Open Access Article
Open Access ArticleAutomation of cell culture assays using a 3D-printed servomotor-controlled microfluidic valve system†
Steffen
Winkler
 a,
Jannik
Menke
a,
Katharina V.
Meyer
a,
Jannik
Menke
a,
Katharina V.
Meyer
 a,
Carlotta
Kortmann
a and
Janina
Bahnemann
a,
Carlotta
Kortmann
a and
Janina
Bahnemann
 *b
*b
aInstitute of Technical Chemistry, Leibniz University Hannover, Hannover, Germany
bInstitute of Physics, University of Augsburg, Universitätsstraße 1, 86159 Augsburg, Germany. E-mail: janina.bahnemann@uni-a.de
First published on 7th November 2022
Abstract
Microfluidic valve systems show great potential to automate mixing, dilution, and time-resolved reagent supply within biochemical assays and novel on-chip cell culture systems. However, most of these systems require a complex and cost-intensive fabrication in clean room facilities, and the valve control element itself also requires vacuum or pressure sources (including external valves, tubing, ports and pneumatic control channels). Addressing these bottlenecks, the herein presented biocompatible and heat steam sterilizable microfluidic valve system was fabricated via high-resolution 3D printing in a one-step process – including inlets, micromixer, microvalves, and outlets. The 3D-printed valve membrane is deflected via miniature on-chip servomotors that are controlled using a Raspberry Pi and a customized Python script (resulting in a device that is comparatively low-cost, portable, and fully automated). While a high mixing accuracy and long-term robustness is established, as described herein the system is further applied in a proof-of-concept assay for automated IC50 determination of camptothecin with mouse fibroblasts (L929) monitored by a live-cell-imaging system. Measurements of cell growth and IC50 values revealed no difference in performance between the microfluidic valve system and traditional pipetting. This novel design and the accompanying automatization scripts provide the scientific community with direct access to customizable full-time reagent control of 2D cell culture, or even novel organ-on-a-chip systems.
1. Introduction
Acting on the micro- to nanometer scale, microfluidic systems have enabled researchers to precisely study or manipulate cells, viruses, and/or proteins by using microbioreactors,1 microfluidic cell separators,2 micromixers,3 integrated biosensors,4 and/or novel organoid/organ-on-a-chip systems.5,6 Unfortunately, relatively low accessibility and the considerable costs of clean room facilities (which are required for standard lithographic fabrication of microfluidic chips) have heretofore prevented this technology from being imported to industrial application on a large scale.7,8 3D printing of microfluidics has started to rise in popularity – since that approach results in drastically reduced chip prototyping times and decreased acquisition costs – and printing resolution has advanced to the point where printing on the micrometer or even nanometer scale is now feasible.9–11 The tantalizing promise of rapid prototyping, considerably higher complexity in the third dimension, customizability, and the ability to use and incorporate several different materials (including biocompatible,12,13 cell adherable14 or heat-resistant polymers13) have all piqued the interest of researchers.Automated liquid control for multiplexing and assaying within microfluidic systems is often accomplished by using microvalves that are directly integrated during the fabrication process to create microfluidic valve systems. Such systems have demonstrated to be capable of effectively managing assay operations in high-throughput by using microvalve arrays.15 However, compared to established pipetting robots and high-throughput screening in the pharmaceutical industry, microfluidic valve systems are typically not superior in throughput, but in performing complex protocols with programmed queues of reactants. For instance, such systems have been used to automate and parallelize on-chip cell seeding and facilitate the cultivation of human umbilical vein endothelial cells (HUVECs) in a modular plug-and-play microfluidic valve system.16 While pipetting systems are usually fixed to a certain plate design, microfluidic valve systems are more versatile and can be plugged to other microfluidic systems that have complex architectures at the micro-scale. For instance, one microfluidic platform was used to immobilize tumor organoids inside microgrooves and was in turn combined with a second microfluidic system for combinatorial and dynamic drug screening.17
On the other hand, there are high-end pipetting systems available that have been combined with live-cell-imaging systems via robotics and automated incubators to fully automate cell culture handling and monitoring.18,19 However, apart from the high costs, these systems are severely limited to standard well-plates and perfusion of sophisticated 3D cell culture systems is not possible. In contrast, microfluidic valve systems are highly customizable and can be interfaced with microfluidically controlled 3D cell culture systems, such as organ-on-a-chip systems and vascularized (3D-printed) hydrogels, which have the potential to become important research platforms for drug discovery and tissue engineering in the near future.6,20–22
The type and operation principles of the microvalves utilized are essential, since they pre-define many features of the microfluidic valve system (e.g., footprint, manufacturability, fabrication costs, operating hardware, complexity in set-up and control, etc.). Typically, they include a flexible membrane – such as Quake-style,23 doormat24 or plunger25 valves – that is deformed for channel closure or opening.26 While microvalves can be actuated in many different ways (e.g., mechanically, magnetically, electrostatically, acoustically, thermally or piezoelectrically, etc.), pneumatic actuation is most commonly used for microfluidic automatization.27 In that approach, pressure or the creation of a vacuum for membrane deformation is distributed to the valves via a microfabricated control channel layer on top of the flow channel layer. In the simplest case, one control channel operates a single flow channel. More advanced systems for increased throughput use microfluidic multiplexing, where the number of total valves is increased to reduce the number of total control channels to 2 log2 of n flow channels.28 However, aside from the fact that most pneumatically driven microfluidic valve systems are difficult to fabricate, further major drawbacks include a more complicated set-up and control. While the microfluidic chip relies on additional pneumatic control channels and pressure/vacuum inlet ports, the whole system requires extra tubing, external solenoid valves, and at least one pressure/vacuum source – all of which combine to substantially increase complexity, cost, system footprint, and the overall statistical risk of failure. Accordingly, the actual use of these systems is still generally limited to a small fraction of micro-engineers.
In an attempt to address these limitations, a 3D-printed microfluidic valve system for spatiotemporal reagent control that is operated by miniature servomotors has been developed. This compact on-chip microvalve control mechanism is connected to a Raspberry Pi computer, which enables automatization and allows the system to function as a portable, remotely controllable, and low-cost device. As a novelty, inlets, outlets, micromixer, microvalves and even valve membranes were entirely fabricated in a single part via 3D printing using a biocompatible and autoclavable material. All of these elements were easily plugged to the servomotors, thereby eliminating the need for the sort of complex fabrication and/or set-up procedures typically required by conventional microfluidic valve systems. While a sufficient mixing accuracy and valve robustness is shown, its applicability for programmable assaying in cell culture is demonstrated as a proof-of-concept. Ultimately, in view of the rapid customization abilities already well known through published 3D models, we envision this device as a dynamic reagent control system suitable for use with more complex microfluidic cell culture systems, like organ-on-a-chip devices.
2. Materials and methods
2.1. Fabrication, post-processing and sterilization of 3D-printed parts
All 3D-printed parts were designed using SolidWorks 2020 (Dassault Systems Deutschland GmbH, Stuttgart, Germany) and are published as .sldprt computer-aided design (CAD) files with this work. The microfluidic chip and printed adapters were fabricated using a high-resolution MultiJet 3D printer (ProJet® MJP 2500 Plus, 3D Systems, Rock Hill, SC, USA). The 3D printing material VisiJet® M2S-HT90 and the VisiJet® M2 Sup were used as build material and support material, respectively. The detailed chemical composition of the build material is not provided by the manufacturer. However, from the safety data sheet it can be concluded that the raw material contains several hazardous chemicals, while the printed polymerized build material is specified as polyacrylate.29 The material is suitable for cell culture applications as it is heat resistant for sterilization and biocompatible with L929 cells according to the manufacturer's USP class VI and a recent study in accordance with the international standard ISO 10993-12:2021(E).13,30 For removal of the support material, parts printed using the ProJet® MJP 2500 Plus were post-processed in accordance with the protocols identified and described in recent publications.13 However, the protocols were slightly modified here by flushing each microchannel in the beginning of each post-processing step. For cell culture applications, the chip was connected to the fittings and tubes as described in chapter 0, and then heat-steam sterilized.2.2. Platform assembly
The 3D-printed system was controlled via a custom Python script using the open-source software Python 3.5.3 (Python Software Foundation, Delaware, USA) and Thonny 3.1.0 (Institute of Computer Science of University Tartu, Tartu, Estonia), which was run on a Raspberry Pi 3 Model B V1.2 (Raspberry Pi Foundation, Cambridge, UK) with a Raspbian GNU/Linux 9 operating system (Raspberry Pi Foundation, Cambridge, UK). A pulse-width modulation (PWM) controller (SparkFun Servo pHAT for Raspberry Pi; SparkFun Electronics Inc., Niwot, USA) was mounted directly on the Raspberry Pi and connected to four 17 × 6.2 × 16 mm Goteck GS-D1083 Micro Servos (Dong Yang Model Technology Co., Ltd., Huizhou, China). Furthermore, a HLS8L-DV3V-S-C relay (Ningbo Helishun Electron Co., Ltd., Ningbo, China) was interposed between the servos and the board in order to enable a shutdown of the servos. Aladdin AL1000 syringe pumps (Word Precision Instruments LLC, Sarasota, USA) were connected to the Raspberry Pi via USB, and to the chip via standard chromatography PTFE tubing (Ø 0.5 mm) and fittings. Prior to running experiments, the 3D-printed microfluidic valve chip was prepared by inserting M5x6 setscrews into the threads above each valve, which were very gently hand-tightened to insure a snug fit. Each of these contained a 3D-printed adapter as the connection of the servomotors to the M5 setscrews. In turn, servos were connected to a 3D-printed housing using standard M1 screws, and were set to the starting position at servo position = 60° and mounted on the adapters. Closure of the valve occurred at the servo position = 90° and opening of the value occurred at −15°, respectively. The servos were switched off when not being operated in order to avoid a permanent load on the servo during the closed state of the valve.2.3. Long-term robustness
For the long-term stability determinations, the valves were successively and repeatedly opened and closed by the servos as a stress test over a total time period of 4–5 days. For each opening event, one valve was opened while the rest of the valves remained closed, and the valve was then rinsed off for 10 s at 500 μL min−1 with ddH2O using an Ismatec IPC peristaltic pump (Cole-Parmer GmbH, Wertheim, Germany). Including the time taken to open/close the valve, the time from the opening of the first valve to the opening of the second valve was estimated by the script to be 10.12 s. Failure of the valve or servo was determined by observing leakage from the valve diaphragm or insufficient closing/opening of the valve, resulting in discharge at an incorrect outlet. The entire experiment was permanently recorded by camera and the number of actuations until failure was determined as a quotient of the elapsed time until failure and the time between two opening events. Experiments were performed with four distinct valves/servos (n = 4) each for a normally post-processed chip, an additionally heat-steam sterilized chip (121 °C, 30 min), and a heat-steam sterilized and incubated chip (ddH2O, 37 °C, 4 weeks). The script and the Ismatec pump commands – including the automatization script and variables that were used – can be found as .py files in the ESI.†2.4. Rinsing volume determinations
At first, one channel was filled with an Allura Red AC (Merck KGaA, Darmstadt, Germany) food dye stock solution in ddH2O with an absorbance of 20 a.u. at 494 nm after opening a single valve. Then, this solution was constantly removed by ddH2O using an Aladdin AL-1000 syringe pump (Waukesha-Pearce Industries, South Main, USA) at a flow rate of 500 μL min−1, and each drop with an average volume of 26.2 ± 0.3 μL was individually collected at the outlet in standard 0.2 mL PCR tubes. If necessary, the droplets were diluted to the proven linear absorbance range at ≤2 a.u. and absorbance of each solution was determined in a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, USA). The droplet containing 100% food dye solution was measured three times and used for normalization. The whole experiment was performed three times (n = 3), with three distinct channels and valves.2.5. Mixing accuracy
At first, all channels were filled with ddH2O. Then, after opening a single valve, one channel was filled with mixtures of ddH2O and Allura Red AC food dye stock solution by variation of the flow rates of two distinct Aladdin AL-1000 syringe pumps. For the experiments using mixtures ranging from 20% to 100% dye, a stock solution with absorbance = 2 a.u. at 494 nm was used. For experiments at higher dilutions (from 1.25–20% dye), a stock solution with absorbance of 20 a.u. at 494 nm was used instead, in order to keep the absorbance above the detection limit of the spectrophotometer. The mixtures were pumped at a total flow rate of 500 μL min−1 with varying rinsing volumes. The absorbance of the dye containing mixtures was determined in a NanoDrop 1000 spectrophotometer. The whole experiment was performed three times (n = 3), with three distinct channels and valves.2.6. Cell culture conditions
L929 cells (DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany, No. ACC2) were routinely cultivated in 75 cm2 cell culture flasks (Corning, CellBind Surface, Corning, NY, USA) in Dulbecco's Modified Eagle's Medium (DMEM; Sigma-Aldrich Chemie GmbH, Steinheim, Germany), supplemented with 10% fetal calf serum (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) and 1% penicillin/streptomycin (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) in a 5% CO2, humidified atmosphere at 37 °C (Heracell 240 incubator, Thermo Fisher Scientific Inc., Waltham, USA). For passaging, cells were harvested at 70–85% confluence using a Trypsin/EDTA solution (Biochrom GmbH, Berlin, Germany).2.7. IC50 determinations of camptothecin
The cytotoxic effect of the anti-cancer drug camptothecin (CPT) was used to induce a concentration-dependent growth rate and to thereby compare the resulting IC50 values of assays created either using the valve system, on one hand, or by manual pipetting, on the other. CPT acts as DNA topoisomerase I inhibitor by preventing DNA replication during S phase, and its toxic effect primarily stems from lethal collision of the DNA cleavage complex with replication forks.31Prior to an assay, cells were seeded in 96-well plates (Sarstedt AG and Co. KG, Nürnbrecht, Germany) at a density of 3500 cells per well in a 100 μL cell culture medium. To ensure a uniform distribution of these cells, the plate was maintained at room temperature for 20 min before transfer to the incubator. After 24 ± 2 h, assays were performed both via manual pipetting and also by using the microfluidic valve system on the same cell culture plate.
For the manually pipetted assay, a 1 mM stock solution of CPT (Merck KGaA, Darmstadt, Germany) in ≥99.7% pure DMSO (Carl Roth GmbH, Karlsruhe, Germany) was prepared. The stock solution was used for creating pre-dilutions in DMSO from which each 1 μL was transferred to the respective wells. Finally, 100 μL of cell culture medium was added to each well, resulting in a total volume of 200 μL and a 0.5% DMSO concentration (including the control without CPT).
In turn, the pre-sterilized and assembled microfluidic valve system was placed on top of the same cell culture plate, with the outlets pointing to their respective wells. Two 10 mL syringes with 0.5% DMSO, containing cell culture medium both with and without 10 μM CPT, were then connected to the chip. The automated microfluidic assay was started, and 100 μL of a respective mixture was added to each well. Rinsing volumes in between each step, as well as other parameters related to these experiments, are summarized in Table S3 and contained in the .py and .xlsx files of the ESI.† All CPT concentrations were performed with three technical replicates for each of these methods.
The plate was transferred to an IncuCyte S3 (Sartorius AG, Göttingen, Germany) live-cell-imaging system that was operated in a 5% CO2, humidified atmosphere at 37 °C, and thereafter it was monitored automatically by the system using phase contrast with a 20× objective creating four pictures per well every two hours. All microscopic images were analyzed automatically using the corresponding software IncuCyte 2021C (Sartorius AG, Göttingen, Germany). For determination of confluence, an image mask was created on three respective images using the parameters in Table S1.† The data of the mean confluence of each well was exported and normalized to the control, and then IC50 values for each time point were determined using logarithmic CPT concentrations in OriginPro 2019b (OriginLab Corporation, Northampton, USA) via non-linear fits using the intrinsic dose response function with weights and the top asymptote fixed to 100%. Finally, the mean IC50 values and standard deviations were calculated from three distinct experiments (n = 3).
3. Results and discussion
3.1. Design and operation principle of the 3D-printed, servomotor-controlled microfluidic valve system
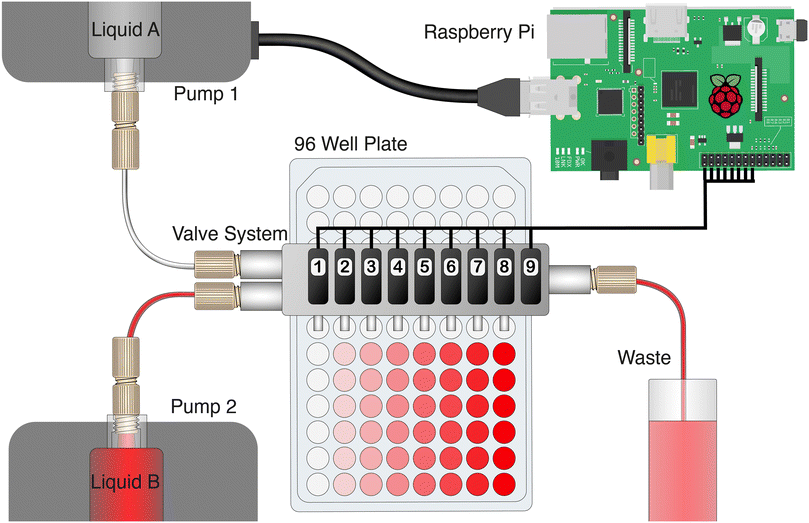
The design of the microfluidic valve system itself is presented in Fig. 1, while the assembly and operation are explained in the videos in the ESI.† The 3D-printed chip contains two inlets, a HC-shaped micromixer, nine plunger microvalves, and nine outlets including the waste outlet. A more detailed illustration of the chip design can be found in a technical drawing in Fig. S1 of the ESI.† The inlets allow waterproof connection to standard chromatography fittings and tubing, and are connected to the channel system with 500 × 500 μm channels.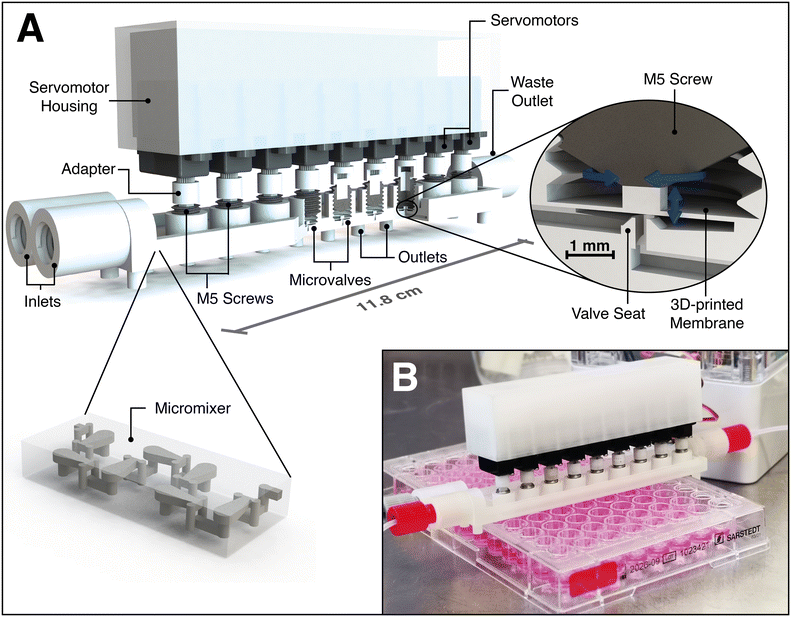 | ||
| Fig. 1 A) CAD illustration of the fully assembled microfluidic valve system. The 3D-printed microfluidic chip includes two inlets compatible to standard HPLC fittings, a micromixer, nine microvalves, eight outlets for mixture distribution, and a waste outlet. The complex 3D structures of the 3D-printed HC-shaped micromixer (published by Enders et al.3) ensure rapid mixing. The valves are operated by 6.2 mm wide servomotors, each of which is connected via a 3D-printed adapter to a M5 setscrew. The membrane is pressed into the valve seat by servo-driven setscrew rotation, resulting in the closure of the valve. All motors are integrated into a servomotor housing to enable quick attachment to the 3D-printed microfluidic chip. B) Picture of the 3D-printed and fully assembled microfluidic valve system during assay operation under the safety cabinet. | ||
The HC-shaped micromixer – based on the design by Enders et al.3 – ensures rapid mixing of the prevailing laminar flow. Therefore, the mixer creates a chaotic flow in combination with the principle of “split-and-recombine” by using complex 3D structures – that can only be produced at great expense with other fabrication techniques. The valve design is a slightly down-scaled redesign of a normally-open plunger valve reported in Au et al.,32 and it consists of both inlet and outlet channels, a valve seat, and a 100 μm thin 3D-printed membrane with a 4 mm Ø circular area. While most microfluidic systems depend on pneumatically controlled valve actuation (which includes additional control channels, control channel connectors, tubing and fittings, external valves, and at least one pressure source), the valve closure of this 3D-printed valve system is managed on chip solely by low-cost miniature servomotors. The motors are connected to M5 setscrews using a 3D-printed adapter, and membrane deflection is realized via rotation of the setscrews inside 3D-printed threads. Finally, all motors are attached to a 3D-printed housing which allows for quick plugging into and/or release from the (pre-sterilized) microfluidic chip (see Fig. S2† for technical drawings). The cost for a single servomotor is $3–10, depending on the supplier, whereas the cost of the 3D printing material is $10 per chip. While the presented microfluidic chip is designed to automate a specific proof-of-concept assay (as described in chapter 0), it can readily be rearranged and/or extended with additional inlets, mixers, valves, or even new functional units.
To facilitate the operation of complex assays, this system was automated as schematically illustrated in Fig. 2. The chip is designed to fit onto a standard 96-well plate, with eight of the outlets pointing into respective wells of one column. Modulation of the pump ratio of two syringe pumps at the chip inlets is used to create different mixtures, while control of the servomotors ensures the correct distribution to designated outlets. A waste outlet allows for rinsing of the main flow channel with a new mixture prior to the delivery to its desired outlet. While the valves are closed via setscrew rotation, membrane deflection during valve opening is achieved only via flow pressure. Since the membranes do not equally deflect during opening – resulting in distinct flow resistances and rates – the valve system is actually operated with a single valve opened at a time. Furthermore, all servos are switched off between two operations in order to prevent a permanent load on the servomotor and potentially induce its early failure. In terms of automatization, the servomotors and the pumps are connected to a compact Raspberry Pi single-board computer and are thereby controlled using a customized Python script. The Python script includes variables for various alternating parameters – including starting times, pump channels, pump rates, valve numbers, and incubation times. All of these parameters can be very quickly adapted for matching new assay protocols or modified chip designs.
3.2. Characterization of the microfluidic valve system
After inducing continuous and repetitive actuation of the valves over the course of 4–5 days as a stress test, with at least 5806 opening and closing events occurring per valve, no failure of either the valve membrane or the threads was observed in any of the tested chips. Accordingly, it is concluded that this system is well-suited for extended use in humid and warm conditions enabling cell culture assay automatization for several weeks. Furthermore, the stability of the polymer membrane might actually be increased at these conditions, since the deformability of the membrane is expected to improve at moderate to high temperatures. Nor was any distribution of the liquid to an unintended outlet ever observed during these tests – leading to the conclusion that sufficient force of the servos and stability of the 3D-printed threads has also been established. Surprisingly, the servos of the system were actually identified as the relative weak spot, since 8 out of the 12 servos malfunctioned after approximately 1500–4200 opening and closing events (as summarized in Table S2†). This weakness may be shored up by using higher quality servos or motors, instead of the lowest priced servos that were used in our experiments. Nonetheless, the stability of all parts is deemed to be generally sufficient for both short-term and long-term use, and we conclude that this valve system enables the automatization of simple chemical or biochemical assays or even – due to its compatibility with heat steam sterilization – full-time reagent control for mammalian cell cultures directly inside an incubator.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), 99.8% (1
100), 99.8% (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and 99.9% (1
500) and 99.9% (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
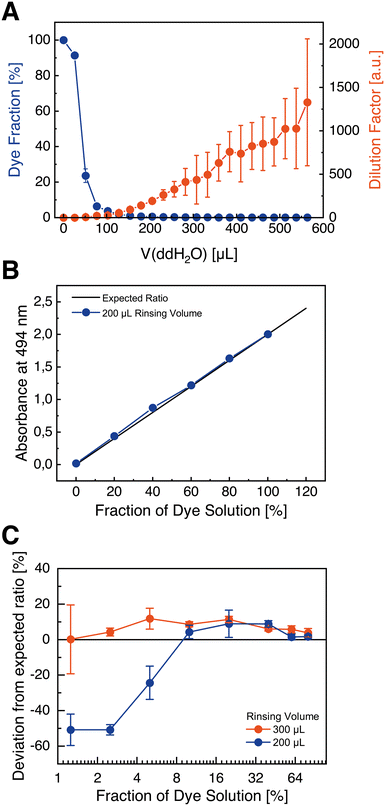
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) of the dye is achieved by around 150 μL, 300 μL, and 500 μL rinsing volumes, respectively. In conclusion, the exchange of a present mixture by another one using 300 μL rinsing volume is likely to be sufficient for most applications – but if necessary, the rinsing volume may be increased for assays using highly active substances or large concentration ranges.
1000) of the dye is achieved by around 150 μL, 300 μL, and 500 μL rinsing volumes, respectively. In conclusion, the exchange of a present mixture by another one using 300 μL rinsing volume is likely to be sufficient for most applications – but if necessary, the rinsing volume may be increased for assays using highly active substances or large concentration ranges.
As presented in Fig. 3B, mixtures of red dye solution and ddH2O show an expected linear dependency with mean deviations of maximum 10% from the ideal ratios measured at the chip outlet after pumped volumes of 200 μL. This accuracy is assumed to be sufficient for many present assays in research. However, at low ratios (such as 1.25%, 2.5%, or 5%), mixtures contain a significantly decreased amount of dye with about 50% deviation from the ideal ratios (Fig. 3C). Nonetheless, this deviation decreases for higher rinsing volumes (such as 300 μL), indicating that at low flow rates of the dye stock solution the dye is not adequately delivered to the system. This may be explained by the high-pressure differential that exists between the two pumps. As a consequence, for assays with a higher concentration range, an increased rinsing volume for low fractions of a substance is recommended.
3.3. Proof-of-concept cell culture cytotoxicity assay
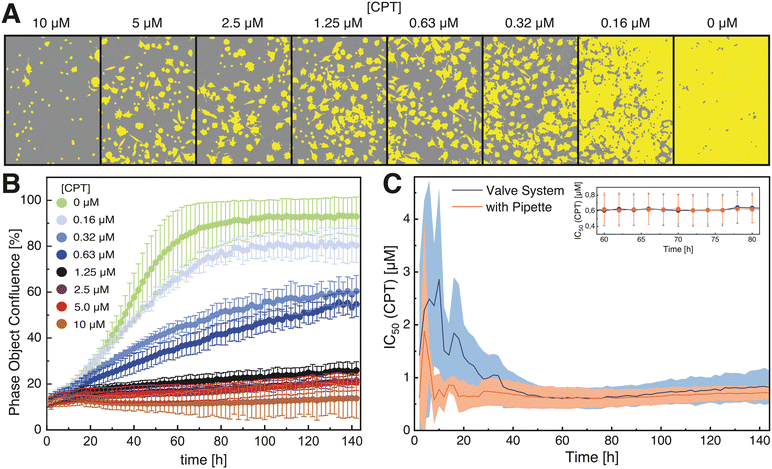
To evaluate the systems performance in a real-world application, IC50 determination of the cytotoxin camptothecin (CPT) with mouse fibroblasts (L929) was performed as a proof-of-concept (POC) and the results were compared to results obtained via traditional manually pipetting. L929 fibroblasts were used as they are a cell line recommended by standard organizations such as the International Standard Organization (ISO) or the United States Pharmacopeia (USP)33,34 for cytotoxicity and biocompatibility assays, while CPT is a well-known anti-cancer drug and serves as a widely used positive control for cytotoxicity and apoptosis.35The results of the proof-of-concept assay are presented in Fig. 4, as a function of the cell growth that was monitored and analyzed by live-cell-imaging. After three days of cultivation, a clear concentration-dependent increase in confluence was observed (Fig. 4A). As expected, the growth curve of cells in the control wells lacking CPT show a lag (∼0–20 h), an exponential (∼20–60 h) and a saturation phase (∼80–140 h) (Fig. 4B), while cells in wells containing high CPT concentrations show no growth at all. For comparison of the valve system and the standard pipetting technique, the IC50 was used as reference value. Since the imaging system allows the time-resolved quantification of the confluence, IC50 values were calculated for each time point (illustrated in Fig. 4C). The IC50 values – including the corresponding standard deviations – decreased over time and became constant after about 50 h. This result is neither unusual nor unexpected, since cells require a certain amount of time to grow and establish a concentration-dependent difference in confluence. At very long cultivation times, IC50 values increase due to the control reaching 100% confluence, and thus become unreliable. Importantly, at constant IC50 values between 60–80 h – where the toxic effect has been established and the control does not reach 100% confluence – the average IC50 values observed in this time period were highly similar, with 0.62 ± 0.19 μM and 0.62 ± 0.20 μM for the pipetted assay and for the assay automated by the microfluidic valve system, respectively. Thus, we conclude that the valve system does ensure a sufficient accuracy for cellular dose–response experiments, and that the deviations in mixing accuracy of the systems do not significantly affect the results. The key result was confirmation that the presented system affords researchers a real opportunity for cell culture assay automatization within the safety cabinet or even directly inside the incubator, using a portable point-of-use device that can be rapidly adapted for the intended use.
4. Conclusions
In contrast to pipetting robots (which represent the current “gold standard” in liquid handling), microfluidic valve systems facilitate far greater spatiotemporally-controlled multiplexing at a compact design. Due to the on-chip valve control by servomotors in the herein presented system, features of pneumatically controlled actuation – such as control channels, connection ports, tubing, external valves, and a vacuum/pressure supply – have been essentially rendered obsolete. The presented 3D-printed microfluidic valve system demonstrated a sufficient accuracy and long-term robustness to allow for its realistic real-world application in a majority of assays. Since a similar performance of the microfluidic valve system and manual pipetting in a proof-of-concept IC50 cell culture assay was observed, this proof-of-concept demonstrates that this system is appropriate and feasible for use in cell culture automatization. Importantly, the effort required to reconfigure this system is negligible compared with other manufacturing techniques, which renders direct integration of microfluidic cell culture chambers (or even organ-on-chip systems) realizable – thereby effectively enabling 24/7 operation even within unsterile environments. Finally, the integration of the microfluidic valve systems into live-cell-imaging systems illustrates a highly promising fusing of liquid handling and cell microscopy, and, in combination with machine-learning and online AI-based image processing, potentially opens the door for fully automated process optimization of adherent cell cultivation in the future.Author contributions
Conceptualization, J. B., S. W.; methodology, S. W., J. M., K. V. M. and C. K.; investigation, S. W. and J. M.; data curation, S. W. and J. M.; writing-original draft preparation, S. W.; writing-review and editing, S. W., K. M. and J. B.; visualization, S. W.; supervision, J. B.; project administration, J. B.; funding acquisition, J. B.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support of the German Research Foundation (DFG) via the Emmy Noether Programme (346772917). The open access publication of this article was supported by the University of Augsburg.References
- S. M. Bjork and H. N. Joensson, Curr. Opin. Biotechnol., 2019, 55, 95–102 CrossRef CAS PubMed.
- A. Enders, J.-A. Preuss and J. Bahnemann, Micromachines, 2021, 12, 1060 CrossRef PubMed , https://pubmed.ncbi.nlm.nih.gov/34577708/.
- A. Enders, I. G. Siller, K. Urmann, M. R. Hoffmann and J. Bahnemann, Small, 2019, 15, e1804326 Search PubMed.
- J.-A. Preuß, P. Reich, N. Bahner and J. Bahnemann, Adv. Biochem. Eng./Biotechnol., 2020, 174, 43–91 CrossRef PubMed , https://pubmed.ncbi.nlm.nih.gov/32313965/.
- F. Yu, W. Hunziker and D. Choudhury, Micromachines, 2019, 10(3), 165 CrossRef PubMed.
- Q. Wu, J. Liu, X. Wang, L. Feng, J. Wu, X. Zhu, W. Wen and X. Gong, J. Geophys. Res. Planets, 2020, 19, 9, DOI:10.1186/s12938-020-0752-0.
- S. Winkler, A. Grünberger and J. Bahnemann, Adv. Biochem. Eng./Biotechnol., 2021, 355–380 CrossRef PubMed , https://pubmed.ncbi.nlm.nih.gov/33495924/.
- H. Andersson and A. van den Berg, Lab Chip, 2006, 6, 467–470 RSC , https://pubmed.ncbi.nlm.nih.gov/33495924/.
- C. M. B. Ho, S. H. Ng, K. H. H. Li and Y.-J. Yoon, Lab Chip, 2015, 15, 3627–3637 RSC , https://pubs.rsc.org/en/content/articlehtml/2015/lc/c5lc00685f?casa_token=9FJcAVRcU9AAAAAA:lKIAGIOiWTjdLcxOYVEnZZLp-wmacOKxmkTqYWOlc1ILeGL3OkN2LNBCjwl_B5HVYG8KiYL1gorG.
- A. V. Nielsen, M. J. Beauchamp, G. P. Nordin and A. T. Woolley, Annu. Rev. Anal. Chem., 2020, 13, 45–65 CrossRef PubMed.
- K.-S. Lee, D.-Y. Yang, S. H. Park and R. H. Kim, Polym. Adv. Technol., 2006, 17, 72–82 CrossRef CAS.
- I. G. Siller, A. Enders, T. Steinwedel, N.-M. Epping, M. Kirsch, A. Lavrentieva, T. Scheper and J. Bahnemann, Materials, 2019, 12, 2125 CrossRef CAS PubMed , https://www.researchgate.net/publication/334180960_Real-Time_Live-Cell_Imaging_Technology_Enables_High-Throughput_Screening_to_Verify_in_Vitro_Biocompatibility_of_3D_Printed_Materials.
- S. Winkler, K. V. Meyer, C. Heuer, C. Kortmann, M. Dehne and J. Bahnemann, Eng. Life Sci., 2022, 1–10 Search PubMed , https://www.researchgate.net/publication/359636650_In_vitro_biocompatibility_evaluation_of_a_heat-resistant_3D_printing_material_for_use_in_customized_cell_culture_devices.
- I. G. Siller, A. Enders, P. Gellermann, S. Winkler, A. Lavrentieva, T. Scheper and J. Bahnemann, Biomed. Mater., 2020, 15, 55007 CrossRef CAS PubMed , https://www.researchgate.net/publication/341034633_Characterization_of_a_customized_3D-printed_cell_culture_system_using_clear_translucent_acrylate_that_enables_optical_online_monitoring.
- T. A. Thorsen, BioTechniques, 2004, 36, 197–199 CrossRef CAS PubMed.
- A. R. Vollertsen, D. de Boer, S. Dekker, B. A. M. Wesselink, R. Haverkate, H. S. Rho, R. J. Boom, M. Skolimowski, M. Blom, R. Passier, A. van den Berg, A. D. van der Meer and M. Odijk, Microsyst. Nanoeng., 2020, 6, 107 CrossRef CAS PubMed , https://www.nature.com/articles/s41378-020-00216-z.
- B. Schuster, M. Junkin, S. S. Kashaf, I. Romero-Calvo, K. Kirby, J. Matthews, C. R. Weber, A. Rzhetsky, K. P. White and S. Tay, Nat. Commun., 2020, 11, 5271 CrossRef CAS PubMed , https://www.nature.com/articles/s41467-020-19058-4.
- G. Pegoraro and T. Misteli, Trends Genet., 2017, 33, 604–615 CrossRef CAS PubMed.
- B. Larson, L. Hussain and J. Schroeder, in Immuno-Oncology. Cellular and Translational Approaches, ed. S.-L. Tan, Springer US; Imprint Humana, New York, NY, 1st edn, 2020, pp. 103–115 Search PubMed.
- R. Kodzius, F. Schulze, X. Gao and M. R. Schneider, Genes, 2017, 8(10), 266 CrossRef PubMed.
- U. Blache and M. Ehrbar, Adv. Wound Care, 2018, 7, 232–246 CrossRef PubMed.
- R. Xie, W. Zheng, L. Guan, Y. Ai and Q. Liang, Small, 2020, 16, e1902838 CrossRef PubMed.
- M. A. Unger, H. P. Chou, T. Thorsen, A. Scherer and S. R. Quake, Science, 2000, 288, 113–116 CrossRef CAS PubMed.
- K. Hosokawa and R. Maeda, J. Micromech. Microeng., 2000, 10, 415–420, DOI:10.1088/0960-1317/10/3/317/meta?casa_token=F4Ps4OEkkd0AAAAA:kRtujr-NOvIACvDGOZgxNaBEoTLWgGEo4fBNYf2P1Z13SDDGgCoHqzYV-dXPf_9AuE6_aQ9i.
- J. Y. Baek, J. Y. Park, J. I. Ju, T. S. Lee and S. H. Lee, J. Micromech. Microeng., 2005, 15, 1015–1020, DOI:10.1088/0960-1317/15/5/017/meta?casa_token=ejX2YH_4cf8AAAAA:FdJnb4BJJivc-5z1e4ZeShVDX8VY5EbM10EOsscnAeMHVAS5fmlqbryTkILQ-dfAbUDvfabF.
- A. K. Au, H. Lai, B. R. Utela and A. Folch, Micromachines, 2011, 2, 179–220 CrossRef , https://www.mdpi.com/2072-<?pdb_no 666X?>666X<?pdb END?>/2/2/179/htm#b39-micromachines-02-00179.
- J.-Y. Qian, C.-W. Hou, X.-J. Li and Z.-J. Jin, Micromachines, 2020, 11(2), 172 CrossRef PubMed.
- T. Thorsen, S. J. Maerkl and S. R. Quake, Science, 2002, 298, 580–584 CrossRef CAS PubMed.
- 3D Systems, Safety Data Sheet: VisiJet M2S-HT90. Available online: (accessed on 16 August 2021), https://infocenter.3dsystems.com/materials/sites/default/files/sds-files/professional/VisiJet_M2S/HT_90/24245-S12-02-A%2CSDS%20GHS%2CEnglish%2CVisiJet%20M2S-HT90.pdf Search PubMed.
- 3D Systems, 3D Systems Corporation Regulatory Information Sheet: VisiJet M2S-HT90. Available online: (accessed on 17 October 2022), https://printer-docs-public.s3.amazonaws.com/sites/default/files/sds-files/professional/VisiJet_M2S/HT_90/24245-S12-02-A%2CSDS%20GHS%2CEnglish%2CVisiJet%20M2S-HT90.pdf.
- L. F. Liu, S. D. Desai, T. K. Li, Y. Mao, M. Sun and S. P. Sim, Ann. N. Y. Acad. Sci., 2000, 922, 1–10 CrossRef CAS PubMed.
- A. K. Au, N. Bhattacharjee, L. F. Horowitz, T. C. Chang and A. Folch, Lab Chip, 2015, 15, 1934–1941 RSC , https://pubs.rsc.org/en/content/articlelanding/2015/lc/c5lc00126a.
- International Organization for Standardization, ISO 10993-12:2021 Biological Evaluation of Medical Devices, ISO copyright office, Venier, Switzerland, 2021 Search PubMed.
- The United States Pharmacopeial Convention, United States Pharmacopoeia 44 - NF 39 - <87> Biological Reactivity Tests, In Vitro, 2022 Search PubMed.
- C. D. Willey, E. S.-H. Yang and J. A. Bonner, in Clinical radiation oncology, ed. J. E. Tepper, J. A. Bogart and L. L. Gunderson, Elsevier, Philadelphia, PA, 2016, pp. 63–79.e4 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc00629d |
| This journal is © The Royal Society of Chemistry 2022 |