Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
MagPure chip: an immunomagnetic-based microfluidic device for high purification of circulating tumor cells from liquid biopsies†
Lucie
Descamps
 a,
Jessica
Garcia
b,
David
Barthelemy
b,
Emmanuelle
Laurenceau
c,
Léa
Payen
b,
Damien
Le Roy
*d and
Anne-Laure
Deman
*a
a,
Jessica
Garcia
b,
David
Barthelemy
b,
Emmanuelle
Laurenceau
c,
Léa
Payen
b,
Damien
Le Roy
*d and
Anne-Laure
Deman
*a
aInstitut des Nanotechnologies de Lyon, INL UMR5270, Université Claude Bernard Lyon 1, Villeurbanne, France. E-mail: anne-laure.deman-him@univ-lyon1.fr
bLaboratoire de Biochimie et Biologie Moléculaire, CICLY UR3738, Groupe Hospitalier Sud, Hospices Civils de Lyon, Pierre Bénite, France
cInstitut des Nanotechnologies de Lyon, INL UMR5270, Ecole Centrale de Lyon, Ecully, France
dInstitut Lumière Matière, ILM UMR5306, Université Claude Bernard Lyon 1, Villeurbanne, France
First published on 14th September 2022
Abstract
The isolation of circulating tumor cells (CTCs) directly from blood, as a liquid biopsy, could lead to a paradigm shift in cancer clinical care by providing an earlier diagnosis, a more accurate prognosis, and personalized treatment. Nevertheless, CTC-specific challenges, including their rarity and heterogeneity, have hampered the wider use of CTCs in clinical studies. Microfluidic-based isolation technologies have emerged as promising tools to circumvent these limitations but still fail to meet the constraints of high purity and short processing time required to ensure compatibility with clinical follow-up. In this study, we developed an immunomagnetic-based microfluidic device, the MagPure chip, to achieve the negative selection of CTCs through the depletion of white blood cells (WBCs) and provide highly purified samples for subsequent analysis. We demonstrate that the MagPure chip depletes all magnetically labeled WBCs (85% of WBCs were successfully labeled) and ensures a CTC recovery rate of 81%. In addition, we show its compatibility with conventional biological studies, including 2D and 3D cell culture, as well as phenotypic and genotypic analyses. Finally, we successfully implemented a two-step separation workflow for whole blood processing by combining a size-based pre-enrichment system (ClearCell FX1®) with the MagPure chip as a subsequent purification step. The total workflow led to high throughput (7.5 mL blood in less than 4 h) and high purity (947 WBCs per mL remaining, 99.99% depletion rate), thus enabling us to quantify CTC heterogeneity in size and tumor marker expression level. This tumor-marker-free liquid biopsy workflow could be used in a clinical context to assess phenotype aggressiveness and the prognosis rate.
Introduction
Early cancer diagnosis and personalized cancer medicine are target objectives to reduce cancer burden.1 In the early stage of the disease, the small size of the primary tumor and the lack of symptoms are obstacles for early screening. But when identified early, cancer is more likely to respond to therapy, therefore leading to a greater survival probability.2 In addition, it became clear over time that the “one drug fits all” treatment model was limited, and is being replaced by personalized medicine where treatment selection for each cancer patient is becoming individualized or customized. The study of circulating tumor cells (CTCs) could help tackle these challenges. CTCs are tumoral cells, which detached from the primary tumor and invaded the biological fluid circulation such blood and lymph. Then, these CTCs can extravasate and colonize distant sites, leading to secondary tumor(s). Being accessible in the bloodstream, the detection, enumeration, and characterization of CTCs can provide clinical information on the tumor stage and can be used in cancer diagnosis and disease prognosis.3–7 On the other hand, CTCs are reliable surrogate biomarkers for treatment efficacy monitoring, enabling a personalized therapeutic approach.8–11 The selection of CTCs directly from a blood assay, referred to as “liquid biopsy”, has therefore attracted large interest in recent years. Contrary to tissue biopsy, liquid biopsy enables a low-invasive and real-time monitoring of the disease evolution, tumor heterogeneity, and response to treatment.12 In recent years, based on focused genomic assay, ctDNA analysis was the major testing of the tumor burden with time. Living CTCs are more complete representative of the tumor burden, containing proteins, metabolites and RNA. Thus, the CTC study may lead to changes in the paradigm of cancer diagnosis and management by achieving earlier diagnosis and more personalized treatment.However, CTC isolation is a great technical challenge limiting their wider implementation in clinical studies for several reasons. First, CTCs are rare; there are approximatively 1 to 1000 CTCs among ca. 107 white blood cells (WBCs) and ca. 109 red blood cells (RBCs) in 1 mL of blood. Furthermore, they present a phenotypical heterogeneity, which results from the epithelial-to-mesenchymal transition (EMT) that they can undergo. It leads to a decrease of epithelial markers (EpCAM, CK) and the appearance of a mesenchymal phenotype,13 which makes the use of tumor makers more complex and limits the effectiveness of affinity-based separation. Besides, some CTCs have a size similarity with WBCs, reducing size-based sorting effectiveness.14 Finally, CTC isolation should result in viable cells to perform downstream analysis such as cell culture, phenotype and genotype investigations, as well as chemoresistance studies. The CellSearch™ system (Veridex, USA) was the first instrument available for CTC isolation and remains the only Food and Drug Administration (FDA)-approved assay for CTC enumeration used to predict cancer patient outcome. The method relies on the enrichment of CTCs expressing epithelial cell adhesion molecule (EpCAM) with immunomagnetic particles. Although this system is considered a “gold standard” for CTC detection, the detection sensitivity highly depends on epithelial markers, consequently cells lacking these markers would be missed.15 In addition to limited recovery, low purity has been reported since CTCs are enriched with a high background of contaminating WBCs,16 limiting downstream characterization.
Over the past decade, microfluidic devices have emerged as promising tools to address these limitations.17 Their micrometric dimensions and laminar flow nature enable precise cell manipulation and single-cell study, in a cost-effective and versatile manner. The handling of small quantities of volume also facilitates the analysis of such rare samples and speeds up processes. In particular, microfluidic-based isolation technologies are based on the differences between CTCs and other normal cells, either in physical properties (size, deformability, density or electric charges) or biological properties (surface marker expression). Studies are still widely conducted to obtain the best performances for CTC isolation, including high throughput, purity, recovery, and clinical relevance. Physical-based separation usually provides very high throughput but limited purity due to CTC size heterogeneity. On the contrary, biological-based separation is very specific due to the antigen–antibody bonding, but requires a labeling step. Thus, both approaches have pros and cons. Several microfluidic technologies, awaiting FDA clearance, have been commercialized for physical-based CTC isolation, such as Parsortix® (ANGLE plc, UK),18 ClearCell® FX1 (Biolidics, Singapore),19 and VTX-1 (Vortex Biosciences, USA).20 Despite the tremendous work achieved in CTC isolation devices, physicians and biologists do not routinely use them since there are still CTC-specific challenges to overcome. Thus, CTC size and tumor marker heterogeneities account for the difficulty to reach high purity and perform subsequent analysis. Indeed, a sample with less than 1000 WBCs per mL would be expected to allow extended downstream analyses, including highly sensitive mutation analysis technologies such as digital PCR and next-generation sequencing.21,22 Moreover, the initial volume of processed total blood as well as the separation time must be compatible with clinical follow-up. Indeed, the blood sample volume should be large enough to collect a sufficient number of CTCs and depict the tumor characteristics (e.g. heterogeneity) while the separation time should be limited as much as possible to not alter the cells for their subsequent analysis. Finally, the system must provide easy-handling for the end-users to ensure its integration in routine clinical studies. All these requirements have hindered the development of CTC isolation devices.
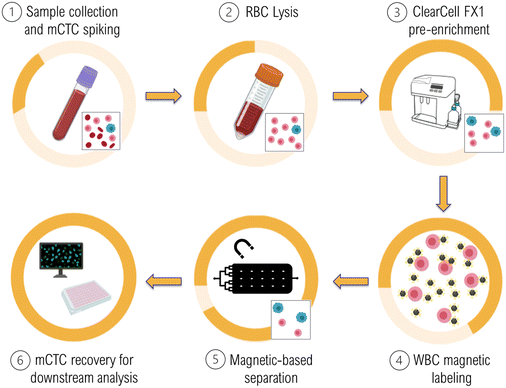
To meet these challenges, we have worked to combine two separation steps. A first step based on cell size to remove a large part of the WBCs followed by an immunomagnetic separation step providing a fine sorting of the CTCs and remaining WBCs. The first separation step is performed by the commercial ClearCell FX1® instrument which represents a plug-and-play system for biologists and has therefore been introduced in clinical studies.23 Nevertheless, this pre-enrichment step fails to meet the constraint of high purity and short processing time required to ensure the compatibility with certain downstream analyses (cell culture, phenotypic and genotypic studies, etc.). We therefore implemented a subsequent purification step through the development of a magnetic-based purification chip, MagPure chip, aiming at improving purity by depleting the remaining WBCs, and therefore facilitating downstream analysis. The immunomagnetic approach was selected for this second step because of its appealing features in the context of CTC isolation, such as selectivity, specificity, and CTC collection. It relies on magnetophoresis, the migration of magnetic particles or magnetically labelled cells when subjected to a non-uniform magnetic field, which offers contactless and robust cell manipulation.24 Since the magnetic force is a function of the generated magnetic field gradient, it is crucial to produce high magnetic field gradients, which can be achieved by reducing the size of the magnetic field source.25 Challenges remain regarding the complexity of microfabrication of such microscale magnetic sources, and their integration with polymer-based microfluidic devices. Recently, the magnetic composite polymer strategy has emerged as a real breakthrough for compatible and cost-effective integration of magnetic materials into polymer-based microfluidic devices.26 The composite strategy consists in doping the polymer matrix with magnetic particles or filaments, conferring magnetic properties to composite polymers. This powder-based approach therefore represents a quite straightforward and cost-effective alternative to film-based approaches.27–29
Here, we present the characteristics of our two-step sorting protocol. First, we describe the MagPure chip, which is based on the composite approach, including the performances of the negative selection using immunomagnetic nanoparticles targeting WBCs to deplete them, and demonstrate the compatibility of the chip with routine biological studies (cell culture, phenotypic and genotypic analyses) after CTC recovery. Then, we present the performances of the two-step protocol to process 7.5 mL of total blood in only three to four hours. The first step relies on a size-based separation technology (ClearCell FX1®), and is followed by the immunomagnetic MagPure chip. We investigated long-term cell viability and targeted cancer stem-like cell marker ALDH1.
Experimental section
Device fabrication and integration
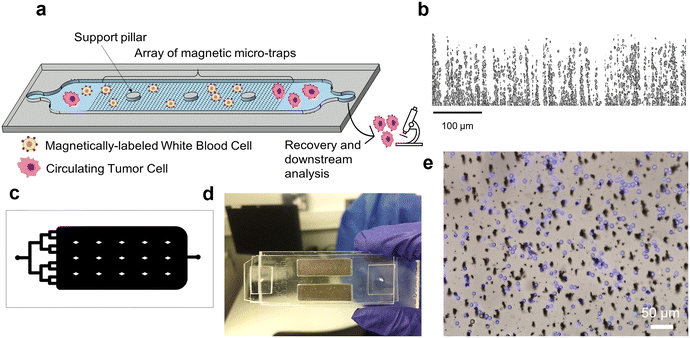
A schematic of the MagPure chip principle is reported in Fig. 1a. Magnetic micro-traps were obtained by the composite approach which consists in mixing a powder composed of NdFeB particles (MQFP-B, 0.5–7 μm size, Magnequench International, Inc., Singapore) with PDMS polymer (Sylgard Silicone Elastomer, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 base and curing agent mixing ratio) at a concentration of 4 wt% NdFeB. As previously reported,30,31 the fabrication steps can be divided as follows: the mixture containing NdFeB micro-particles and uncured PDMS was poured into a 100 μm-thick Kapton mold stuck to a silanized glass slide. The composite was then cured at 70 °C for 2 h in a magnetic field of 300 mT supplied by a bulk NdFeB magnet (60 × 30 × 15 mm3, magnetization along the shortest dimension) to allow PDMS cross-linking and NdFeB particle self-assembly. After Kapton mold removal, the thickness of the composite membrane was increased to 1 mm by pouring liquid PDMS and curing the ensemble at 70 °C for 2 h. Finally, the NdFeB@PDMS membrane was peeled off the glass substrate and NdFeB microstructures were magnetized using a homemade magnetizing system (two magnets of dimensions 50.8 × 50.8 × 25.4 mm3, spaced 2.5 mm from each other) that produced a field of 1.2 T.
1 base and curing agent mixing ratio) at a concentration of 4 wt% NdFeB. As previously reported,30,31 the fabrication steps can be divided as follows: the mixture containing NdFeB micro-particles and uncured PDMS was poured into a 100 μm-thick Kapton mold stuck to a silanized glass slide. The composite was then cured at 70 °C for 2 h in a magnetic field of 300 mT supplied by a bulk NdFeB magnet (60 × 30 × 15 mm3, magnetization along the shortest dimension) to allow PDMS cross-linking and NdFeB particle self-assembly. After Kapton mold removal, the thickness of the composite membrane was increased to 1 mm by pouring liquid PDMS and curing the ensemble at 70 °C for 2 h. Finally, the NdFeB@PDMS membrane was peeled off the glass substrate and NdFeB microstructures were magnetized using a homemade magnetizing system (two magnets of dimensions 50.8 × 50.8 × 25.4 mm3, spaced 2.5 mm from each other) that produced a field of 1.2 T.
The microstructure of the fabricated micro-traps was characterized by X-ray tomography in the volume of the composite (Fig. 1b). The inner structure of a 1 mm3 NdFeB@PDMS membrane was observed using the EasyTomNano μCT tomograph (RX Solutions), with a resolution of 0.3 μm. The detailed operation procedure can be found in ref. 30 and 31. Final images of 1700 × 1700 × 400 voxel, i.e. 510 × 510 × 120 μm3, were obtained and processed with ImageJ to characterize NdFeB particles' spatial organization in the volume of the composite membrane.
The magnetic micro-trap array was finally integrated into a microfluidic capture chamber (see Fig. 1c). The chamber mold (40 × 20 × 0.1 mm3) was obtained by UV-lithography (UV-KUB 2, Kloe) through exposition of two layers of a 50 μm dry photoresist (Eternal Materials Co., Etertec®, Kaohsiung City, Taiwan) deposited on a glass slide. The channel mold was irreversibly bonded to the composite membrane using O2 plasma bonding (Plasma Cleaner PDC-002-HPCE, Harrick Plasma). The MagPure chip was mounted on top of two permanent milli-magnets (25 × 8 × 2 mm3, remanent magnetization BR ∼1.4 T, Supermagnete) to attract target cells flowing in the upper part of the 100 μm-high channel as described in ref. 31 and 32; and therefore overcome the limited interaction distance of micro-magnets.33 The milli-magnets (housed in a homemade plastic holder) were spaced by 4 mm and positioned at 2 cm from the chamber input, as shown in Fig. 1d.
Device operation
Samples were injected using a pressure controller (FLOW EZ™, Fluigent, France). The MagPure Chip was connected with 508 μm inner diameter PFTE tubing (1/16′′ OD). The chip input was connected to the FLOW EZ™ pressure controller while the output was secured with tape into a clean 1.5 mL Eppendorf tube for sample collection. Since the injection is controlled in pressure, the corresponding pressure (in mbar) for the desired flow rate (e.g. in mL h−1) was first estimated by calculation, then experimentally assessed by measuring the collected liquid volume after a certain injection time. Before sample injection, the microfluidic device was sterilized by flushing 70% ethanol for 10 min, followed by 1% Pluronic F-108 (Sigma-Aldrich, USA) flushing for 20 min to remove ethanol and coat the channel walls to prevent non-specific cell adhesion. To monitor sample injection and cell sorting, the microfluidic chip was placed under a Zeiss fluorescence microscope (Zeiss Imager D1) equipped with ZEN blue imaging software.Cell culture and preparation
A549 and MCF-7 cell lines were obtained from American Type Culture Collection (ATCC). A549 cells (CCL-185™), which originate from a lung adenocarcinoma, were cultivated in F12-K medium (ATCC), while MCF-7 cells (HTB-22™), originating from a breast cancer, were cultivated in Dulbecco's modified Eagle medium (DMEM, Life Technologies). Both media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. Cells were cultured at 37 °C under a humidified atmosphere of 5% CO2. Cells were grown in T25 flasks to pre-confluence and detached from flasks for experiments using 1 mL of 0.25% trypsin-ethylene diamine tetra acetic acid (EDTA) solution at 37 °C. After cell resuspension in medium, 10 μL was taken to perform cell counting in a KOVA® slide. A549 (lung cancer) and MCF-7 (breast cancer) cells lines were then spiked in a blood sample to mimic CTCs, they can therefore be referred to as mCTCs. To track mCTC sorting, cancer cells were stained with CellTracker™ Green (Life Technologies) in 0.2% Pluronic (1 μL per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) and incubated for 45 min in the CO2 incubator.
000 cells) and incubated for 45 min in the CO2 incubator.
Blood sample preparation
All experiments involving healthy blood samples were performed in compliance with the relevant laws and institutional guidelines of France and approved by the French national blood collection institution, named “Etablissement Français du Sang” (EFS). All healthy volunteers provided informed consent for blood samples to be collected. Blood samples were collected from healthy volunteers into EDTA tubes (BD Vacutainer). Red blood cells were first removed using a lysis buffer (Biolidics limited®, CBB-F016003), following the manufacturer's protocol. Lysis buffer was added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 v/v ratio (blood/lysis buffer) and incubated for 10 min at room temperature (RT) and then centrifuged at 500 × g for 10 min. After centrifugation, the supernatant, containing lysed red blood cells, is discarded and the pellet, containing WBCs, is resuspended in 1 mL of Dulbecco's phosphate buffered saline (DPBS 1X, Lonza Bioscience). Finally, WBCs were enumerated using a Türk's solution (which destroys RBCs and platelets) in a 1
4 v/v ratio (blood/lysis buffer) and incubated for 10 min at room temperature (RT) and then centrifuged at 500 × g for 10 min. After centrifugation, the supernatant, containing lysed red blood cells, is discarded and the pellet, containing WBCs, is resuspended in 1 mL of Dulbecco's phosphate buffered saline (DPBS 1X, Lonza Bioscience). Finally, WBCs were enumerated using a Türk's solution (which destroys RBCs and platelets) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v ratio (WBC/Türk) to stain the nuclei of WBCs and facilitate their counting.
10 v/v ratio (WBC/Türk) to stain the nuclei of WBCs and facilitate their counting.
The capture experiments were performed with mimicking patient blood samples. Cancer cells were spiked in the WBC-containing sample, in concentration mimicking the ClearCell size-based separation system output. More information on the ClearCell FX1 instrument procedure is reported in S1, ESI.† Added cancer cells within the injection sample should be in a sufficient number to study recovery efficiency and perform compatibility studies of the magnetic chip with downstream analyses, while taking into account the rarity of CTCs. Thus, a final mimicking sample composed of 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 WBCs and 20
000 WBCs and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mCTCs was prepared.
000 mCTCs was prepared.
White blood cell magnetic labeling
WBCs were magnetically labeled with antibody-conjugated superparamagnetic nanoparticles (MasterBeads Carboxylic Acid 0215, Ademtech SA, Pessac, France). The nanoparticles (NP) were 500 nm in diameter and composed of a magnetic core (approximately 70% iron oxide) encapsulated by a hydrophilic polymer shell with carboxyl groups on its surface. Covalent attachment of human-anti-CD45-DL650 and human-anti-CD15-AlexaFluo647 fluorescent antibodies (purchased from R&D Systems, Minneapolis, MO, USA) to the nanoparticles' surface was performed following the manufacturer's protocol. The magnetic labeling conditions, from NP concentration, to incubation temperature, duration and agitation, were firstly optimized. The description of the optimization procedure is detailed in S2, ESI† (see Tables S1 and S2). Finally, an average of 85% of WBCs were successfully labeled.Recovery and capture efficiency calculation
To facilitate the visualization and discrimination of WBCs and mCTCs, the former were stained with Hoechst 33342 (Ready Flow Reagent™, ThermoFisher Scientific, USA), while the latter were marked with CellTracker™ Green. After processing within the MagPure chip, cells were collected from the chip output and centrifuged for 5 min at 300g. The aim is to concentrate the cells in a smaller volume to get a more precise cell counting. After centrifugation, half of the supernatant is removed, and the cell pellet is resuspended in the remaining volume until getting a homogenized suspension. Cells were then counted with a KOVA® slide under a fluorescence microscope (Zeiss Imager D1) and cells with a GFP signal were identified as cancer cells, while other cells only expressing a DAPI signal were identified as WBCs. Two to three counts were performed to get the most precise estimation of WBC capture and mCTC recovery efficiencies. The capture efficiency was calculated by ((NWBC,input − NWBC,output)/NWBC,input) × 100%. The recovery efficiency was calculated by (NmCTC,output/NmCTC,input) × 100%. The reproducibility of capture and recovery was studied by performing each experiment at a fixed flow rate at least five times.Cell viability and integrity characterization
First, the viability of collected A549 cancer cells was investigated after the magnetophoretic separation using a Live/Dead assay. Cancer cells were stained with 2 μM calcein-AM and 4 μM EthD-1 (LIVE/DEAD® Viability/Cytotoxicity Kit, Invitrogen™, L3224) for 30 min in the incubator. Death control cells were exposed to 10% Triton-PBS solution for 10 min. The discrimination of live and dead populations was assessed thanks to fluorescent signal measurement within live and death control A549 wells. Finally, the cell fluorescent signal was measured: collected cancer cells with a calcein-AM+/EthD-1− staining pattern were counted as live cells, whereas cells with calcein-AM−/EthD-1+ staining patterns were counted as dead cells.The integrity of recovered A549 cancer cells was determined using an immunofluorescence assay. Cells were stained with DAPI for nucleus staining, anti-CD45/anti-CD15/anti-CD41 antibodies for WBC targeting, and Phalloidin for F-actin cytoskeleton staining. Recovered A549 cancer cells could be differentiated from WBCs based on phalloidin-positive (phalloidin+), CD45/CD15/CD41-negative (CD45-/CD15-/CD41-) and DAPI positive (DAPI+).
Downstream 2D and 3D cell culture
After purification within the magnetic chip, isolated cells were cultured in (i) a Falcon® 96-well flat bottom microplate to study the ability of recovered mCTCs to re-adhere and proliferate; and in (ii) a Corning® 96-well round bottom microplate to study their ability to form spheroids. Corning® round bottom microplates are ultra-low adherent (ULA) plates which favor spherical three-dimensional aggregation of mCTCs composed of proliferating cancer cells and therefore better mimic cellular organization in human tumors. For both culture conditions, isolated cells were first centrifuged (300g, 5 min) and resuspended in culture medium (filtered for 3D culture) at a concentration of 50 A549/μL. This value was chosen so as to be compatible with the 96-well plate format requiring an initial seeding density of ∼104 cells and a medium volume of approximatively 100–200 μL. About 5000 of recovered mCTCs were spiked in a well with 150 μL of medium and incubated at 37 °C with 5% CO2. The medium was renewed every two days. In particular, the medium was completely removed for 2D cell culture as mCTCs are adhering to the well bottom so there is no risk of cell detachment (discarding the remaining WBCs at the same time) while for 3D cell culture, half of it was carefully removed and replaced with a fresh medium since mCTCs are in suspension within the well. Control (unprocessed) A549 cells were also seeded under the same conditions. Three replicates were performed for reproducibility. Cell growth was then monitored over days with an automated microscope equipped with a microplate reader (LionHeart™, BioTek).Downstream phenotypic characterization (IF)
After magnetic purification, the output sample containing enriched mCTCs (A549 cells) and remaining WBCs was concentrated through centrifugation (300g for 5 min) and mounted on a poly-L-lysine coated glass slide. The spotted cells were fixed with 4% paraformaldehyde for 10 min at RT and permeabilized with 0.1% Triton in PBS for 15 min at RT. After washing, cells were then blocked with an in-house saturation solution mix (5% fetal bovine serum, 1% bovine serum albumin, and 5% Fc receptor blocking reagent from Miltenyi Biotec in PBS) for 30 min at RT in the dark. Finally, cells were immunostained overnight with primary antibodies: (i) anti-CD45 antibody (rat anti-human, MA5-17687, ThermoFisher), AlexaFluor647-conjugated anti-CD15 antibody (mouse anti-human, 562369, BD Bioscience), and AlexaFluor647-conjugated anti-CD41 antibody (mouse anti-human, 303726, BioLegend) for white blood cell staining; (ii) anti-ALDH1 antibody (rabbit anti-human, 702728, ThermoFisher) for mCTC (A549) staining. The day after, cells were incubated with DAPI (4′,6-diamidino-2-phenylindole, 62248, ThermoFisher) and secondary antibodies, including AlexaFluor647-conjugated anti-rat antibody (A-21247, ThermoFisher) (targeting rat anti-human CD45) and AlexaFluor488-conjugated anti-rabbit antibody (11800074, ThermoFisher) (targeting rabbit anti-human ALDH1). The slide was scanned using the Lionheart™ fluorescence microscope equipped with 4× and 20× objectives. In particular, three filters were used to detect the fluorescent signal: DAPI for nucleus staining, CY5 for CD45, CD15, and CD41, and GFP for ALDH1.Downstream genotypic characterization (FISH)
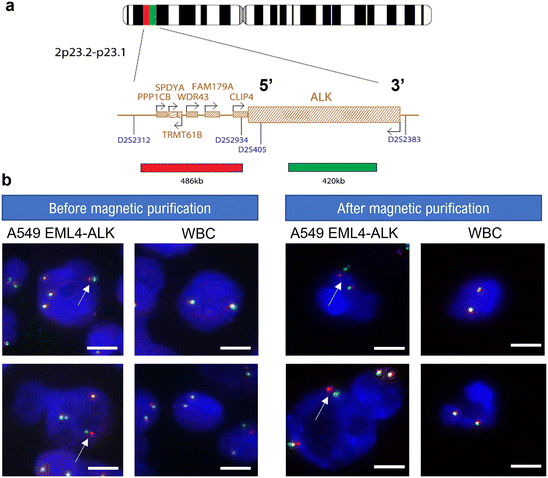
To study the EML4-ALK fusion, the EML4-ALK fusion-A549 cell line was purchased from ATCC (CCL-185IG™). ATCC CCL-185IG can be a useful model to study the tyrosine kinase signaling pathway, and to screen ALK inhibitors. There are multiple EML4-ALK fusion variants, and the ATCC CCL-185IG cell line contains the most prevalent one, the variant 1 (E13; A20), in which EML4 intron 13 is fused with ALK intron 20. FISH experiments were performed using the Aquarius® kit (CytoCell, OGT) which contains a DAPI counterstain and the ALK Breakapart probe, consisting of a green 420 kb probe, which spans the majority of the ALK gene and a red 486 kb probe, which is telomeric to the ALK gene. Fluorescence images were taken with the PANNORAMIC Scan II (3DHISTECH Ltd), equipped with FITC, Texas Red, and DAPI filters.Two-step workflow for mCTC isolation from whole blood samples
Briefly, 7.5 mL blood samples were collected from healthy volunteers and A549 cells (mCTCs) were spiked in the samples at a concentration of ∼2500 mCTCs mL−1. Then, RBCs were lysed and removed from the samples. Cells were then processed within the commercial size-based separation system, ClearCell FX1 using program 3 for 30 min or program 1 for 60 minutes (the last one increasing the purity level). After this first pre-enrichment step, collected cells are incubated with magnetic nanoparticles coated with anti-CD45 and anti-CD15 antibodies to magnetically label the remaining WBCs. Next, samples containing magnetically labeled WBCs and A549 cells were processed through the MagPure chip to perform a purification step by depleting WBCs. Finally, A549 cells were recovered at the chip output for downstream characterization. In particular, here we investigated long-term cell viability and conducted subsequent phenotypic studies targeting the cancer stem-like cell marker, ALDH1. The detailed operation procedure of the two-step workflow is reported in S3, ESI.†Results and discussion
A. MagPure chip performances
Next, the sorting microfluidic chamber has been sized to fit with the number of WBCs to trap, while preserving the compact format of a standard glass slide (25 × 75 mm2). Since the median number of WBCs remaining at the output of the size-based separation ClearCell FX1 system was estimated at 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 WBCs (internal data), and the maximum number could reach 1
000 WBCs (internal data), and the maximum number could reach 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for certain patients, the trapping density should therefore be at least 106 micro-traps. Finally, the chamber dimensions were fixed at 40 × 20 mm2, providing a trapping density as high as 1.2 × 106 traps. Besides the high trapping capacity and compact format, the microfluidic chamber features should provide an optimized liquid filling and limit air bubble formation, which was achieved by the parallel input microchannel design. Finally, the integrated diamond-shaped support pillars prevented the chamber roof from collapsing due to the large ratio (>10) between the chamber width and height.
000 for certain patients, the trapping density should therefore be at least 106 micro-traps. Finally, the chamber dimensions were fixed at 40 × 20 mm2, providing a trapping density as high as 1.2 × 106 traps. Besides the high trapping capacity and compact format, the microfluidic chamber features should provide an optimized liquid filling and limit air bubble formation, which was achieved by the parallel input microchannel design. Finally, the integrated diamond-shaped support pillars prevented the chamber roof from collapsing due to the large ratio (>10) between the chamber width and height.
This simple and low-cost composite approach allows permanent magnetic microstructures to be obtained with new aspect ratios and with performances comparable to the best ones reported in the literature (Fig. 2), which were obtained with more conventional manufacturing methods.28,34,36–44 A description of reported studies can be found in S4, ESI† (see Table S3).
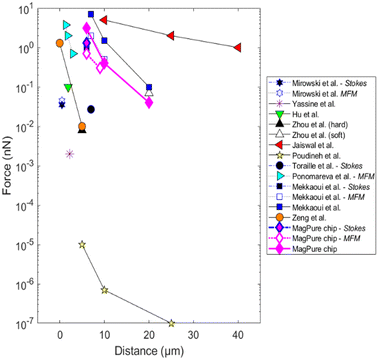 | ||
| Fig. 2 Comparison of the magnetic performances of the fabricated composite micro-magnets with other microscale magnetic sources from the literature. The graph represents the forces exerted on the center of the magnetic target as a function of the distance to the micro-source. When not specified, the magnetic force was determined by numerical studies. A description of the magnetic micro-sources and magnetic force measurement methods can be found in Table S3.† | ||
Here, the fabricated micro-magnets that line the bottom of the microchannel can produce magnetic forces of a few nN on model microbeads (average diameter 12 μm, 1 vol% Fe3O4 in polystyrene). The generated forces were independently determined by simulation as well as experimentally by magnetic force microscopy and Stokes' drag fore measurements.31 Finally, the capture of labeled WBCs on micro-traps is reported in Fig. 1e.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mCTCs (2500 mCTCs per mL) to take into account the low number of CTCs in blood samples (high bound of the clinical range) while providing a sufficient number of isolated cells to study the compatibility of the MagPure chip with routine downstream analyses. The mCTCs were spiked into samples containing 300
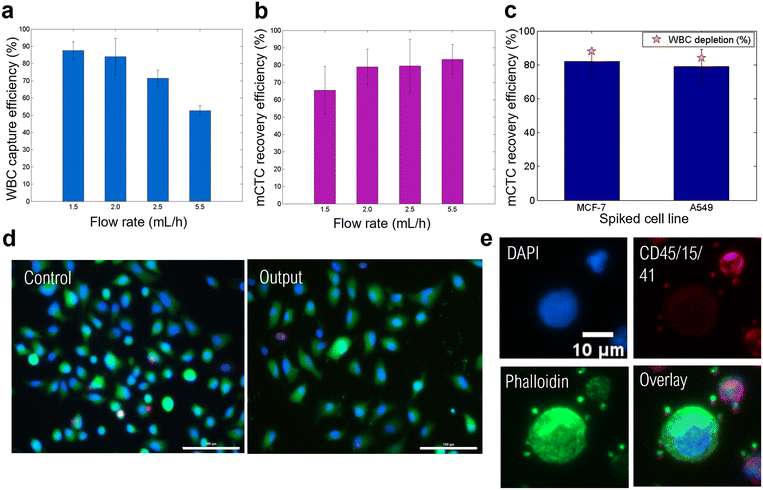
000 mCTCs (2500 mCTCs per mL) to take into account the low number of CTCs in blood samples (high bound of the clinical range) while providing a sufficient number of isolated cells to study the compatibility of the MagPure chip with routine downstream analyses. The mCTCs were spiked into samples containing 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 WBCs (median ClearCell FX1 output) which were magnetically-labeled WBCs. The WBC capture efficiency and the A549 cell recovery efficiency at different flow rates are shown in Fig. 3a and b, respectively. WBC capture efficiencies reached 88, 84, 72, and 53% at 1.5, 2.0, 2.5, and 5.5 mL h−1, respectively. The WBC labeling protocol that we implemented allows us to statistically label about 85% of WBCs. We therefore achieved a capture rate of magnetically labeled WBCs close to 100% at flow rates of 1.5 and 2 mL h−1. A549 cell recovery efficiencies reached 66, 79, 80, and 83% at 1.5, 2.0, 2.5, and 5.5 mL h−1, respectively. It can be observed that increasing the flow rate decreases the capture efficiency but improves the mCTC recovery. Indeed, the capture efficiency decrease can be explained by the higher drag force competing with the magnetic force. Concerning the CTCs, at flow rates of 1.5 and 2 mL h−1, approximatively 20% of CTCs were lost in the system. We observed that few CTCs (<1%) got stuck in the trapping chamber with WBCs. The remaining were probably not injected in the system because of their quick sedimentation in the injection tube. This is consistent with the fact that their recovery is enhanced by increasing the flow rate, as a faster injection time limits cell sedimentation in the input reservoir prior to injection. An injection time of approximatively 15–20 min for flow rates comprised between 2 and 2.5 mL h−1 was achieved. In order to avoid cell sedimentation, the injection system could be improved by providing regular agitation to the input reservoir containing the cell sample.
000 WBCs (median ClearCell FX1 output) which were magnetically-labeled WBCs. The WBC capture efficiency and the A549 cell recovery efficiency at different flow rates are shown in Fig. 3a and b, respectively. WBC capture efficiencies reached 88, 84, 72, and 53% at 1.5, 2.0, 2.5, and 5.5 mL h−1, respectively. The WBC labeling protocol that we implemented allows us to statistically label about 85% of WBCs. We therefore achieved a capture rate of magnetically labeled WBCs close to 100% at flow rates of 1.5 and 2 mL h−1. A549 cell recovery efficiencies reached 66, 79, 80, and 83% at 1.5, 2.0, 2.5, and 5.5 mL h−1, respectively. It can be observed that increasing the flow rate decreases the capture efficiency but improves the mCTC recovery. Indeed, the capture efficiency decrease can be explained by the higher drag force competing with the magnetic force. Concerning the CTCs, at flow rates of 1.5 and 2 mL h−1, approximatively 20% of CTCs were lost in the system. We observed that few CTCs (<1%) got stuck in the trapping chamber with WBCs. The remaining were probably not injected in the system because of their quick sedimentation in the injection tube. This is consistent with the fact that their recovery is enhanced by increasing the flow rate, as a faster injection time limits cell sedimentation in the input reservoir prior to injection. An injection time of approximatively 15–20 min for flow rates comprised between 2 and 2.5 mL h−1 was achieved. In order to avoid cell sedimentation, the injection system could be improved by providing regular agitation to the input reservoir containing the cell sample.
The size-independent separation performed with the MagPure chip was also demonstrated by studying the recovery efficiency of MCF-7 cancer cells which present a slightly larger average cell diameter than A549 cells (average diameter 19 μm against 16 μm).45 Experiments were performed at 2 mL h−1. Comparison with A549 results is shown in Fig. 3c. Similar recovery efficiencies were achieved by the magnetic chip, reaching 82 ± 8% and 79 ± 10% for MCF-7 and A549 cells, respectively. Besides, WBC capture efficiencies were also consistent: 88 ± 9% and 84 ± 11% for spiked MCF-7 and A549 cells, respectively. Thus, the magnetic chip enables size-independent sorting, in combination with tumor marker-independent separation, therefore showing the versatility of the MagPure chip. Finally, we set the optimal flow rate at 2 mL h−1, leading to a WBC depletion efficiency of ∼86 ± 10%. This separation step can be conducted in only 20 min.
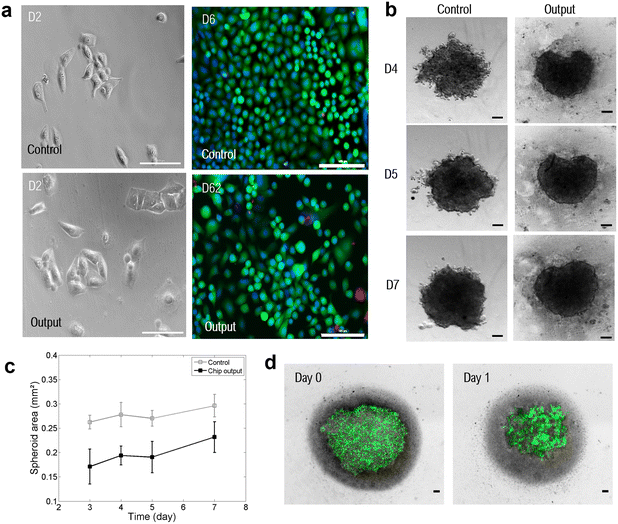
We also checked that the magnetic separation within the chip did not harm the cells. To do so, we performed viability and integrity studies. First, the viability of collected A549 cancer cells was investigated after the magnetophoretic separation using a Live/Dead assay. After processing the cell sample through the MagPure chip at a flow rate of 2 mL h−1, cancer cells were collected and cultured in a 96 well-plate in culture medium for 48 h until reaching pre-confluence. Recovered cells were compared with control A549 cells; the latter were kept in the incubator the whole time and seeded in the same concentration but without any WBCs. Finally, cancer cells with a calcein-AM+/EthD-1− staining pattern were counted as alive cells, whereas cells with calcein-AM−/EthD-1+ staining patterns were counted as dead cells. Representative fluorescence images of collected cancer cells and control A549 cells are shown in Fig. 3d. The magnetic chip enabled preserving an 81% cell viability (∼1000 analyzed fluorescence cancer cells) after 48 h of cell culture. The viability of control A549 cells reached 91%, this difference might be due to their ideal incubation conditions, while recovered A549 cells were processed at room temperature and subjected to prolonged interactions with WBCs. Nevertheless, processed A549 cells were collected with a very good viability.
Cancer cell integrity was determined using an immunofluorescence assay. Cell samples were processed through the MagPure chip at a flow rate of 2 mL h−1 and collected cells, as well as control cells, were stained with DAPI for nucleus staining, anti-CD45/anti-CD15/anti-CD41 antibodies for WBC targeting, and phalloidin for F-actin cytoskeleton staining (Fig. 3e). Indeed, cell integrity is related to F-actin filament which is a major component of the cytoskeleton and is involved in fundamental cellular processes, such as cell division, morphogenesis, and migration.46 Recovered A549 cancer cells could be differentiated from WBCs based on phalloidin-positive (phalloidin+), CD45/CD15/CD41-negative (CD45-/CD15-/CD41-) and DAPI positive (DAPI+). Interestingly, WBCs showed a lower expression of phalloidin which results from dynamic polymerization and depolymerization of F-actin.47 These processes account for WBC actin cytoskeleton plasticity, and therefore WBC motility,48 which is crucial to their role in defensive mechanisms of the body. Several morphological characteristics of control and recovered cancer cells were established from DAPI and phalloidin fluorescence signal measurements, these being cell nucleus and cytoplasm sizes, nuclear-cytoplasmic ratio (N:C ratio), and circularity (see S5, Fig. S1, ESI†). All in all, this immunofluorescence assay allowed us to assess the preservation of isolated cancer cell integrity after processing through the MagPure chip.
In addition to 2D cell culturing, recovered cancer cells were cultured in 3D using ultra-low attachment (ULA) 96-well plates with a round bottom to investigate cancer cell spheroid formation. Spheroids, which refer to three-dimensional aggregates of cells, have emerged as better models to mimic the 3D conformation of the tumor structure.49 Indeed, spheroids have a characteristic layer-like structure consisting of a necrotic core, an inner layer of quiescent cells, and a layer of proliferating cells. As a result, a spheroid has gradients in nutrients, secretions, and oxygen along the spheroid radius. In this regard, spheroid cultures more accurately recapitulate in vivo physiological conditions than standard 2D cultures. Previous studies reported the resistance of spheroids to chemotherapy50–53 and photodynamic therapy,54,55 as well as the differential expression of various genes (associated with cell survival, proliferation, differentiation, and resistance to therapy) in cells grown as spheroids as compared to 2D monolayers.56–58 Thus, spheroid formation is highly relevant for the establishment of drug resistance and testing of novel therapeutic targets. In this context, the compatibility of the magnetic chip with subsequent 3D cell culture was studied and the A549 cancer cell line was used for cancer cell spheroid formation. After processing through the MagPure chip, recovered cells were centrifuged and resuspended in filtered culture medium at a concentration of 50 A549 per μL. Representative images of formed spheroids over days are reported in Fig. 4b. Recovered A549 cancer cells self-agglomerated to form compact spheroids. In particular, the evolution of spheroid growth over days was studied by calculating the spheroid area for both recovered cells after magnetic purification and control cells, as a comparison (Fig. 4c). It can be observed that the formed spheroids present a similar growth pattern to the control group, with a growth reduction between 4 and 5 days of culture. It demonstrates spheroid compaction, a cellular arrangement naturally occurring during spheroid formation. Indeed, at first cells form loose aggregates, but direct cell–cell contact results in upregulated cadherin expression, which promotes strong adhesion of initial cell aggregates. After this delay phase during which homophilic binding occurs between cadherins of peripheral cells, cells are compacted into solid aggregates to form spheroids due to this homophilic cadherin–cadherin binding.59,60 Finally, at day 7, a 20% spheroid area increase was observed, therefore demonstrating spheroid growth. Thus, spheroid formation can be described as a three-step process: (i) initial cell aggregation, highlighted in Fig. 4d, followed by (ii) spheroid compaction and, finally, (iii) spheroid growth.61 The spheroid area for the chip output is lower than the control group's one which could be explained by the variability in cell counting and spiking (few microliters compared to milliliter volume). Consequently, even in the presence of few remaining WBCs that were not depleted in the MagPure chip, as well as excess magnetic nanoparticles (dark background visible behind spheroid), collected A549 cells were able to form spheroids and proliferate. Finally, long-term culture of spheroids was also investigated. Recovered A549 cells were cultured for 20 days, and evolution of the spheroid area was monitored (see S6, Fig. S2, ESI†). The spheroid area reached 0.7 mm2 after two weeks before decreasing. Indeed, from a certain point, the necrotic core starts sending extracellular signals stopping cell proliferation and leading to apoptosis. The purification step performed within the MagPure chip enabled successful spheroid formation and monitoring for several weeks. Drug sensitivity testing could therefore be considered as another downstream application on recovered cancer cells. Finally, we studied the chip compatibility with fluorescence in situ hybridization (FISH) for molecular profiling of individual cells at the chromosomal scale through hybridization of DNA probes on entire chromosomes or single unique sequences. FISH is a gold standard technique for probing genetic aberrations such as gene rearrangements (translocations, inversions) and changes in gene copy number associated with cancer.62 In many cancers, such chromosomal abnormalities often indicate lower survival rates and poor treatment efficacy.63 In particular, the anaplastic lymphoma kinase (ALK) genetic abnormality, located on the short arm of chromosome 2, is a key oncogenic driver, especially in non-small cell lung cancer.64 Thus, FISH is routinely performed on lung cancer tissues to detect EML4 and ALK gene fusion which occurs in 3–7% of NSCLC patients.65 This detection is crucial since specific tyrosine kinase inhibitors (ALK inhibition) are recommended as the first-line standard therapy for these patients (according to the US FDA), improving both the patient's quality of life and overall survival compared to traditional chemotherapy.66,67 For this experiment, the EML4-ALK Fusion-A549 (A549 EML4-ALK) cell line was employed to detect this intrachromosomal translocation by FISH. A549 EML4-ALK cells with WBCs were first processed through the MagPure chip for purification and FISH was subsequently performed. The 5′ ALK probe was labeled with a red fluorophore and the 3′ ALK probe with a green one (Fig. 5a). Representative images showing molecular FISH analysis in both WBCs and recovered A549 EML4-ALK cells and comparison with unprocessed (control) cells can be found in Fig. 5b. ALK-negative cells (i.e. control WBCs) show an overlapping of 5′ (red) and 3′ (green) signals, producing a fused 5′3′ signal (yellow signal). ALK-positive cells (spiked A549 EML4-ALK cells) can be identified by a split of the 5′ (red) and 3′ (green) signals. Molecular analysis by FISH performed before and after the purification step within the magnetic chip enabled the preservation of EML4-ALK gene fusion to be demonstrated in recovered A549 EML4-ALK cells. From this result it can be deduced that (i) recovered cancer cells were viable and structurally intact, and (ii) remaining WBCs did not affect the analysis of recovered cancer cells. Thus, the MagPure chip provides purified samples that can be readily investigated via standard genetic analysis, which is usually very difficult to conduct due to the large number of contaminated WBCs. The compatibility of the magnetic chip technology with the FISH assay has therefore been assessed for the detection of ALK gene arrangement in mCTCs.
B. Liquid biopsy-based workflow
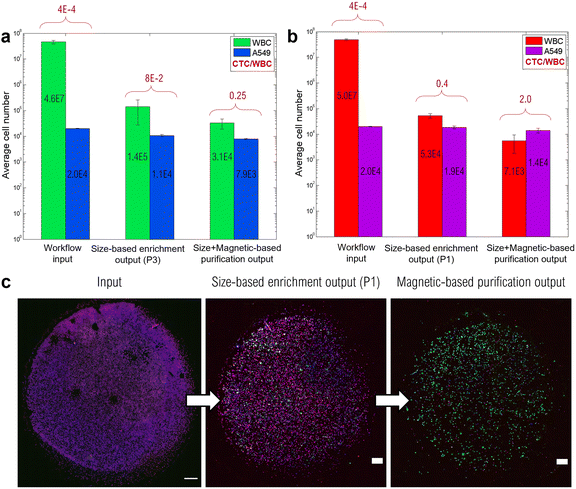
Furthermore, we determined the enrichment performances of the two-step workflow by assessing the number of WBCs and A549 cells after each workflow step (KOVA® slide counting). The CTC/WBC ratio was then calculated for each step in order to underline the benefit of the two-step separation process. In particular, two running programs provided by the ClearCell FX1 instrument were investigated. The CTC size cutoff of 14 μm (program P1) can be lowered by altering the flow ratios at the output (program P3). This will result in a faster processing (30 min for P3 against 60 min for P1) but also in a higher number of background WBCs. More information on these two programs can be found in S1, ESI.† Workflow performances are summarized in Fig. 7a and b, including both P3 and P1 running programs, respectively. First, ClearCell program P3 (Fig. 7a) enabled the WBC number to be reduced by a factor 300, from an initial number of 4.6.107 WBCs to a remaining number of 1.4.105. The mCTC recovery rate of the size-based enrichment step using the P3 program reached 53.4 ± 5.4%, leading to a CTC/WBC ratio of 8%. Adding the MagPure chip for further purification resulted in a 3 times higher CTC/WBC ratio, which reached 25%. The combination of the two separation methods led to a total depletion rate of 99.93 ± 0.04%, with a final number of ∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 WBCs. The achieved low number of background WBCs can therefore facilitate subsequent phenotypic and genotypic studies. ClearCell program P1 (Fig. 7b) achieved a greater WBC depletion, with ∼50
000 WBCs. The achieved low number of background WBCs can therefore facilitate subsequent phenotypic and genotypic studies. ClearCell program P1 (Fig. 7b) achieved a greater WBC depletion, with ∼50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 remaining WBCs (against 140
000 remaining WBCs (against 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with P3), resulting in a CTC/WBC ratio of 40%. Finally, the combination of the two sorting methods, P1 and MagPure chip, resulted in a depletion rate as high as 99.986 ± 0.004%, with a final number of 7100 WBCs, and a recovery of 71 ± 15%. It is worth mentioning that the whole workflow could be conducted in less than 4 hours, which shows the suitability of this workflow in a clinical context, and ensures cell viability preservation for subsequent analysis and culture.
000 with P3), resulting in a CTC/WBC ratio of 40%. Finally, the combination of the two sorting methods, P1 and MagPure chip, resulted in a depletion rate as high as 99.986 ± 0.004%, with a final number of 7100 WBCs, and a recovery of 71 ± 15%. It is worth mentioning that the whole workflow could be conducted in less than 4 hours, which shows the suitability of this workflow in a clinical context, and ensures cell viability preservation for subsequent analysis and culture.
Although CTC counts in patient samples usually range between 1 and 1000 per mL of blood (2500 mCTCs per mL were spiked here), the outstanding CTC/WBC ratio obtained after the MagPure chip paves the way for downstream single-cell studies.
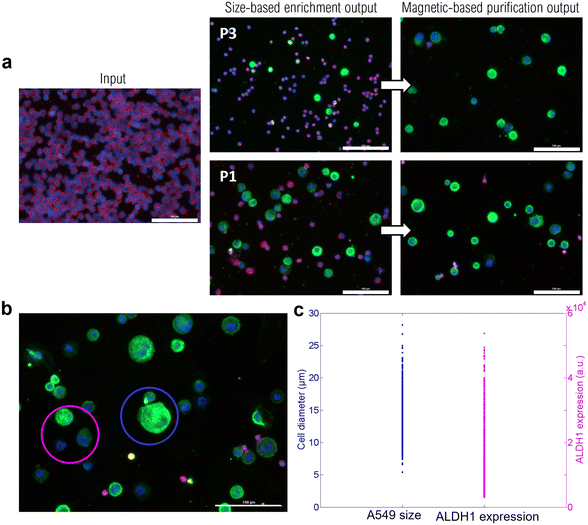
As an example, single-cell RNA sequencing can explore drug resistance mechanisms and help elucidate inter- and intra-patient heterogeneity.68 Although it presents significant challenges including preservation of RNA quality and single-cell mRNA amplification,69 it could fill the gap in the personalized medicine approach. The high purity achieved after this two-step separation is highlighted in Fig. 7c, where an overview of the immunolabeled cell spots at various stages of the workflow is depicted. After processing through the two-step workflow, cells were recovered for the phenotypic study by immunofluorescence (IF) staining. IF staining has remained the universal gold standard to distinguish recovered CTCs among background cells. In particular, it allows morphological and phenotypical studies by targeting specific molecules within cells. A549 cancer cells were targeted by the anti-ALDH1 antibody and AlexaFluor-488 fluorophore (GFP) while WBCs were labeled with anti-CD45, anti-CD15, and anti-CD41 antibodies conjugated to AlexaFluor-647 (CY5). For both, the cell nucleus was stained using DAPI. In particular, to assess the benefit of the additional magnetic-based purification step with the MagPure chip, the output of the first size-based enrichment step through ClearCell FX1 was divided into two: one half for the direct subsequent immunofluorescence assay, the other for processing through the MagPure chip and then the immunofluorescence assay. This way, the outputs of ClearCell FX1 and the magnetic chip can be directly compared. What can be observed in Fig. 7c is that the size-based enrichment step enables a first WBC depletion in comparison with the input. Nevertheless, the benefit of the additional magnetic-based purification step appears even more clearly since mainly green dots (A549 cells) are present in the spot and excess background WBCs were for the most part removed. The purity was determined by calculating the ratio of the number of A549 cancer cells (CY5-/GFP+) to the number of WBCs (CY5+/GFP). After ClearCell FX1 enrichment, a purity of 41% was found and this value reached 84% after the MagPure chip.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 a.u. ALDH1 is a marker of cancer stem-like cells, and its expression is associated with an aggressive phenotype and an augmented epithelial-mesenchymal transition,70,71 which can result in poor prognosis. Thus, being able to quantify the ALDH1 expression level could help determining patient prognosis.
000 a.u. ALDH1 is a marker of cancer stem-like cells, and its expression is associated with an aggressive phenotype and an augmented epithelial-mesenchymal transition,70,71 which can result in poor prognosis. Thus, being able to quantify the ALDH1 expression level could help determining patient prognosis.
Besides, the observed size overlap between A549 cells and WBCs (in the diameter range of 5–15 μm) reveals the challenges in size-based sorting methods (see S8, Fig. S4, ESI†). The additional immumomagnetophoretic-based purification step allowed for further WBC depletion, reducing their number by ∼7.5 (from average 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 WBCs after P1 to 7100 after the MagPure chip). This study highlights that, in addition to providing purified and viable cell samples, the MagPure chip enables the recovery of CTCs independently of their size or marker expression. These criteria are highly requested given the reported heterogeneity of CTCs.72–74
000 WBCs after P1 to 7100 after the MagPure chip). This study highlights that, in addition to providing purified and viable cell samples, the MagPure chip enables the recovery of CTCs independently of their size or marker expression. These criteria are highly requested given the reported heterogeneity of CTCs.72–74
| MagRC | μ-MixMACS chip | iFCS | CTC-iChip | Nagrath's group | This work | |
|---|---|---|---|---|---|---|
| a Recovery of the MagPure chip only. | ||||||
| Separation method | Magnetic | Magnetic | Magnetic | Size + magnetic | Size + magnetic | Size + magnetic |
| EpCAM-dependent | Yes | No | No | No | Yes | No |
| Throughput (mL h−1) | 0.5 | 18 | 12 | 9.6 | 3 | 3.5 |
| Blood volume (mL) | 10 | 5 | ~10 | 10 | 1 | 7.5 |
| Total processing time (h) | 96 | 6 | NA | 6–7 | NA | 3–4 |
| Recovery | 93% | 94% | 99% | 98% | 90% | 71% (79%)a |
| Depletion | 99.98% | 99.63% | 99.95% | NA | NA | 99.99% |
| Remaining WBCs per mL | 2000 | NA | 533 | 445 | 42.4 | 947 |
| Downstream analysis | IF, phenotype and protein profiling, RT-qPCR | IF | IF, cell culture | IF, imaging flow cytometry | IF, qRT-PCR | 2D/3D cell culture, IF, FISH |
| Ref. | 11, 72 | 75 | 14 | 73 | 76 | — |
Conclusion
We have developed a magnetophoretic-based microfluidic chip, the MagPure chip, for tumor marker- and size-independent isolation of CTCs via negative depletion of WBCs. This study first dealt with the optimization of the magnetophoretic trapping intrinsic performances of the chip, through enhancement of the WBC magnetic labeling, improvement of the microfluidic chamber integrating permanent micro-magnets, and determination of the operation fluidic conditions. Notably, the developed magnetic device does not require complex and expensive fabrication processes like conventional systems.The adopted composite approach enables the straightforward integration of dense arrays of permanently magnetized micro-traps that generate large magnetic forces, leading to compact magnetic devices. The developed magnetophoretic chip achieved an average WBC depletion efficiency of 87% (corresponding to the average ratio of labeled WBCs) and an average mCTC recovery rate of 81%. Furthermore, the MagPure chip compatibility with conventional biological studies, including 2D and 3D cell culture, as well as phenotypic and genotypic analyses, was demonstrated. Indeed, processing through the MagPure chip ensured the preservation of recovered cancer cell viability and integrity. After conducting these studies on model blood samples with spiked cancer cell lines, the magnetic chip was combined with a size-based separation system, the ClearCell FX1, to benefit from both technologies' advantages. It is worth mentioning that the developed purification step through the MagPure chip could be combined to any pre-enrichment step. The designed two-step workflow led to high-throughput and high purity, with a final mCTC recovery efficiency of 70% and a WBC depletion rate as high as 99.99%, with an average contaminating number of WBCs per mL of 947 after the ClearCell P1 enrichment program and magnetic purification. The separation protocol is performed on 7.5 mL of blood and takes between 3 and 4 hours. The highly purified sample thus obtained enabled downstream analysis such as long-term cell culture and phenotypic analysis. Such subsequent studies are crucial to relate to clinical decisions and personalized medicine strategies. This work demonstrated the value of emerging microfluidic and magnetophoretic technologies for characterizing liquid biopsy samples, an important research topic that will lead to changes in the paradigm of cancer diagnosis and management. Future work will consist in assessing the two-step workflow performances in a clinical context by isolation of CTCs from patient samples.
Author contributions
L. D. designed the MagPure chip and performed the cell sorting and downstream analysis experiments. A.-L. D. and D. L. R. conceived the project and supervised the research. L. P. provided the ClearCell FX1 commercial system. D. B. and L. P. provided the blood samples. J. G. and D. B. assisted in the biological experiments. L. D. analysed the data. All of the authors discussed the results. L. D., D. L. R., and A.-L. D. prepared the original draft manuscript with input from all the authors. L. D., E. L., L. P., D. L. R., and A.-L. D. reviewed and edited the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge funding for this work from LUTON, PACK AMBITION RECHERCHE AuRA (grant #1701103701-40890). The authors acknowledge the support staff from Nanolyon technological platform. All the staff of the CIRCAN team from the Hospices Civils de Lyon is also gratefully acknowledged for their help on biological sample preparation.Notes and references
- American Cancer Society, 2021.
- M. Samandari, M. G. Julia, A. Rice, A. Chronopoulos and A. E. del Rio Hernandez, Transl. Res., 2018, 201, 98–127 CrossRef.
- S. Ju, C. Chen, J. Zhang, L. Xu, X. Zhang, Z. Li, Y. Chen, J. Zhou, F. Ji and L. Wang, Biomark. Res., 2022, 10, 1–25 CrossRef.
- F. Tanaka, K. Yoneda, N. Kondo, M. Hashimoto, T. Takuwa, S. Matsumoto, Y. Okumura, S. Rahman, N. Tsubota, T. Tsujimura, K. Kuribayashi, K. Fukuoka, T. Nakano and S. Hasegawa, Clin. Cancer Res., 2009, 15, 6980–6986 CrossRef CAS PubMed.
- J. A. Thiele, K. Bethel, M. Králíčková and P. Kuhn, Annu. Rev. Pathol.: Mech. Dis., 2017, 12, 419–447 CrossRef CAS PubMed.
- J. G. Moreno, M. C. Miller, S. Gross, W. J. Allard, L. G. Gomella and L. W. M. M. Terstappen, Urology, 2005, 65, 713–718 CrossRef.
- M. Cristofanilli, D. F. Hayes, G. T. Budd, M. J. Ellis, A. Stopeck, J. M. Reuben, G. V. Doyle, J. Matera, W. J. Allard, M. C. Miller, H. A. Fritsche, G. N. Hortobagyi and L. W. M. M. Terstappen, J. Clin. Oncol., 2005, 23, 1420–1430 CrossRef PubMed.
- J. Den Toonder, Lab Chip, 2011, 11, 375–377 RSC.
- C. Alix-Panabières and K. Pantel, Cancer Discovery, 2016, 6, 479–491 CrossRef PubMed.
- D. Quandt, H. D. Zucht, A. Amann, A. Wulf-Goldenberg, C. Borrebaeck, M. Cannarile, D. Lambrechts, H. Oberacher, J. Garrett, T. Nayak, M. Kazinski, C. Massie, H. Schwarzenbach, M. Maio, R. Prins, B. Wendik, R. Hockett, D. Enderle, M. Noerholm, H. Hendriks, H. Zwierzina and B. Seliger, Oncotarget, 2017, 8, 48507–48520 CrossRef PubMed.
- M. Labib, Z. Wang, S. U. Ahmed, R. M. Mohamadi, B. Duong, B. Green, E. H. Sargent and S. O. Kelley, Nat. Biomed. Eng., 2021, 5, 41–52 CrossRef CAS.
- E. Lin, T. Cao, S. Nagrath and M. R. King, Annu. Rev. Biomed. Eng., 2018, 20, 329–352 CrossRef CAS PubMed.
- M. E. Francart, J. Lambert, A. M. Vanwynsberghe, E. W. Thompson, M. Bourcy, M. Polette and C. Gilles, Dev. Dyn., 2018, 247, 432–450 CrossRef PubMed.
- W. Zhao, Y. Liu, B. D. Jenkins, R. Cheng, B. N. Harris, W. Zhang, J. Xie, J. R. Murrow, J. Hodgson, M. Egan, A. Bankey, P. G. Nikolinakos, H. Y. Ali, K. Meichner, L. A. Newman, M. B. Davis and L. Mao, Lab Chip, 2019, 19, 1860–1876 RSC.
- B. Mostert, J. Kraan, J. Bolt-de Vries, P. van der Spoel, A. M. Sieuwerts, M. Schutte, A. M. Timmermans, R. Foekens, J. W. M. Martens, J.-W. Gratama, J. A. Foekens and S. Sleijfer, Breast Cancer Res. Treat., 2011, 127, 33–41 CrossRef CAS PubMed.
- D. A. Smirnov, D. R. Zweitzig, B. W. Foulk, M. C. Miller, G. V. Doyle, K. J. Pienta, N. J. Meropol, L. M. Weiner, S. J. Cohen, J. G. Moreno, M. C. Connelly, L. W. M. M. Terstappen and S. M. O'Hara, Cancer Res., 2005, 65, 4993–4997 CrossRef CAS PubMed.
- L. Descamps, D. Le Roy and A. Deman, Int. J. Mol. Sci., 2022, 23, 1981 CrossRef CAS.
- M. C. Miller, P. S. Robinson, C. Wagner and D. J. O'Shannessy, Cytometry, Part A, 2018, 93, 1234–1239 CrossRef PubMed.
- Y. Lee, G. Guan and A. A. Bhagat, Cytometry, Part A, 2018, 93, 1251–1254 CrossRef.
- E. Sollier-Christen, C. Renier, T. Kaplan, E. Kfir and S. C. Crouse, Cytometry, Part A, 2018, 93, 1240–1245 CrossRef CAS PubMed.
- C. Alix-Panabières and K. Pantel, Nat. Rev. Cancer, 2014, 14, 623–631 CrossRef PubMed.
- S. Morganti, P. Tarantino, E. Ferraro, P. D. Amico, G. Viale, D. Trapani, B. Achutti and G. Curigliano, Crit. Rev. Oncol. Hematol., 2019, 133, 171–182 CrossRef PubMed.
- J. Garcia, Évaluation du patrimoine tumoral circulant dans la prise en charge thérapeutique des patients atteints de cancer broncho-pulmonaire, PhD thesis, Université de Lyon, 2019 Search PubMed.
- A. Munaz, M. J. A. Shiddiky and N. T. Nguyen, Biomicrofluidics, 2018, 12, 031501 CrossRef PubMed.
- O. Cugat, J. Delamare and G. Reyne, IEEE Trans. Magn., 2003, 39, 3607–3612 CrossRef.
- L. Descamps, D. Le Roy, C. Tomba and A. Deman, Magnetochemistry, 2021, 7, 100 CrossRef CAS.
- P. Chen, Y. Y. Huang, K. Hoshino and J. X. J. Zhang, Sci. Rep., 2015, 5, 1–9 Search PubMed.
- M. Poudineh, E. H. Sargent and S. O. Kelley, ACS Appl. Mater. Interfaces, 2017, 9, 25683–25690 CrossRef CAS PubMed.
- C. M. Xu, M. Tang, J. Feng, H. F. Xia, L. L. Wu, D. W. Pang, G. Chen and Z. L. Zhang, Lab Chip, 2020, 20, 1418–1425 RSC.
- L. Descamps, S. Mekkaoui, M.-C. Audry, A.-L. Deman and D. Le Roy, AIP Adv., 2020, 10, 15215 CrossRef CAS.
- L. Descamps, M.-C. Audry, J. Howard, S. Mekkaoui, C. Albin, D. Barthelemy, L. Payen, J. Garcia, E. Laurenceau, D. Le Roy and A.-L. Deman, Cell, 2021, 10, 1734 CrossRef CAS.
- L. Descamps, S. Mekkaoui, M. C. Audry, E. Laurenceau, J. Garcia, L. Payen, A. L. Deman and D. Le Roy, 24th Int. Conf. Miniaturized Syst. Chem. Life Sci. (uTAS), 2020, pp. 442–443 Search PubMed.
- N. M. Dempsey, D. Le Roy, H. Marelli-Mathevon, G. Shaw, A. Dias, R. B. G. Kramer, L. Viet Cuong, M. Kustov, L. F. Zanini, C. Villard, K. Hasselbach, C. Tomba and F. Dumas-Bouchiat, Appl. Phys. Lett., 2014, 104, 262401 CrossRef.
- S. Mekkaoui, L. Descamps, M. C. Audry, A. L. Deman and D. Le Roy, Langmuir, 2020, 36, 14546–14553 CrossRef CAS PubMed.
- S. Mekkaoui, D. Le Roy, M.-C. Audry, J. Lachambre, V. Dupuis, J. Desgouttes and A.-L. Deman, Microfluid. Nanofluid., 2018, 22, 119 CrossRef.
- L. Zeng, X. Chen, J. Du, Z. Yu, R. Zhang, Y. Zhang and H. Yang, Nanoscale, 2021, 13, 4029 RSC.
- E. Mirowski, J. Moreland, A. Zhang, S. E. Russek and M. J. Donahue, Appl. Phys. Lett., 2005, 86, 1–3 CrossRef.
- O. Yassine, C. P. Gooneratne, D. A. Smara, F. Li, H. Mohammed, J. Merzaban and J. Kosel, Biomicrofluidics, 2014, 8, 034114 CrossRef CAS PubMed.
- X. Hu, S. R. Goudu, S. R. Torati, B. Lim, K. Kim and C. Kim, Lab Chip, 2016, 16, 3485–3492 RSC.
- R. Zhou, Q. Yang, F. Bai, J. A. Werner, H. Shi, Y. Ma and C. Wang, Microfluid. Nanofluid., 2016, 20, 1–12 CrossRef.
- R. Zhou and C. Wang, Microfluid. Nanofluid., 2016, 20, 1–11 CrossRef.
- D. Jaiswal, A. T. Rad, M. P. Nieh, K. P. Claffey and K. Hoshino, J. Magn. Magn. Mater., 2017, 427, 7–13 CrossRef CAS.
- L. Toraille, K. Aïzel, É. Balloul, C. Vicario, C. Monzel, M. Coppey, E. Secret, J. M. Siaugue, J. Sampaio, S. Rohart, N. Vernier, L. Bonnemay, T. Debuisschert, L. Rondin, J. F. Roch and M. Dahan, Nano Lett., 2018, 18, 7635–7641 CrossRef CAS PubMed.
- S. Ponomareva, A. Dias, B. Royer, H. Marelli, J. F. Motte, D. Givord, F. Dumas-Bouchiat, N. M. Dempsey and F. Marchi, J. Micromech. Microeng., 2019, 29, 015010 CrossRef CAS.
- W. Zhao, R. Cheng, B. D. Jenkins, T. Zhu, N. E. Okonkwo, C. E. Jones, M. B. Davis, S. K. Kavuri, Z. Hao, C. Schroeder and L. Mao, Lab Chip, 2017, 17, 3097–3111 RSC.
- O. Sirenko, J. Hesley, I. Rusyn and E. F. Cromwell, Assay Drug Dev. Technol., 2014, 12, 43–54 CrossRef CAS PubMed.
- C. Zhu and R. Skalak, Biophys. J., 1988, 54, 1115–1137 CrossRef CAS PubMed.
- Y. Samstag, J. Leukocyte Biol., 2003, 73, 30–48 CrossRef CAS PubMed.
- K. Moshksayan, N. Kashaninejad, M. E. Warkiani, J. G. Lock, H. Moghadas, B. Firoozabadi, M. S. Saidi and N. T. Nguyen, Sens. Actuators, B, 2018, 263, 151–176 CrossRef CAS.
- Z. Wen, Q. Liao, Y. Hu, L. You, L. Zhou and Y. Zhao, Braz. J. Med. Biol. Res., 2013, 46, 634–642 CrossRef CAS PubMed.
- A. Zuchowska, K. Kwapiszewska, M. Chudy, A. Dybko and Z. Brzozka, Electrophoresis, 2017, 38, 1206–1216 CrossRef CAS PubMed.
- X. Liu, H. Lin, J. Song, T. Zhang, X. Wang, X. Huang and C. Zheng, Micromachines, 2021, 12(6), 681 CrossRef PubMed.
- J. Koch, D. Mönch, A. Maaß, C. Gromoll, T. Hehr, T. Leibold, H. J. Schlitt, M.-H. Dahlke and P. Renner, PLoS One, 2021, 16, 1–13 Search PubMed.
- Y. C. Chen, X. Lou, Z. Zhang, P. Ingram and E. Yoon, Sci. Rep., 2015, 5, 1–12 Search PubMed.
- R. W. K. Wu, E. S. M. Chu, J. W. M. Yuen and Z. Huang, J. Photochem. Photobiol., B, 2020, 210, 111987 CrossRef CAS PubMed.
- P. Longati, X. Jia, J. Eimer, A. Wagman, M.-R. Witt, S. Rehnmark, C. Verbeke, R. Toftgård, M. Löhr and R. L. Heuchel, BMC Cancer, 2013, 13, 95 CrossRef CAS PubMed.
- S. Chandrasekaran, J. R. Marshall, J. A. Messing, J.-W. Hsu and M. R. King, PLoS One, 2014, 9, 1–12 Search PubMed.
- Y. Shichi, N. Sasaki, M. Michishita, F. Hasegawa, Y. Matsuda, T. Arai, F. Gomi, J. Aida, K. Takubo, M. Toyoda, H. Yoshimura, K. Takahashi and T. Ishiwata, Sci. Rep., 2019, 9, 1–10 CrossRef CAS PubMed.
- R. Z. Lin and H. Y. Chang, Biotechnol. J., 2008, 3, 1172–1184 CrossRef CAS PubMed.
- X. Cui, Y. Hartanto and H. Zhang, J. R. Soc., Interface, 2017, 14, 20160877 CrossRef PubMed.
- I. Smyrek, B. Mathew, S. C. Fischer, S. M. Lissek, S. Becker and E. H. K. Stelzer, Biol. Open, 2019, 8, bio037051 CAS.
- M. Ilie, E. Long, C. Butori, V. Hofman, C. Coelle, V. Mauro, K. Zahaf, C. H. Marquette, J. Mouroux, P. Paterlini-Bréchot and P. Hofman, Ann. Oncol. Off. J. Eur. Soc. Med. Oncol., 2012, 23, 2907–2913 CrossRef CAS PubMed.
- M. A. Gertz, M. Q. Lacy, A. Dispenzieri, P. R. Greipp, M. R. Litzow, K. J. Henderson, S. A. Van Wier, G. J. Ahmann and R. Fonseca, Blood, 2005, 106, 2837–2840 CrossRef CAS PubMed.
- M. Soda, Y. L. Choi, M. Enomoto, S. Takada, Y. Yamashita, S. Ishikawa, S. I. Fujiwara, H. Watanabe, K. Kurashina, H. Hatanaka, M. Bando, S. Ohno, Y. Ishikawa, H. Aburatani, T. Niki, Y. Sohara, Y. Sugiyama and H. Mano, Nature, 2007, 448, 561–566 CrossRef CAS PubMed.
- E. Sánchez-Herrero, M. Provencio and A. Romero, Adv. Lab. Med., 2020, 1, 20190019 Search PubMed.
- B. J. Solomon, D.-W. Kim, Y.-L. Wu, K. Nakagawa, T. Mekhail, E. Felip, F. Cappuzzo, J. Paolini, T. Usari, Y. Tang, K. D. Wilner, F. Blackhall and T. S. Mok, J. Clin. Oncol., 2018, 36, 2251–2258 CrossRef CAS.
- J. Zhou, J. Zheng, X. Zhang, J. Zhao, Y. Zhu, Q. Shen, Y. Wang, K. Sun, Z. Zhang, Z. Pan, Y. Shen and J. Zhou, BMC Cancer, 2018, 18, 10 CrossRef.
- Z. Liu, Y. Huang, W. Liang, J. Bai, H. Feng, Z. Fang, G. Tian, Y. Zhu, H. Zhang, Y. Wang, A. Liu and Y. Chen, Lab Chip, 2021, 21, 2881–2891 RSC.
- Y. Li, S. Wu and F. Bai, Semin. Cell Dev. Biol., 2018, 75, 88–97 CrossRef CAS PubMed.
- J. Yao, Q. Jin, X. D. Wang, H. J. Zhu, Q. C. Ni and H. Fan, Med., 2017, 96, 1–6 Search PubMed.
- M. F. Chen, P. T. Chen, M. S. Lu and W. C. Chen, Mol. Carcinog., 2018, 57, 78–88 CrossRef CAS PubMed.
- M. Poudineh, P. M. Aldridge, S. Ahmed, B. J. Green, L. Kermanshah, V. Nguyen, C. Tu, R. M. Mohamadi, R. K. Nam, A. Hansen, S. S. Sridhar, A. Finelli, N. E. Fleshner, A. M. Joshua, E. H. Sargent and S. O. Kelley, Nat. Nanotechnol., 2017, 12, 274–281 CrossRef CAS PubMed.
- F. Fachin, P. Spuhler, J. M. Martel-Foley, J. F. Edd, T. A. Barber, J. Walsh, M. Karabacak, V. Pai, M. Yu, K. Smith, H. Hwang, J. Yang, S. Shah, R. Yarmush, L. V. Sequist, S. L. Stott, S. Maheswaran, D. A. Haber, R. Kapur and M. Toner, Sci. Rep., 2017, 7, 1–11 CrossRef CAS PubMed.
- C. Renier, E. Pao, J. Che, H. E. Liu, C. A. Lemaire, M. Matsumoto, M. Triboulet, S. Srivinas, S. S. Jeffrey, M. Rettig, R. P. Kulkarni, D. Di Carlo and E. Sollier-Christen, npj Precis. Oncol., 2017, 1, 15 CrossRef PubMed.
- T. Y. Lee, K. A. Hyun, S. I. Kim and H. I. Jung, Sens. Actuators, B, 2017, 238, 1144–1150 CrossRef CAS.
- R. M. Jack, M. M. G. Grafton, D. Rodrigues, M. D. Giraldez, C. Griffith, R. Cieslak, M. Zeinali, C. K. Sinha, E. Azizi, M. Wicha, M. Tewari, D. M. Simeone and S. Nagrath, Adv. Sci., 2016, 3, 1–8 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc00443g |
| This journal is © The Royal Society of Chemistry 2022 |