HUVEC CM (Fig. S8†). Furthermore, we cultured SHED with either HUVECs or plexin-B1KD-HUVECs with or without Sema4D treatment in a microfluidic device. There is no difference in terms of total vessel length and the percentage of SM22α+SHED covered vessel structures between the HUVECs and plexin-B1KD-HUVECs groups without Sema4D treatment. Sema4D significantly increased the total vessel length and the percentage of SM22α+SHED covered vessel structures in HUVEC formed vessels. These effects were significantly reduced when Sema4D was added in plexin-B1KD-HUVECs cultures (Fig. 5C and S9†). Taken together, these findings confirmed that the Sema4D enhanced the recruitment of SHED through acting on endothelial plexin-B1.3.4 The recruitment of SHED by Sema4D is mediated through endothelial derived PDGF-BB signaling
As we figured out that the role of Sema4D in the recruitment of SHED for vascular stabilization is mainly exerted indirectly through endothelial derived factors, it is of interest to know what specific growth factors are responsible here. Up to date, endothelial cell secreted PDGF-BB is the best characterized angiogenic factor in recruiting mural cells. In addition, basic fibroblast growth factor (b-FGF) is known to act synergistically with PDGF-BB in stimulating proliferation, migration and differentiation of pericytes.35,36 Among other growth factors, transforming growth factor β1 (TGF-β1) is reported to induce mural cell differentiation, and heparin binding epidermal growth factor (HB-EGF) is known to play a role in increasing the proliferation of pericytes.3,19 Angiopoietin-like-4 (ANGPTL4) is also reported to modulate the pericyte coverage and control vascular permeability.37According to our results, Sema4D significantly increased (p < 0.01) the expression of secretory PDGF-BB in HUVECs at 24 h as shown by ELISA (Fig. 6A) although no difference was observed in the mRNA and total protein levels (Fig. 6B–D). There was no detectable expression of PDGF-BB in SHED. Plexin-B1 protein expression in endothelial cells was not changed (Fig. 6C and D) with Sema4D treatment despite the significantly increased (p < 0.05) mRNA levels (Fig. 6B).
ANGPTL4 was not detected in both cell types (data are not shown as the expression levels were below the detectable range of the ELISA). No significant increase in the expression of b-FGF, VEGF, HB-EGF and TGF-β1 was detected in either HUVECs or SHED following Sema4D treatment (Fig. S10†).
PDGF-BB, of which the primary source is endothelial cells, is the most important angiogenic factor that facilitates the stabilization of the nascent vessels by inducing differentiation and recruitment of mural cells.3,38,39 Genetic disruption of PDGF-BB or PDGFRβ in mice has resulted in pericyte loss, which has further led to abnormalities such as excessive vascularization and microaneurysms.40
In order to investigate the role of PDGF-BB/PDGFR-β in the recruitment of SHED, PDGFR-β inhibitor (CP-673451) was added to Sema4D-treated HUVEC CM in trans-well assay. Sema4D-treated HUVEC CM could significantly enhance the migration of SHED, which was blocked by PDGFR-β inhibitor (p < 0.01, Fig. 7A). Furthermore, we cultured HUVECs and SHED with or without Sema4D and PDGFR-β inhibitor treatment in a microfluidic device. We did not observe any difference in terms of vessel formation and SM22α+SHED coverage between HUVECs + SHED group and HUVECs + SHED + PDGFR-β inhibitor group, which is in accordance with the results of a previous study.41 In contrast, Sema4D significantly increased the total vessel length and the percentage of SM22α+SHED covered vessel structures, which was significantly blocked by PDGFR-β inhibitors (p < 0.01, Fig. 7B). This further confirmed that the recruitment of SHED is mediated through PDGF-BB/PDGFR pathway, which is consistent with the findings of the previous studies.3,39
It has also been reported that mural cells can be induced from Flk1+ mesoderm cells by using PDGF-BB.42 Furthermore, tumor cell-derived PDGF-BB could induce mesenchymal stem cell-pericyte transition.39 Another endothelial cell-derived factor—b-FGF is known to induce the proliferation, migration and differentiation of many cell types–including vSMCs and pericytes.36 Besides, b-FGF is shown to synergize with PDGF-BB to increase the vascular stability and improvement in hind-limb ischemia.43 The exact mechanism, under which the differentiation of SHED into mural cells is increased, however, is not clear and therefore, further studies are needed in this regard.
4. Conclusion
The interaction between endothelial and mural cells is essential for vascular development and maturation.42,44 In this regard, the recruitment of mural cells to nascent vessels formed by endothelial cells is a critical prerequisite45–47 for achieving stable functional vasculature. In the current study, using a 3D microfluidic device, for the first time, we demonstrated the role of Sema4D–plexin-B1 signaling in the recruitment of SHED as mural cells to stabilize the EC formed vascular tubes. The administration of Sema4D to EC compartment of the microfluidic device resulted in enhanced recruitment of SM22α+SHED to support the EC formed vasculature.Taken together, the capacity of the 3D microfluidic device to recreate in vivo 3D physiological microenvironment and ability to control the addition of cells and chemical factors in a spatiotemporal manner enabled us to examine the role of Sema4D–plexin-B1 signaling in mediating endothelial–SHED cross talk in vascular stabilization. Particularly, the use of 3D microfluidic device provided an excellent platform that enables reproducing the experimental conditions and results as well as imaging the live cells and immunofluorescent labelled cells in high resolution. The results of this study further highlight the potential role of Sema4D as a target growth factor in fabrication of engineered tissue constructs with mature vessels.
Author contributions
L. Zhang, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; Y. Han: contributed to data acquisition and analysis, critically revised the manuscript; Q. Chen: contributed to data acquisition and analysis, critically revised the manuscript; W. L. Dissanayaka, contributed to conception, design, data analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.Data availability
Data for this paper are available at HKU Data Repository at DOI: https://doi.org/10.25442/hku.20278737.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research was supported by the Research Grants Council, Hong Kong (General Research Fund; 17117619).References
- J. Rouwkema and A. Khademhosseini, Trends Biotechnol., 2016, 34, 733–745 CrossRef CAS PubMed.
- S. Ben-Shaul, S. Landau, U. Merdler and S. Levenberg, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2955–2960 CrossRef CAS PubMed.
- S. S. Kemp, K. N. Aguera, B. Cha and G. E. Davis, Arterioscler., Thromb., Vasc. Biol., 2020, 40, 2632–2648 CrossRef CAS PubMed.
- T. Takebe, N. Koike, K. Sekine, M. Enomura, Y. Chiba, Y. Ueno, Y. W. Zheng and H. Taniguchi, Transplant. Proc., 2012, 44, 1130–1133 CrossRef CAS PubMed.
- D. C. Darland and P. A. D'Amore, J. Clin. Invest., 1999, 103, 157–158 CrossRef CAS PubMed.
- H. Gerhardt and C. Betsholtz, Cell Tissue Res., 2003, 314, 15–23 CrossRef PubMed.
- S. Kim and H. Von Recum, Tissue Eng., Part B, 2008, 14, 133–147 CrossRef CAS PubMed.
- M. Miura, S. Gronthos, M. Zhao, B. Lu, L. W. Fisher, P. G. Robey and S. Shi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5807–5812 CrossRef CAS PubMed.
- J. H. Kim, G. H. Kim, J. W. Kim, H. J. Pyeon, J. C. Lee, G. Lee and H. Nam, Mol. Cells, 2016, 39, 790 CrossRef CAS PubMed.
- S. P. Chapoval, Mol. Med., 2018, 24, 1–20 CAS.
- K. Lontos, J. Adamik, A. Tsagianni, D. L. Galson, J. M. Chirgwin and A. Suvannasankha, Front. Endocrinol., 2018, 9, 322 CrossRef PubMed.
- K. T. Maleki, M. Cornillet and N. K. Björkström, Clin. Immunol., 2016, 163, 52–59 CrossRef CAS PubMed.
- I. Oinuma, Y. Ito, H. Katoh and M. Negishi, J. Biol. Chem., 2010, 285, 28200–28209 CrossRef CAS PubMed.
- J. R. Sierra, S. Corso, L. Caione, V. Cepero, P. Conrotto, A. Cignetti, W. Piacibello, A. Kumanogoh, H. Kikutani and P. M. Comoglio, J. Exp. Med., 2008, 205, 1673–1685 CrossRef CAS PubMed.
- E. S. Ch'ng and A. Kumanogoh, Mol. Cancer, 2010, 9, 251 CrossRef PubMed.
- P. Conrotto, D. Valdembri, S. Corso, G. Serini, L. Tamagnone, P. M. Comoglio, F. Bussolino and S. Giordano, Blood, 2005, 105, 4321–4329 CrossRef CAS PubMed.
- I. Zuazo-Gaztelu, M. Pàez-Ribes, P. Carrasco, L. Martín, A. Soler, M. Martínez-Lozano, R. Pons, J. Llena, L. Palomero, M. Graupera and O. Casanovas, Cancer Res., 2019, 79, 5328–5341 CrossRef CAS PubMed.
- Y. H. Hsu, M. L. Moya, C. C. Hughes, S. C. George and A. P. Lee, Lab Chip, 2013, 13, 2990–2998 RSC.
- J. G. Xu, S. Y. Zhu, B. C. Heng, W. L. Dissanayaka and C. F. Zhang, Stem Cell Res. Ther., 2017, 8, 10 CrossRef PubMed.
- J. R. Basile, A. Barac, T. Zhu, K. L. Guan and J. S. Gutkind, Cancer Res., 2004, 64, 5212–5224 CrossRef CAS PubMed.
- H. Zhou, Y. Yang, N. O. Binmadi, P. Proia and J. R. Basile, Exp. Cell Res., 2012, 318, 1685–1698 CrossRef CAS PubMed.
- J. Kim, M. Chung, S. Kim, D. H. Jo, J. H. Kim and N. L. Jeon, PLoS One, 2015, 10(7), e0133880 CrossRef PubMed.
- S. Bang, S. R. Lee, J. Ko, K. Son, D. Tahk, J. Ahn, C. Im and N. L. Jeon, Sci. Rep., 2017, 7, 8083 CrossRef PubMed.
- Y. Han, T. Gong, C. Zhang and W. Dissanayaka, J. Dent. Res., 2020, 99, 804–812 CrossRef CAS PubMed.
- W. Yuan, Y. Lv, M. Zeng and B. M. Fu, Microvasc. Res., 2009, 77, 166–173 CrossRef CAS PubMed.
- B. Gökçinar-Yagci, D. Uçkan-Çetinkaya and B. Çelebi-Saltik, Stem Cell Rev. Rep., 2015, 11, 549–559 CrossRef PubMed.
- F. J. Huang, W. K. You, P. Bonaldo, T. N. Seyfried, E. B. Pasquale and W. B. Stallcup, Dev. Biol., 2010, 344, 1035–1046 CrossRef CAS PubMed.
- E. Avolio, V. V. Alvino, M. T. Ghorbel and P. Campagnolo, Pharmacol. Ther., 2017, 171, 83–92 CrossRef CAS PubMed.
- T. Yamazaki and Y. S. Mukouyama, Front. Cardiovasc. Med., 2018, 5, 78 CrossRef PubMed.
- R. H. Adams and K. Alitalo, Nat. Rev. Mol. Cell Biol., 2007, 8, 464–478 CrossRef CAS PubMed.
- M. Wanjare, S. Kusuma and S. Gerecht, Stem Cell Rep., 2014, 2, 746 CrossRef CAS PubMed.
- J. Xu, T. Gong, B. C. Heng and C. F. Zhang, FASEB J., 2017, 31, 1775–1786 CrossRef CAS PubMed.
- U. Ozerdem and W. B. Stallcup, Angiogenesis, 2004, 7, 269–276 CrossRef CAS PubMed.
- S. P. Parthiban, W. He, N. Monteiro, A. Athirasala, C. M. França and L. E. Bertassoni, Sci. Rep., 2020, 10, 21579 CrossRef CAS PubMed.
- K. Hosaka, Y. Yang, M. Nakamura, P. Andersson, X. Yang, Y. Zhang, T. Seki, M. Scherzer, O. Dubey and X. Wang, Cell Discovery, 2018, 4, 1–14 CrossRef CAS PubMed.
- Z. Galzie, A. R. Kinsella and J. A. Smith, Biochem. Cell Biol., 1997, 75, 669–685 CrossRef CAS PubMed.
- E. G. Perdiguero, A. Galaup, M. Durand, J. Teillon, J. Philippe, D. M. Valenzuela, A. J. Murphy, G. D. Yancopoulos, G. Thurston and S. Germain, J. Biol. Chem., 2011, 286, 36841–36851 CrossRef CAS PubMed.
- N. H. Davies, C. Schmidt, D. Bezuidenhout and P. Zilla, Tissue Eng., Part A, 2012, 18, 26–34 CrossRef CAS PubMed.
- K. Dhar, G. Dhar, M. Majumder, I. Haque, S. Mehta, P. J. Van Veldhuizen, S. K. Banerjee and S. Banerjee, Mol. Cancer, 2010, 9, 1–12 CrossRef PubMed.
- M. Enge, M. Bjarnegård, H. Gerhardt, E. Gustafsson, M. Kalén, N. Asker, H. P. Hammes, M. Shani, R. Fässler and C. Betsholtz, EMBO J., 2002, 21, 4307–4316 CrossRef CAS PubMed.
- K. Haase, M. R. Gillrie, C. Hajal and R. D. Kamm, Adv. Sci., 2019, 6, 1900878 CrossRef CAS PubMed.
- J. Yamashita, H. Itoh, M. Hirashima, M. Ogawa, S. Nishikawa, T. Yurugi, M. Naito, K. Nakao and S.-I. Nishikawa, Nature, 2000, 408, 92–96 CrossRef CAS PubMed.
- R. Cao, E. Brakenhielm, R. Pawliuk, D. Wariaro, M. J. Post, E. Wahlberg, P. Leboulch and Y. Cao, Nat. Med., 2003, 9, 604–613 CrossRef CAS PubMed.
- A. N. Stratman, S. A. Pezoa, O. M. Farrelly, D. Castranova, L. E. Dye, M. G. Butler, H. Sidik, W. S. Talbot and B. M. Weinstein, Development, 2017, 144, 115–127 CAS.
- D. O. Traktuev, S. Merfeld-Clauss, J. Li, M. Kolonin, W. Arap, R. Pasqualini, B. H. Johnstone and K. L. March, Circ. Res., 2008, 102, 77–85 CrossRef CAS PubMed.
- R. K. Jain, Nat. Med., 2003, 9, 685–693 CrossRef CAS PubMed.
- G. Bergers and S. Song, Neuro-Oncology, 2005, 7, 452–464 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc00632d |
| This journal is © The Royal Society of Chemistry 2022 |