Open Access Article
Open Access ArticleUV-DIB: label-free permeability determination using droplet interface bilayers†
Robert
Strutt‡
 ab,
Felix
Sheffield‡
ab,
Felix
Sheffield‡
 ab,
Nathan E.
Barlow
ab,
Anthony J.
Flemming
c,
John D.
Harling
d,
Robert V.
Law
ab,
Nicholas J.
Brooks
ab,
Laura M. C.
Barter
ab and
Oscar
Ces
ab,
Nathan E.
Barlow
ab,
Anthony J.
Flemming
c,
John D.
Harling
d,
Robert V.
Law
ab,
Nicholas J.
Brooks
ab,
Laura M. C.
Barter
ab and
Oscar
Ces
 *ab
*ab
aDepartment of Chemistry, Molecular Sciences Research Hub, Imperial College London, Shepherd's Bush, London, W12 0BZ, UK. E-mail: o.ces@imperial.ac.uk
bInstitute of Chemical Biology, Molecular Sciences Research Hub, Imperial College London, Shepherd's Bush, London, W12 0BZ, UK
cSyngenta, Jealott's Hill International Research Centre, Bracknell, Berkshire RG42 6EY, UK
dMedicinal Chemistry, GlaxoSmithKline, Stevenage, SG1 2NY, UK
First published on 2nd February 2022
Abstract
Simple diffusion of molecular entities through a phospholipid bilayer, is a phenomenon of great importance to the pharmaceutical and agricultural industries. Current model lipid systems to probe this typically only employ fluorescence as a readout, thus limiting the range of assessable chemical matter that can be studied. We report a new technology platform, the UV-DIB, which facilitates label free measurement of small molecule translocation rates. This is based upon the coupling of droplet interface bilayer technology with implemented fiber optics to facilitate analysis via ultraviolet spectroscopy, in custom designed PMMA wells. To improve on current DIB technology, the platform was designed to be reusable, with a high sampling rate and a limit of UV detection in the low μM regime. We demonstrate the use of our system to quantify passive diffusion in a reproducible and rapid manner where the system was validated by investigating multiple permeants of varying physicochemical properties across a range of lipid interfaces, each demonstrating differing kinetics. Our system permits the interrogation of structural dependence on the permeation rate of a given compound. We present this ability from two structural perspectives, that of the membrane, and the permeant. We observed a reduction in permeability between pure DOPC and DPhPC interfaces, concurring with literature and demonstrating our ability to study the effects of lipid composition on permeability. In relation to the effects of permeant structure, our device facilitated the rank ordering of various compounds from the xanthine class of compounds, where the structure of each permeant differed by a single group alteration. We found that DIBs were stable up to 5% DMSO, a molecule often used to aid solubilisation of pharmaceutical and agrochemical compounds. The ability of our device to rank-order compounds with such minor structural differences provides a level of precision that is rarely seen in current, industrially applied technologies.
Introduction
Biological systems are contained and subdivided by phospholipid membranes. All cells are surrounded by a plasma membrane, eukaryotic cells further include membrane-enclosed organelles such as mitochondria and chloroplasts. The lipid composition of these membranes (which compartmentalise drug targets) will vary among and within species and can be affected by environmental factors such as nutrition, disease and age.1–8 Additionally there are tissues which, while being cellularised, comprise a near contiguous macroscopic membrane such as the blood–brain barrier of mammals or the Casparian strip in plant roots.9,10 Understanding the engineering rules that control how a molecule can translocate across a cellular membrane remains critical in the development of novel molecular entities.Drug translocation across a cell membrane can occur via multiple transport routes, including (i) passive diffusion through the lipids of the membrane, (ii) transporter protein–drug interactions, and (iii) liposomal delivery through lipid fusion.11 Passive diffusion across lipid bilayers (where ∼50% of the eukaryotic phospholipid material is estimated to be phosphatidylcholine (PC) based)12 is facilitated by a concentration gradient of the permeant across the membrane. This particular pathway is understood to be a major indicator of oral bioavailability, as well as foliar uptake,13–15 with many current assays placing focus on this particular transport route.16–18 Therefore, biologically active chemistry, will interact with membranes whose varying properties will likely affect their efficacy, selectivity, and safety in patients and/or the environment. These affects are critical to the development of sustainable chemistry in agriculture as well as to personalised medicine in pharmaceuticals. For all this, up to now heavy reliance has been placed on simple correlations with the physical properties of active ingredients, e.g. the Lipinski ‘rule of five’ and equivalents, or on crude measures of membrane permeation such as PAMPA.17,19,20
Pure lipid based systems are rapidly emerging as viable methods to screen diffusion of molecular entities.21 Lipid–drug interactions are viewed as important indicators of biocompatibility for liposomal drug delivery22 as well as implicated in the mechanism of non-specific efflux by proteins such as p-glycoprotein, thus having large effects on an entities activity.23 Modelling this entire process from a kinetic perspective is therefore amenable to understanding each component of the permeability process either in vitro or in vivo. A droplet interface bilayer (DIB) presents a lipid membrane (of tuneable composition and symmetry) separated by two aqueous compartments in which the contents of each can be controlled (Fig. 1b and c). As shown previously by our lab, DIBs provide a useful platform to interrogate simple diffusion (Fig. 1c),24 where the potential of this technique in an agrochemical context was demonstrated.25 In the literature, some of these platforms have provided valuable information, such as the permeability of a fluorescent antibiotic,26 and a chemotherapy drug.27 However, continued reliance on fluorescence based readouts has meant that the range of screenable candidates,28,29 and therefore the scope of these as widespread assays is limited.30 Fluorescent chemical structures often have a low rotational bond number and if tagged to a compound can have a high molecular weight. This limits the sampleable chemical space which in turn affects the range of measured permeabilities derived from these interfaces. A reliance on image analysis approaches often requires a high degree of technical expertise, coupled with a rigorous experimental and characterisation process. Furthering this argument, recent advancements have seen the development of molecular entities outside Lipinski rules.31 A novel, widely applicable approach to delineate the impact of simple diffusion in vitro is therefore required as an initial screen in the development of novel entities. Whilst similarly developed platforms have satisfied in part the following improvements,32–36 a system is yet to emerge which miniaturizes the label free permeability problem with two equal volume, controllable compartments, where non-invasive detection in the μM range with a low recording time interval can be performed.
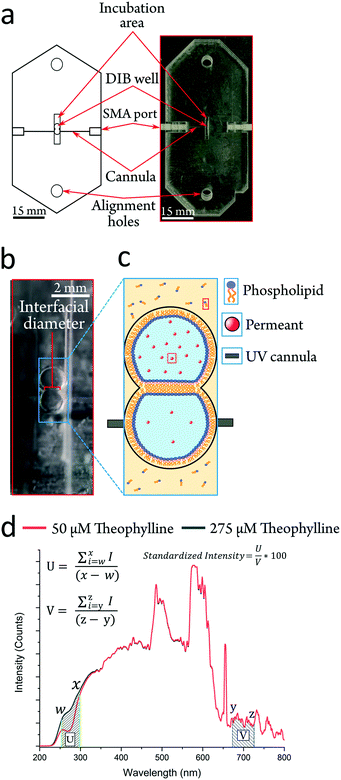
Herein we present a highly facile, cheap and reusable platform, which can screen the lipid bilayer permeability of any UV absorbent compound with an appreciable water solubility (Fig. 1a). The UV-DIB features UV cannulas (responsive from ∼225–1000 nm (Fig. 1d)), embedded into acrylic layers, aligned across a compartment to facilitate a DIB permeation system. Our method is highly flexible in the design iteration process and implementable in rapid prototyping, where fabrication is estimated to take ∼2 h from conception to generation. Our method is ‘label-free’ in that detection relies upon the central molecular structure of the permeant. The readiness of our experimental procedure to characterise permeability is demonstrated and it is shown that the design principles of our fabrication method can be applied in a chip format, which can monitor label-free concentrations. Passive permeability assessment was performed with a total of 9 permeants with varying relevance to either therapeutic or agrochemical use to demonstrate the applicability of the device in industrially relevant research fields. Seven of these nine permeants exist in a narrow chemical space with systematic functional group changes in the subset. This demonstrates our ability to probe the nuances of the permeant structure–permeability relationship. We further present the capacity of the method by uncovering novel, relevant trends relating to the biophysics of DIBs as well as exploring the limits of model membrane permeability.
Results
Generation of UV-detecting permeability chips
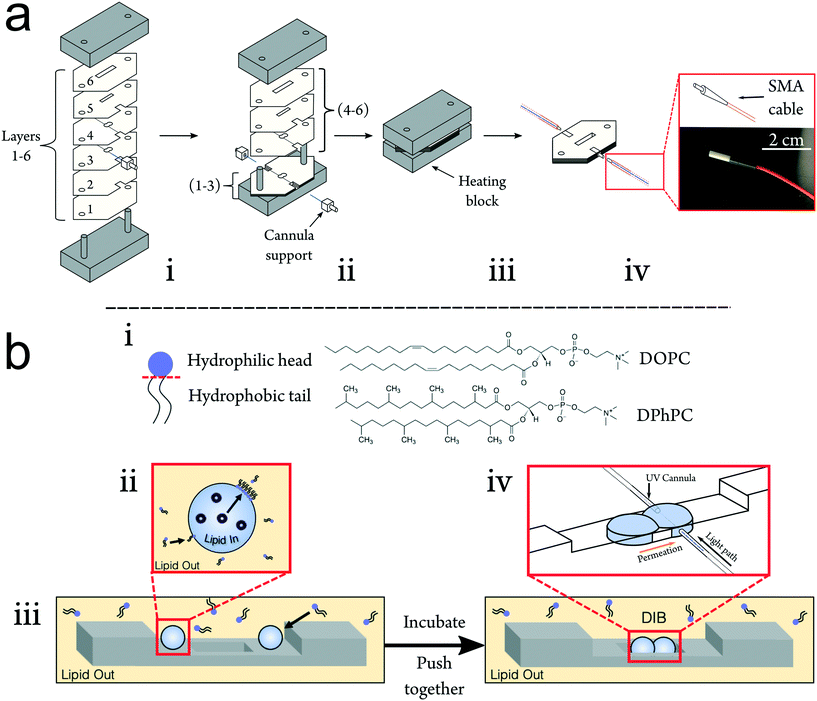
To improve upon the current standard for DIB permeability assessment the UV-DIB is: reusable, capable of recording with a small time interval, able to determine concentration from UV spectra in the low μM regime and reasonably facile to use. In consideration of these factors and scalability, the platform was constructed from poly(methyl methacrylate) (PMMA), frequently employed in microfluidic devices.37,38 PMMA is cheap, has high transparency and can be easily cut with a laser cutter at the desired μm resolution (smallest feature size of chip: ∼200 μm). Previous work has utilised PMMA as a suitable substrate to facilitate droplet arrangement.39,40 DIBs using nanoliter volumes are typically formed within well-like assemblies where the DIB itself is free standing in oil. By cutting features in layers of varying thicknesses and stacking these on top of one another (Fig. 2ai), wells that can directly house a DIB architecture can be assembled. This approach reduced the conformational error between DIB formations, hence yielding a high reproducibility in terms of DIB morphology. Moreover, the method allows for precise etching of the PMMA surface, facilitating cannula alignment across the DIB chamber. Layers of cut material can be bonded together by applying heat and pressure beyond the PMMA glass transition temperature (Tg).22,41,42 For our fabrication we found the optimal conditions to be 170 degrees °C heating applied with 0.5 N m pressure for 45 minutes. It is worth noting that this process should be optimised to the height of the chip, at which reduced chip height may result in deformation/fracturing of the upper and lower layers.We demonstrate the construction of 3D chip architectures with distinct spaces for droplet incubation and measurement (Fig. 2biii). When compared to soft lithography and 3D printing, this chip fabrication scheme reduces cost, skill requirement and fabrication time. We estimate the fabrication time per chip to be approximately 1 hour, where the only specialist equipment required is a laser cutter. It is also worth noting that this approach is clean room free. Acrylic bonding is an established method for chip formation and fiber optics have been previously implemented within PMMA chips.37 However up until now, these approaches have not been coupled to DIB technologies to enable non-invasive label-free detection.43,44 In the UV-DIB, a UV-transmitting cannula, featuring an attachment point for an SMA cable is aligned across a tailored chamber space as illustrated in Fig. 1b and 2 – here shown to support a DIB between two 0.9 μL droplets. In the presented device, the path length between cannula ends was engineered to be 1 mm, across the measured droplet. As is shown in Fig. 2biv, the cannula is flush against the side of the DIB well with the droplet in contact with this same face. The heat press stage sealed the cannulas within the chip and showed no leakage of oil between chip layers. As the chip was sealed, it was completely reusable where both aqueous and oil phases could easily be removed via pipette. As far as we are aware, PMMA presents an inert surface and our experiments did not indicate that any of the compounds studied interacted directly with the PMMA.
Method for conducting permeability assays
To measure the passive diffusion of 9 different test compounds (paracetamol, thiamethoxam and range of xanthine derivatives), an experimental procedure was developed (Fig. 3). All compounds showed significant absorption in the UV region from 250–290 nm whilst exhibiting minimal variation in the visible regions of the spectra (denoted U and V regions in Fig. 1d). This was exploited to standardize the intensity of each spectrum into a single measurement. All compounds studied showed a strong linear calibration region (mean R2 = 0.99 ± 0.01) in the μM concentration regime (ESI† Table S2). Per permeant, the linear region was then used to determine a starting acceptor and donor concentration to establish a concentration gradient (for each permeant this is listed in ESI† Table S2). An initial concentration in the acceptor (at t = 0 conc >0) compartment was employed for two reasons; significant noise was observed at concentrations below the chosen initial acceptor concentration (toward 0), attributed to the limit of detection of our spectrometer (LOD ∼ 25 μM – ESI† Fig. S4). Secondly, it was assumed that a fraction of the compound may solubilise in the lipid monolayer/bilayer, potentially altering the intrinsic behaviour of the membrane.36,45 The same standardized intensity was present for large droplets representative of the fused DIB volume, single droplets representative of one side of the DIB and equal concentration droplet DIBs (ESI† Fig. S3 for theophylline controls). Leakage of permeant concentration from the aqueous into the oil phase would mitigate our permeation model as the oil would act as a sink for the initial donor concentration. To confirm that this was not occurring, 1.8 μl droplets containing a high concentration of the permeant were recorded. Each permeant had an appreciable water solubility, where significant leakage of each permeant into the oil phase was initially assumed to be minimal.46 A leakage assay was performed for each permeant, with all experiments demonstrating negligible loss of mass into the oil phase, as noted by a continuous UV transmission over time (Fig. 3b and ESI† Fig. S3, S4, S6 and S7). It is worth noting the ease of this method in determining oil leakage for more hydrophobic compounds. These experiments were also used for comparative analysis to permeation plots (ESI† Fig. S3 and S6).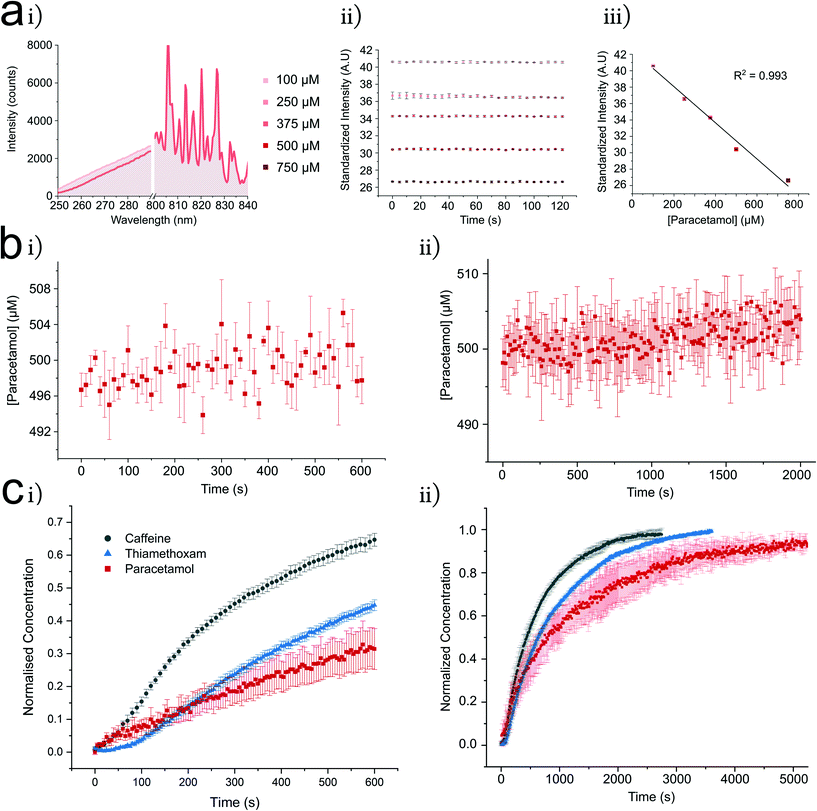 | ||
| Fig. 3 The UV DIB platform allows characterisation of label free simple diffusion across a lipid membrane. Shown here are example characterisations of lipid in and out systems using caffeine, paracetamol and thiamethoxam. a i U (250–290 nm) and V, non-absorbent (800–840 nm) regions (see Fig. 1d for U and V regions) for paracetamol 0.9 μL droplets, illustrate the variance in the U region at 100 to 500 μM with minor fluctuation in the V region – hence enabling the spectra to be converted into a single point measurement. ii Monitoring each paracetamol concentration shows a linear distribution across 120 s (n = 3), iii the mean value of plots in ii are used for a calibration curve displaying linearity between 100 and 750 μM (standard deviation (SD) reported for each point). b Leakage assessment of 500 μM paracetamol from a 1.8 μL droplet into hexadecane oil on the t = 600 s (n = 3) (i) and t = 2000 s (n = 3) timescales (ii) data converted into concentration using the calibration curve shown in 3a. Low deviation from the initial concentration in the leakage assays across both timescales indicates that paracetamol remains in the aqueous phase. c DIBs formed from 0.9 μL droplets show compound permeation is controlled toward acceptor droplets denoted by exponential character concentration time curves, t = 600 s (n = 8) (i), all permeants interpreted with method outlined in a and b. Longer distributions (ii) show each compound reaching equilibrium (n = 3). This experimental procedure was conducted for all of the presented compounds in Table 1, for more information on experimental details see ESI† Table S2. | ||
Typical concentration changes in the acceptor DIB droplet are shown in Fig. 3c, here shown with 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) using the lipid in and out formation method. In Fig. 4c, the concentration of each compound increases to the expected equilibrium concentration. Where possible, this was further confirmed by rupturing the DIB at equilibrium and observing the same standardized intensity. Reaching equilibrium strongly reflects the observed leakage profiles in which retention of the compound in the oil or lipid was minimal. In each case an exponential increase is observed in line with Fick's first law of diffusion (eqn (1)). Our chip guides the droplets into arrangement, controlling the DIB area. Within the well, the DIB area was measured by microscopy (ESI Fig. S1†). Occasionally, a delay was observed between droplet contact at t = 0 and the exponential character of the concentration change (Fickian behaviour). Within DIB literature it is known that ‘zip up’ time refers to drainage of any residual oil between monolayer leaflets.47,48 The clear exponential character of the curves supports assumption of continuous DIB area following zip up. When determining permeability from concentration–time curves, care was taken to extract this coefficient from data after zip up of the DIB had occurred. Interestingly, within our DIB formation method, the zip up time for DOPC with caffeine (∼50 s) appears to be much reduced as compared to estimations of 250 s for a comparative system in Lee et al.35 It is important to note that there is a difference in bulk oil phase between our system and that reported by Lee et al. We also observed this phenomenon in the case of thiamethoxam, where the zip-up time appeared to be ∼100 s. In the case of hydrophilic compounds it seems unlikely that they will affect the bilayer formation mechanism, particularly given permeant concentrations in the μM regime.
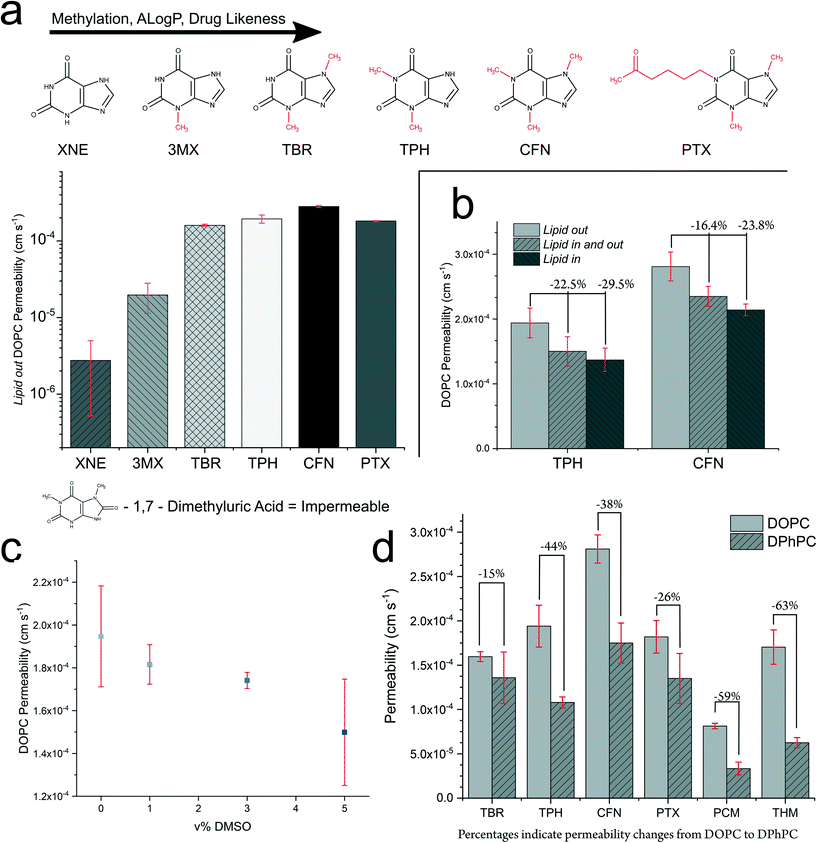 | ||
| Fig. 4 The UV-DIB can be used to perform a variety of experiments exploring transport phenomena across DIB membranes with diversely structured permeants. a Permeability assessed within systematically altered chemical space. Increasing drug favourable properties results in an increase in DOPC permeability. A clear cut off in the permeability is observed for xanthine (XNE) and 3 methyl xanthine (3MX). Note, that this data is plotted from Table 1, with a log scale applied to the y-axis. b DOPC permeability assessed for theophylline (TPH) and caffeine (CFN) show impact of DIB formation mechanism. Percentages indicate permeability change relative to lipid out. c The effect of DMSO v% on DOPC lipid out permeability to theophylline. d Exploration of the dependence of chemical structure on permeability change from DOPC to DPhPC. All experiments report mean from 3–8 repeats, error bars indicate 1 standard deviation from the mean, shorthand labels for permeants from Table 1. | ||
For the UV-DIB, an applicable model is proposed for interpreting a permeability coefficient from the monitored droplet. Following our observation of mass balance for the chosen permeants within our system (Fig. 3b, ESI† Fig. S3, S4, S6 and S7), a derivation of Fick's first law of diffusion, (which is included in the ESI† and is similar to one utilised by Lee et al.)35 provided determination of a permeability constant for each experiment, given by:
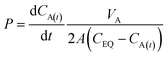 | (1) |
| CA(t) = (CA(t=0) − CEQ)e−kt + CEQ | (2) |
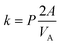 | (3) |
Permeability studies within the UV-DIB
Having determined a suitable method for calculating permeability constants, we next showcased the capacity of the system with a series of novel experiments. Table 1 presents the lipid out DOPC permeability of each studied permeant, spanning 3 orders of magnitude. One of the permeants, 1,7-dimethhyluric acid, was deemed impermeable.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D ph 7.4 calculated with MarvinSketch
D ph 7.4 calculated with MarvinSketch
| Permeant | P (×10−5 cm s−1) | P − SD (×10−5 cm s−1) | MW (Da) | HBA/HBD | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D (pH 7.4) D (pH 7.4) |
|---|---|---|---|---|---|
| Xanthine (XNE) | 0.276 | 0.224 | 152 | 3/3 | −0.372 |
| 3-Methylxanthine (3MX) | 1.97 | 0.842 | 166 | 3/2 | −1.15 |
| Theobromine (TBR) | 16.0 | 0.560 | 180 | 3/1 | −0.781 |
| Theophylline (TPH) | 19.4 | 2.35 | 180 | 3/1 | −0.952 |
| Caffeine (CFN) | 28.1 | 0.920 | 194 | 3/0 | −0.557 |
| Pentoxifylline (PTX) | 18.2 | 0.184 | 278 | 4/0 | 0.234 |
| 1,7-Dimethyluric acid | Impermeable | n/a | 196 | 5/4 | −1.21 |
| Paracetamol (PCM) | 8.13 | 0.310 | 151 | 2/2 | 0.910 |
| Thiamethoxam (THM) | 17.0 | 1.94 | 292 | 7/0 | 1.07 |
For our studied permeants, we sought to understand a chemical structure–permeability relationship within a reduced physicochemical space. To achieve this, the xanthine series was selected for its commercial availability, diversity in structures and therapeutic relevance. It can be seen that most of the compounds are readily permeable (<×10−4 cm s−1) across a DOPC bilayer. Xanthine to caffeine in Table 1 represents systematic changes to the xanthine chemical structure, in which the influence of functional groups can be explored. It is worth noting that theophylline, theobromine, caffeine and pentoxifylline are recognised therapeutic agents. Fig. 4a, shows that as the xanthine ring system is methylated, permeation increases. This increase in permeability occurs within a narrow MW window (152–278 Da) and interestingly the two lowest MW compounds (which are not recognised therapeutics) were the least permeable within this set. Between xanthine and 3-methylxanthine, the p value is 0.0104 and between 3-methylxanthine and theobromine, the p value is <0.0001 (two-tailed t-tests) indicating successful rank ordering of compounds across a large permeability range. The permeability decreases from caffeine (trimethylated) to pentoxifylline as one of the methyl groups is replaced with a 2-hexanone group. The HBA/HBD count changes systematically within this series(Table 1 and Fig. 4a). From xanthine to caffeine, the HBD count is inversely proportional to the permeability. 1-7-Dimethyluric acid is likely too hydrophilic to bypass the DIB and no significant difference was found between the leakage and permeation profiles (ESI† Fig. S6). Interestingly, thiamethoxam retains a high permeability with a HBA total of 7.
It is known that DMSO% can affect permeability, we explored the possibility of our assay for use with compounds otherwise insoluble in PBS requiring co-solvation with DMSO.49,50 This was important to assess if the UV-DIB is to have access to similar chemical scope as PAMPA. Theophylline is a water soluble therapeutic which acted as our control permeant. Solutions containing theophylline and varying v/v% DMSO were prepared. From 0–5% DMSO, we observe an apparent linear reduction in the permeability of a DOPC DIB to theophylline (Fig. 4c). We also noted a slight reduction in DIB success rate, although could form stable DIBs up to 5%. Most industrially relevant assays are performed at 1% v/v DMSO in cell culture, as to otherwise not induce cytotoxicity. Of importance, is that no statistical significance was observed in the permeability values for 0 and 1% DMSO.
We also studied the effect of lipid formation mechanism. Within DIB literature two, aqueous – oil monolayer formation mechanisms exist – that of lipid in, where lipid coating occurs from ruptured vesicles51 and that of lipid out – where oil-dissolved lipid arranges from the oil phase.52 We sought to investigate the effect of lipid location (and therefore monolayer formation mechanism) on permeability. For lipid in, a vesicle ‘sink’ exists for the permeant to interact with. This may introduce a secondary permeation process between the vesicle and the bulk aqueous phase, thus requiring equilibration between these two compartments prior to measuring permeation across the DIB. Furthermore, the location of lipid can affect monolayer formation.53 We assessed lipid in, lipid out and lipid in and out using theophylline and caffeine as permeants (Fig. 4b). With lipid in droplets, we noticed an increase in light scattering, noted by decreased signal intensities relative to the same concentration of permeant in a lipid out only droplet (ESI† Fig. S5). The lipid out permeability was higher than the comparative lipid in system. This trend was consistent for both theophylline and caffeine (two tailed t-test, p = 0.05 – Fig. 4b), however a slight increase in permeability was observed for paracetamol comparing lipid out vs. lipid in and out (ESI† Table S2). Combining both lipid in and lipid out (for theophylline and caffeine) does not demonstrate a significant difference in permeability from employing lipid in individually.
Lipid type is known to influence permeability, where DPhPC – a lipid derived from archaea slows permeation when compared to DOPC. This is due to branched acyl chain groups increasing the lipophilicity of the inner membrane and membrane packing through interdigitation. The lipid fluidity contributes to membrane permeability, recently explored in a fluorescent system,54 however like prior DIB permeation experiments a single permeant was used to explore this. Here we asked whether the chemical structure of the permeant, drastically affected the permeability reduction from DOPC to DPhPC. We observed a reduction in permeability from DOPC to DPhPC independent of chemical structure (Fig. 4d). Interestingly, permeability reduction appears independent of lipid formation mechanism (evidenced by experiments detailed in ESI† Table S2). The % reduction in permeability was not uniform, suggesting that permeant–lipid interactions are potential contributors to the overall reduction in permeability typically attributed to membrane fluidity and lateral pressure. This non uniformity resulted in a different rank ordering of the permeants comparing DOPC vs. DPhPC within the lipid out dataset. Theophylline and theobromine are structural isomers of one another (di-methylation of xanthine ring structure) however demonstrate differences in their % reduction. The methylation in theobromine is more splayed apart on the ring structure, which may accelerate the permeation process in DPhPC relative to theophylline.
We also built membranes of a higher complexity with regards to lipid composition. The use of custom lipid mixtures or plant tissue extracts to form DIBs in our system paves the way for enhanced understanding of the target lipidome in determining agrochemical uptake. A soy polar lipid mixture (40% PC, 16% PE, 11% PI and 33% other) was employed to form DIBs that more closely represent the lipidome of a typical model plant.55–59 These plant-like membranes were then used to perform permeability assays on caffeine and thiamethoxam. In our experiments, we noted a reduction in permeability across the soy membranes, compared to DOPC, independent of the DIB formation method (ESI† Table S2).
Discussion
Fundamental limitations have been apparent when studying simple diffusion through mimetic lipid membranes using techniques such as PAMPA or planar bilayers including; low sampling rate, reliance on fluorescence detection and inaccuracy in bio membrane representation.17,18,24,35,36,60 A central advantage of the PAMPA method, has been its ability to employ UV-spectroscopy. With this technique now readily accessible in DIBs, the chemical space which can be sampled across lipid bilayer interfaces is significantly increased. The capacity of the UV-DIB is highlighted by the study of the xanthine series. This study outlined a chemical structure–permeability relationship within a reduced chemical space, which mimics a typical drug design process. The compounds demonstrating the highest permeabilities are the known therapeutics within the series. As far as we are aware, this is the first time that a DIB platform has been used to systematically study structural motifs and permeation, a technique not possible through approaches utilising fluorescence, a property delicately connected to the permeant structure. The reduced permeability values for theophylline compared to caffeine (Table 1) agree with similar experiments in a simulation of membrane penetraton.61 The nuance of the permeant structure–permeability relationship is further evidenced by the variation in DOPC vs. DPhPC permeation seen for structural isomers (Fig. 4d). Furthermore, data in Fig. 4c shows that DIBs formed with 1% DMSO showed negligible reduction in the permeability, despite this effect being more pronounced at higher DMSO concentrations. As a proof of concept, we observed this trend with thiamethoxam in both lipid out and lipid in and out systems (ESI† Fig. S8). As DIBs were found to be stable under 5% DMSO, it stands to reason that chemical structure–permeability relationships could also be uncovered in drug/agrochemical discovery programmes in which water solubility in the μM regime may not be accessible for each candidate across a development pipeline.We chose the neonicotinoid insecticide thiamethoxam as a model permeant relevant to the agrochemical sector. A study using PAMPA with an egg PC lipid mixture, with two 300 μL compartments previously reported a membrane permeability of 0.66 × 10−6 cm s−1 for thiamethoxam,18 however in our 0.9 μL DIB we report a DOPC permeability of 17.0 × 10−5 cm s−1. A variance of this magnitude for thiamethoxam clearly illustrates the discrepancies observed between lipid bilayer and PAMPA systems. This discrepancy is explored thoroughly by Lee et al. using a planar bilayer system, with which our caffeine and paracetamol permeability values strongly agree despite a difference in bulk oil phase.35 Outside of expected inter lab variability, PAMPA values may be negatively skewed due to an excess of oil in the plate interface.
A further consideration lies in the effects of an unstirred water layer (UWL); a stagnant layer of water adjacent to the membrane surface with a concentration that differs from the bulk solution.62,63 For this reason, the thickness of this boundary layer can largely influence permeation kinetics. It is well documented that by stirring the bulk solution, the thickness of the UWL can be reduced – increasing the effective membrane permeability.17 A stagnant UWL thickness in a PAMPA well plate of up to 3800 μm has been reported.64,65 We have previously calculated a (0.8 μL) DIB UWL thickness of approximately 137 μm under stagnant conditions, providing a thickness much closer to those expected in vivo (estimated to be 40–100 μm in humans and up to 500 μm in some plants).24,65,66 It is worth noting this UWL thickness applies to a membrane separated by equal volume droplets. The use of DIBs thus provides a platform with a tuneable lipid composition as well as a fundamental membrane and unstirred water layer thickness more representative of those found in vitro.
An unclear relationship between Lipinski rule of 5 parameters, oral bioavailability and efficacy is emerging.20,67,68 A typical lipophilicity metric used to predict passive uptake is the octanol–water partition coefficient (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). A trend between sufficient aqueous solubility and lipophilicity metrics have been observed for molecules outside of Lipinski rules, particularly with respect to increased molecular weight (MW).69 Coupled to this, there is growing need for further interrogating the relationship between human oral absorption and simple diffusion across bio-representative lipid membranes. Within our data set, we found one impermeable compound indicating compounds below an A
P). A trend between sufficient aqueous solubility and lipophilicity metrics have been observed for molecules outside of Lipinski rules, particularly with respect to increased molecular weight (MW).69 Coupled to this, there is growing need for further interrogating the relationship between human oral absorption and simple diffusion across bio-representative lipid membranes. Within our data set, we found one impermeable compound indicating compounds below an A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P threshold (ESI† Table S2) may be deemed lipid bilayer impermeable. Permeation was readily observed amongst the di-methylated xanthine species in the series studied. Interestingly, it was the addition of a single carbonyl group to the di-methylated xanthine ring structure (1,7-dimethyluric acid) that attenuated bilayer permeation. This finding indicates the subtle relationship, previously uncovered in PAMPA, between chemical structure and interface permeation. Delineating the role of simple diffusion as a transport mechanism in the entire drug/agrochemical uptake process is thus critical,15 where current investigation in this area is derived from PAMPA. Despite rigorous assay development and employment in research and industry, PAMPA permeability values have indicated loose correlation with human intestinal absorption.70,71
P threshold (ESI† Table S2) may be deemed lipid bilayer impermeable. Permeation was readily observed amongst the di-methylated xanthine species in the series studied. Interestingly, it was the addition of a single carbonyl group to the di-methylated xanthine ring structure (1,7-dimethyluric acid) that attenuated bilayer permeation. This finding indicates the subtle relationship, previously uncovered in PAMPA, between chemical structure and interface permeation. Delineating the role of simple diffusion as a transport mechanism in the entire drug/agrochemical uptake process is thus critical,15 where current investigation in this area is derived from PAMPA. Despite rigorous assay development and employment in research and industry, PAMPA permeability values have indicated loose correlation with human intestinal absorption.70,71
The experiments outlined in Fig. 4b were performed using different lipid locations, impacting the monolayer formation method (lipid in/lipid out). In either of our lipid solutions, we used a buffer and oil lipid concentration (6.36 mM for DOPC) substantially greater than the critical micelle concentration (CMC) for DOPC (∼0.1–10 μM).72 It is worth noting our system satisfies a theoretical estimation of stability.47 For caffeine and theophylline, lipid out presented the fastest permeability, where there were no permeant–vesicle interactions in the aqueous phase. Lipid in and out appears to be slightly higher than lipid in alone, suggesting that the interplay of monolayer formation mechanisms may affect permeability.53 We also report a lower permeability value of paracetamol in a lipid out system when compared to a lipid in and out system (ESI† Table S2). Indeed, a permeating DIB system formed with lipid in may present a more complicated process if permeant structure dependent vesicle interaction is considered. Lipid asymmetry is a central motif of biological membranes,73 where lipid in formation can be used to impart asymmetry in DIBs.74 Recent work has also achieved DIB bilayer asymmetry through a lipid out technique indicating that permeation directionality with respect to bilayer leaflet composition is an important factor in model asymmetric membranes.34 Furthermore, many membrane proteins have been reliably studied in lipid in DIBs, through reconstitution of proteoliposomes.76 These features are central to modelling membrane transport phenomena and are otherwise not achievable in PAMPA.
It is important to impart biomimicry in permeability assays through biologically relevant lipid compositions.75 Bilayer lateral pressure has been highlighted as a key parameter correlating with DIB permeability.54 In light of the results presented in Fig. 4d, lipid–permeant interactions may also be important. In this work, we observed a permeability reduction in DPhPC (relative to DOPC) for the studied permeants, reporting a % reduction between 15 and 63% with a mean reduction of 40.8 ± 18.6%. Our lab has previously shown a comparative 60% reduction in the permeability value for resorufin (fluorescent dye).24 These % reductions do not correlate with MW, however, an apparent trend was observed with A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. Perhaps most importantly within the assayed dataset, the permeability rank ordering of compounds changed between DOPC and DPhPC (Fig. 4d). Our group has also shown that higher orders of compositional complexity within the membrane can be achieved in DIBs. This was demonstrated through measurement of resorufin permeability across custom lipid formulations modelling plasma membranes in tobacco, oats and Arabidopsis thaliana.25 In light of the results in Fig. 4, caffeine permeation was assessed across a soy polar lipid extract interface, noting a 44% decrease in permeability when compared to a pure DOPC interface (ESI† Table S2). The difference in permeability is likely due to the presence of PE & PI headgroups (16 & 11 wt% respectively) increasing the lateral pressure and thus, resistance to permeation.54,77–79
P. Perhaps most importantly within the assayed dataset, the permeability rank ordering of compounds changed between DOPC and DPhPC (Fig. 4d). Our group has also shown that higher orders of compositional complexity within the membrane can be achieved in DIBs. This was demonstrated through measurement of resorufin permeability across custom lipid formulations modelling plasma membranes in tobacco, oats and Arabidopsis thaliana.25 In light of the results in Fig. 4, caffeine permeation was assessed across a soy polar lipid extract interface, noting a 44% decrease in permeability when compared to a pure DOPC interface (ESI† Table S2). The difference in permeability is likely due to the presence of PE & PI headgroups (16 & 11 wt% respectively) increasing the lateral pressure and thus, resistance to permeation.54,77–79
The ability to monitor permeant concentration in two or more droplets simultaneously has previously been conducted reliably using fluorescence-based detection.28,30,32,80 The generation of high throughput data sets with relevant drug compounds will be required to thoroughly probe whether PAMPA permeability rank ordering agrees with pure lipid systems such as DIBs. Droplet networks are formed between multiple aqueous volumes, where network translocation has been studied with fluorescence.81,82 Previous developments in literature have seen the use of a microfluidic UV micro spectroscopy based approach for simultaneous acceptor and donor recording in a two droplet DIB, in the mM range.32 Use of a UV detection system to monitor permeation in more than two droplets has yet to be explored and therefore we believe that a variation of the UV-DIB chip could be employed to address some of the limitations currently in the literature and improve experimental throughput.
Conclusion
In this work, we present a significant advancement in in vitro modelling of permeant interactions with synthetic bilayers. Through a modular, adjustable and facile method we have fabricated chips on which passive membrane permeability assays using industrially relevant permeant concentrations can be conducted. We developed a chip fabrication method in which fiber optic cannulas are embedded into the chip, providing a low cost, low skill and novel approach in label-free assessment of passive permeation across a lipid membrane. By extracting the components of a UV-spectrometer our device is highly modular and tuneable to multiple conditions. The DIB method is shown to be fundamentally equipped for the investigation of transport phenomena through uncovering novel trends relevant to a variety of fields. We hypothesise that our method of simple diffusion characterisation can be exploited in a wide range of applications, including in varied chemical space, opening up the literature to further support DIB permeability studies and probe discrepancies between PAMPA and emerging model lipid membrane systems.24,30,32Methods
UV-DIB chip fabrication
The chip was first designed as a series of layers using CorelDraw Graphics Suite that could be arranged above one another and subsequently bonded together. The layers of varying thickness (0.5 mm or 1 mm) were cut from sheets of PMMA (Clarex 001, Weatherall Equipment and Instruments Ltd, UK) using a laser cutter (VLS 2.30, Universal Laser Systems, Austria). Once cut, chip components were rinsed using 70% v/v ethanol in water before drying each element thoroughly under a stream of nitrogen to remove any debris from the cutting process. A custom brushed aluminium heating block (two plates, 9 × 5 × 1.5 cm, L × W × H) equipped with supports (d = 0.4 cm) was used to stack the layers of PMMA. One of the plates was threaded such that four M6 screws were used to hold the two plates together and allow pressure to be applied evenly (measured using a torque wrench). The supports aided vertical alignment of the design elements prior to UV cannula insertion and thermal bonding.This fabrication method utilized two pre-cut UV cannulas equipped with ceramic SMA connection points (L = 10 mm, Ø = 200 μm, 0.22 NA), purchased from Thorlabs UK embedded into the chip. Layers 1 and 2 (1 mm each) were first aligned on the heating block. Etches on the surfaces of layers 3 and 4 (0.5 mm each) of the chip were introduced to guide the cannulas into alignment. Each of the ceramic connections were also fitted with PMMA supports designed to fit tightly into spaces cut into the chip, further aiding alignment (Fig. 2). Once alignment was complete, layers 4–6 (2.5 mm) were added above and bolts on the heating block were tightened to 0.5 N m−1 to apply uniform to the layers inside with a torque wrench. The press was heated to 170 °C for 45 minutes before removing it from the heating mantle and allowing the press to cool to room temperature. The total thickness of the chip was 5 mm.
Sample preparation
Stock solutions of each permeant were initially formed based on their water solubility, where subsequent serial dilution would provide a range of concentrations to conduct calibration and the permeability assay. Paracetamol, caffeine, theophylline and 1,7-dimethyluric acid were all purchased from Sigma-Aldrich, UK. Xanthine, theobromine and 3-methylxanthine were purchased from Tokyo Chemicals, UK. Pentoxifylline was purchased from Tocris Bioscience, UK. The thiamethoxam used was provided by Syngenta, UK. Phosphate buffered saline 1X from Corning was used to prepare the permeant solutions to a pH of 7.4.Liposomes were prepared by evaporating the solvent from a lipid-in-chloroform stock (DOPC, DPhPC and SPLE purchased from Avanti Polar Lipids, USA) under a stream of nitrogen to form a thin lipid film. The produced lipid films were subsequently desiccated overnight at room temperature to remove residual solvent. Each film was then rehydrated to a lipid concentration of 5 mg mL−1, using the various permeant concentrations formed previously and was vortexed for two minutes to ensure full suspension of the lipid film. The resulting vesicular mixture was then extruded 15 times through a 0.1 μm polycarbonate filter to form the final vesicle-containing aqueous permeant solution.
Due to the conjunction of both lipid in and lipid out methods in our system, lipid oil was also prepared. The lipid oil was prepared by forming a dried lipid film and was subsequently solubilised using hexadecane (purchased from Sigma Aldrich) to a lipid concentration of 5 mg ml−1. The resultant solution was vortexed until the entire film had dissolved.
Calibration and leakage
Spectra were captured using Oceanview software from Ocean Insight with a HR2000+ spectrometer and DH – mini light source (deuterium bulb power = 19 μW). Before collecting a spectrum, an appropriate integration time was selected. All spectra were taken as the average of 3 scans and a boxcar signal filter was applied. Calibration was conducted by first flooding the well with 70 μL of lipid oil (5 mg ml−1), a single 1.8 μL droplet of a given concentration was then injected into the DIB chamber between the two cannulas. A spectrum was captured every 5 s for a total of two minutes (n = 3). The mean intensity for each concentration within the monitoring period was used to form a calibration curve. A leakage assay was performed with a high concentration of the permeant and monitored for a longer time period to assess any changes in intensity with time.Permeability assays and data interpretation
To form the droplet interface bilayer used for permeability assessment, the well of the chip was first flooded with 70 μL of lipid in hexadecane (5 mg ml−1). A 0.9 μL droplet of each of the chosen donor and acceptor concentrations were pipetted into the incubation areas. The membrane area was confirmed via microscopy with subsequent analysis of the diameter via ImageJ. A circular area was calculated from three repeats, SE on this measurement was fed into propagated error equations (see ESI† derivation). Droplets were incubated at room temperature for 5 minutes for lipid monolayer formation in lipid in and out method and 10 min for lipid out and lipid in, before manually manipulating each droplet into the DIB chamber, forcing them into contact. Spectral collection would begin once both droplets were in the chamber and in contact and would occur thereon. Data was processed and analysed with a python script employing the standard libraries. Upon completion of a permeability assay, the chip was washed with 70% v/v ethanol in water to remove any oil and permeant residues ready for reuse. Data was fitted as detailed in the results section for further details see the ESI.†Author contributions
RS and FS contributed equally to this work. RS and FS performed experiments and writing. RS, FS and NB performed data analysis. RS, FS, NB, OC, NJB, RL, AJF, JH and LB designed experiments.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by Engineering and Physical Sciences Research Council (EPSRC) Institute of Chemical Biology Centre for Doctoral Training studentships, awarded to R. S. and F. S. The authors would like to thank James Hindley and David Miller for their useful feedback and review of this work. Grant Codes: (EP/S023518/1) & (EP/R511547/1).References
- A. J. Hulbert, N. Turner, L. H. Storlien and P. L. Else, Dietary fats and membrane function: Implications for metabolism and disease, Biol. Rev. Cambridge Philos. Soc., 2005, 80, 155–169 CrossRef CAS PubMed
.
- A. J. Hulbert, On the importance of fatty acid composition of membranes for aging, J. Theor. Biol., 2005, 234(2), 277–288 CrossRef CAS PubMed
.
- S. K. Abbott, P. L. Else, T. A. Atkins and A. J. Hulbert, Fatty acid composition of membrane bilayers: Importance of diet polyunsaturated fat balance, Biochim. Biophys. Acta, Biomembr., 2012, 1818(5), 1309–1317 CrossRef CAS PubMed
.
- C. Sohlenkamp and O. Geiger, Bacterial membrane lipids: Diversity in structures and pathways, FEMS Microbiol. Rev., 2015, 40, 133–159 CrossRef PubMed
.
- J. Wang, H. R. Juliani, D. Jespersen and B. Huang, Differential profiles of membrane proteins, fatty acids, and sterols associated with genetic variations in heat tolerance for a perennial grass species, hard fescue (Festuca Trachyphylla), Environ. Exp. Bot., 2017, 140, 65–75 CrossRef CAS
.
- A. J. Desai and L. J. Miller, Changes in the plasma membrane in metabolic disease: impact of the membrane environment on G protein-coupled receptor structure and function, Br. J. Pharmacol., 2018, 175, 4009–4025 CrossRef CAS PubMed
.
- T. Harayama and H. Riezman, Understanding the diversity of membrane lipid composition, Nat. Rev. Mol. Cell Biol., 2018, 19, 281–296 CrossRef CAS PubMed
.
- K. Peynshaert, J. Devoldere, A.-K. Minnaert, S. C. De Smedt and K. Remaut, Morphology and Composition of the Inner Limiting Membrane: Species-Specific Variations and Relevance toward Drug Delivery Research, Curr. Eye Res., 2019, 44(5), 465–475 CrossRef CAS PubMed
.
- M. Goldstein, T. Malchi, M. Shenker and B. Chefetz, Pharmacokinetics in Plants: Carbamazepine and Its Interactions with Lamotrigine, Environ. Sci. Technol., 2018, 52(12), 6957–6964 CrossRef CAS PubMed
.
-
J. M. McGregor, N. D. Doolittle, E. Youngers, S. D. Bell and E. A. Neuwelt, Pharmacokinetics of Drug Delivery Past the Blood-Brain Barrier, in Nervous System Drug Delivery: Principles and Practice, Elsevier, 2019, pp. 57–72 Search PubMed
.
- N. J. Yang and M. J. Hinner, Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins, Methods Mol. Biol., 2015, 1266, 29–53 CrossRef CAS PubMed
.
- G. van Meer, D. R. Voelker and G. W. Feigenson, Membrane lipids: where they are and how they behave, Nat. Rev. Mol. Cell Biol., 2008, 9(2), 112–124 CrossRef CAS PubMed
.
- M. Kansy, F. Senner and K. Gubernator, Physicochemical high throughput screening: Parallel artificial membrane permeation assay in the description of passive absorption processes, J. Med. Chem., 1998, 41, 1007–1010 CrossRef CAS PubMed
.
- J. A. Zabkiewicz, Spray formulation efficacy-holistic and futuristic perspectives, Crop Prot., 2007, 26(3), 312–319 CrossRef
.
-
M. Orsi and J. W. Essex, Chapter 4. Passive Permeation Across Lipid Bilayers: a Literature Review, in Molecular Simulations and Biomembranes, Royal Society of Chemistry, Cambridge, 2010, pp. 76–90 Search PubMed
.
- C. A. Larregieu and L. Z. Benet, Distinguishing between the permeability relationships with absorption and metabolism to improve BCS and BDDCS predictions in early drug discovery, Mol. Pharmaceutics, 2014, 11(4), 1335–1344 CrossRef CAS PubMed
.
- P. Berben, A. Bauer-Brandl, M. Brandl, B. Faller, G. E. Flaten and A.-C. Jacobsen,
et al., Drug permeability profiling using cell-free permeation tools: Overview and applications, Eur. J. Pharm. Sci., 2018, 119, 219–233 CrossRef CAS PubMed
.
- S. Hofstetter, A. Beck, S. Trapp and A. Buchholz, How to Design for a Tailored Subcellular Distribution of Systemic Agrochemicals in Plant Tissues, J. Agric. Food Chem., 2018, 66(33), 8687–8697 CrossRef CAS PubMed
.
- Y. Zhang, B. A. Lorsbach, S. Castetter, W. T. Lambert, J. Kister and N. X. Wang,
et al., Physicochemical property guidelines for modern agrochemicals, Pest Manage. Sci., 2018, 74(9), 1979–1991 CrossRef CAS PubMed
.
- M. D. Shultz, Two Decades under the Influence of the Rule of Five and the Changing Properties of Approved Oral Drugs, J. Med. Chem., 2019, 62, 1701–1714 CrossRef CAS PubMed
.
- H. Li, T. Zhao and Z. Sun, Analytical techniques and methods for study of drug-lipid membrane interactions, Rev. Anal. Chem., 2018, 37(1), 1–23 Search PubMed
.
- A. Cern, Y. Barenholz, A. Tropsha and A. Goldblum, Computer-aided design of liposomal drugs: In silico prediction and experimental validation of drug candidates for liposomal remote loading, J. Controlled Release, 2014, 173(1), 125–131 CrossRef CAS PubMed
.
- K. M. Giacomini, S. M. Huang, D. J. Tweedie, L. Z. Benet, K. L. R. Brouwer and X. Chu,
et al., Membrane transporters in drug development, Nat. Rev. Drug Discovery, 2010, 9, 215–236 CrossRef CAS PubMed
.
- N. E. Barlow, G. Bolognesi, S. Haylock, A. J. Flemming, N. J. Brooks and L. M. C. Barter,
et al., Rheological Droplet Interface Bilayers (rheo-DIBs): Probing the Unstirred Water Layer Effect on Membrane Permeability via Spinning Disk Induced Shear Stress, Sci. Rep., 2017, 7(1), 17551 CrossRef PubMed
.
- N. E. Barlow, E. Smpokou, M. S. Friddin, R. Macey, I. R. Gould and C. Turnbull,
et al., Engineering plant membranes using droplet interface bilayers, Biomicrofluidics, 2017, 11(2), 24107 CrossRef CAS PubMed
.
- M. Schaich, J. Cama, K. Al Nahas, D. Sobota, H. Sleath and K. Jahnke,
et al., An Integrated Microfluidic Platform for Quantifying Drug Permeation across Biomimetic Vesicle Membranes, Mol. Pharmaceutics, 2019, 16(6), 2494–2501 CrossRef CAS PubMed
.
- E. Stephenson and K. Elvira, Biomimetic artificial cells to model the effect of membrane asymmetry on chemoresistance, Chem. Commun., 2021, 57(54), 6534–6537 RSC
.
- M. A. Czekalska, T. S. Kaminski, K. Makuch and P. Garstecki, Passive and parallel microfluidic formation of droplet interface bilayers (DIBs) for measurement of leakage of small molecules through artificial phospholipid membranes, Sens. Actuators, B, 2019, 286, 258–265 CrossRef CAS
.
- S. Purushothaman, J. Cama and U. F. Keyser, Dependence of norfloxacin diffusion across bilayers on lipid composition, Soft Matter, 2016, 12(7), 2135–2144 RSC
.
- J. L. Korner, E. B. Stephenson and K. S. Elvira, A bespoke microfluidic pharmacokinetic compartment model for drug absorption using artificial cell membranes, Lab Chip, 2020, 20(11), 1898–1906 RSC
.
- M. R. Naylor, A. M. Ly, M. J. Handford, D. P. Ramos, C. R. Pye and A. Furukawa,
et al., Lipophilic Permeability Efficiency Reconciles the Opposing Roles of Lipophilicity in Membrane Permeability and Aqueous Solubility, J. Med. Chem., 2018, 61(24), 11169–11182 CrossRef CAS PubMed
.
- T. Nisisako, S. A. Portonovo and J. J. Schmidt, Microfluidic passive permeability assay using nanoliter droplet interface lipid bilayers, Analyst, 2013, 138(22), 6793–6800 RSC
.
- S. Bachler, D. Haidas, M. Ort, T. A. Duncombe and P. S. Dittrich, Microfluidic platform enables tailored translocation and reaction cascades in nanoliter droplet networks, Commun. Biol., 2020, 3(1), 769 CrossRef CAS PubMed
.
- S. Bachler, M. Ort, S. D. Krämer and P. S. Dittrich, Permeation Studies across Symmetric and Asymmetric Membranes in Microdroplet Arrays, Anal. Chem., 2021, 93(12), 5137–5144 CrossRef CAS PubMed
.
- Y. Lee, H.-R. Lee, K. Kim and S. Q. Choi, Static and Dynamic Permeability Assay for Hydrophilic Small Molecules Using a Planar Droplet Interface Bilayer, Anal. Chem., 2018, 90(3), 1660–1667 CrossRef CAS PubMed
.
- Y. Lee and S. Q. Choi, Quantitative analysis for lipophilic drug transport through a model lipid membrane with membrane retention, Eur. J. Pharm. Sci., 2019, 15(134), 176–184 CrossRef PubMed
.
- M. M. Islam, A. Loewen and P. B. Allen, Simple, low-cost fabrication of acrylic based droplet microfluidics and its use to generate DNA-coated particles, Sci. Rep., 2018, 8(1), 8763 CrossRef PubMed
.
- T. Trantidou, M. S. Friddin, K. B. Gan, L. Han, G. Bolognesi and N. J. Brooks,
et al., Mask-Free Laser Lithography for Rapid and Low-Cost Microfluidic Device Fabrication, Anal. Chem., 2018, 90(23), 13915–13921 CrossRef CAS PubMed
.
- S. A. Sarles and D. J. Leo, Physical encapsulation of droplet interface bilayers for durable, portable biomolecular networks, Lab Chip, 2010, 10(6), 710 RSC
.
- M. A. Holden, D. Needham and H. Bayley, Functional bionetworks from nanoliter water droplets, J. Am. Chem. Soc., 2007, 129(27), 8650–8655 CrossRef CAS PubMed
.
- R. T. Kelly and A. T. Woolley, Thermal bonding of polymeric capillary electrophoresis microdevices in water, Anal. Chem., 2003, 75(8), 1941–1945 CrossRef CAS PubMed
.
- H. Shadpour, H. Musyimi, J. Chen and S. A. Soper, Physiochemical properties of various polymer substrates and their effects on microchip electrophoresis performance, J. Chromatogr. A, 2006, 1111(2), 238–251 CrossRef CAS PubMed
.
- A. Liga, J. A. S. Morton and M. Kersaudy-Kerhoas, Safe and cost-effective rapid-prototyping of multilayer PMMA microfluidic devices, Microfluid. Nanofluid., 2016, 20(12), 164 CrossRef
.
- D. N. Schafer, E. A. Gibson, E. A. Salim, A. E. Palmer, R. Jimenez and J. Squier, Microfluidic cell counter with embedded optical fibers fabricated by femtosecond laser ablation and anodic bonding, Opt. Express, 2009, 17(8), 6068 CrossRef CAS PubMed
.
- A. K. Ghose, V. N. Viswanadhan and J. J. Wendoloski, Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods, J. Phys. Chem. A, 1998, 102(21), 3762–3772 CrossRef CAS
.
- V. Lukacova, M. Peng, G. Fanucci, R. Tandlich, A. Hinderliter and B. Maity,
et al., Drug-membrane interactions studied in phospholipid monolayers adsorbed on nonporous alkylated microspheres, J. Biomol. Screening, 2007, 12(2), 186–202 CrossRef CAS PubMed
.
- B. Guiselin, J. O. Law, B. Chakrabarti and H. Kusumaatmaja, Dynamic Morphologies and Stability of Droplet Interface Bilayers, Phys. Rev. Lett., 2018, 120(23), 238001 CrossRef CAS PubMed
.
- T. Trantidou, M. Friddin, Y. Elani, N. J. Brooks, R. V. Law and J. M. Seddon,
et al., Engineering Compartmentalized Biomimetic Micro- and Nanocontainers, ACS Nano, 2017, 11(7), 6549–6565 CrossRef CAS PubMed
.
- Y. Lee, P. A. Pincus and C. Hyeon, Effects of Dimethyl Sulfoxide on Surface Water near Phospholipid Bilayers, Biophys. J., 2016, 111(11), 2481–2491 CrossRef CAS PubMed
.
- R. K. Venkatraman and C. R. Baiz, Ultrafast Dynamics at the Lipid-Water Interface: DMSO Modulates H-Bond Lifetimes, Langmuir, 2020, 36(23), 6502–6511 CrossRef CAS PubMed
.
- S. Leptihn, O. K. Castell, B. Cronin, E. H. Lee, L. C. M. Gross and D. P. Marshall,
et al., Constructing droplet interface bilayers from the contact of aqueous droplets in oil, Nat. Protoc., 2013, 8(6), 1048–1057 CrossRef CAS PubMed
.
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. L. Hwang and R. Syeda,
et al., Droplet interface bilayers, Mol. BioSyst., 2008, 4(12), 1191–1208 RSC
.
- G. A. Venkatesan, J. Lee, A. B. Farimani, M. Heiranian, C. P. Collier and N. R. Aluru,
et al., Adsorption Kinetics Dictate Monolayer Self-Assembly for Both Lipid-In and Lipid-Out Approaches to Droplet Interface Bilayer Formation, Langmuir, 2015, 31(47), 12883–12893 CrossRef CAS PubMed
.
- V. Faugeras, O. Duclos, D. Bazile and A. R. Thiam, Membrane determinants for the passive translocation of analytes through droplet interface bilayers, Soft Matter, 2020, 16(25), 5970–5980 RSC
.
- C. P. Rochester, P. Kjellbom and C. Larsson, Lipid composition of plasma membranes from barley leaves and roots, spinach leaves and cauliflower inflorescences, Physiol. Plant., 1987, 71(3), 257–263 CrossRef CAS
.
- H. A. Schwertner and J. B. Biale, Lipid composition of plant mitochondria and of chloroplasts, J. Lipid Res., 1973, 14(2), 235–242 CrossRef CAS
.
- A. H. Thomas, Á. Catalá and M. Vignoni, Soybean phosphatidylcholine liposomes as model membranes to study lipid peroxidation photoinduced by pterin, Biochim. Biophys. Acta, Biomembr., 2016, 1858(1), 139–145 CrossRef CAS PubMed
.
- S. K. Kalra and J. L. Brooks, Lipids of ripening tomato fruit and its mitochondrial fraction, Phytochemistry, 1973, 12(3), 487–492 CrossRef CAS
.
- P. G. Roughan and R. D. Batt, The glycerolipid composition of leaves, Phytochemistry, 1969, 8(2), 363–369 CrossRef CAS
.
- M. Velický, D. F. Bradley, K. Y. Tam and R. A. W. Dryfe, In situ artificial membrane permeation assay under hydrodynamic control: Permeability-ph profiles of warfarin and verapamil, Pharm. Res., 2010, 27(8), 1644–1658 CrossRef PubMed
.
- M. Paloncýová, K. Berka and M. Otyepka, Molecular insight into affinities of drugs and their metabolites to lipid bilayers, J. Phys. Chem. B, 2013, 117(8), 2403–2410 CrossRef PubMed
.
- J. Dainty, Water Relations of Plant Cells, Adv. Bot. Res., 1963, 1(C), 279–326 CAS
.
- T. J. Pedley, Calculation of unstirred layer thickness in membrane transport experiments: A survey, Q. Rev. Biophys., 1983, 16(2), 115–150 CrossRef CAS PubMed
.
- I. J. Hidalgo, K. M. Hillgren, G. M. Grass and R. T. Borchardt, Characterization of the Unstirred Water Layer in Caco-2 Cell Monolayers Using a Novel Diffusion Apparatus, Pharm. Res., 1991, 8(2), 222–227 CrossRef CAS PubMed
.
- A. Avdeef, P. E. Nielsen and O. Tsinman, PAMPA - A drug absorption in vitro model: 11. Matching the in vivo unstirred water layer thickness by individual-well stirring in microtitre plates, Eur. J. Pharm. Sci., 2004, 22(5), 365–374 CAS
.
- J. Dainty, The polar permeability of plant cell membranes to water, Protoplasma, 1963, 57(1–4), 220–228 CrossRef CAS
.
- P. Matsson and J. Kihlberg, How Big Is Too Big for Cell Permeability?, J. Med. Chem., 2017, 60(5), 1662–1664 CrossRef CAS PubMed
.
- W. Yang, P. Gadgil, V. R. Krishnamurthy, M. Landis, P. Mallick and D. Patel,
et al., The Evolving Druggability and Developability Space: Chemically Modified New Modalities and Emerging Small Molecules, AAPS J., 2020, 22(2), 21 CrossRef PubMed
.
- M. R. Naylor, A. M. Ly, M. J. Handford, D. P. Ramos, C. R. Pye and A. Furukawa,
et al., Lipophilic Permeability Efficiency (LPE) reconciles the opposing roles of lipophilicity in membrane permeability and aqueous solubility, J. Med. Chem., 2018, 61(24), 11169–11182 CrossRef CAS PubMed
.
- C. Zhu, L. Jiang, T. M. Chen and K. K. Hwang, A comparative study of artificial membrane permeability assay for high throughput profiling of drug absorption potential, Eur. J. Med. Chem., 2002, 37(5), 399–407 CrossRef CAS PubMed
.
- H. Yu, Q. Wang, Y. Sun, M. Shen, H. Li and Y. Duan, A new PAMPA model proposed on the basis of a synthetic phospholipid membrane, PLoS One, 2015, 10(2), e0116502 CrossRef PubMed
.
- G. A. Venkatesan, G. J. Taylor, C. M. Basham, N. G. Brady, C. P. Collier and S. A. Sarles, Evaporation-induced monolayer compression improves droplet interface bilayer formation using unsaturated lipids, Biomicrofluidics, 2018, 12(2), 024101 CrossRef PubMed
.
- J. H. Lorent, K. R. Levental, L. Ganesan, G. Rivera-Longsworth, E. Sezgin and M. Doktorova,
et al., Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape, Nat. Chem. Biol., 2020, 16(6), 644–652 CrossRef CAS PubMed
.
- N. E. Barlow, H. Kusumaatmaja, A. Salehi-Reyhani, N. Brooks, L. M. C. Barter and A. J. Flemming,
et al., Measuring bilayer surface energy and curvature in asymmetric droplet interface bilayers, J. R. Soc., Interface, 2018, 15(148), 20180610 CrossRef CAS PubMed
.
- J. Korner and K. Elvira, The role of temperature in the formation of human-mimetic artificial cell membranes using droplet interface bilayers (DIBs), Soft Matter, 2021, 17(39), 8891–8901 RSC
.
- M. Allen-Benton, H. E. Findlay and P. J. Booth, Probing membrane protein properties using droplet interface bilayers, Exp. Biol. Med., 2019, 244, 709–720 CrossRef CAS PubMed
.
- D. Huster, A. J. Jin, K. Arnold and K. Gawrisch, Water permeability of polyunsaturated lipid membranes measured by 17O NMR, Biophys. J., 1997, 73(2), 855–864 CrossRef CAS PubMed
.
- M. Palaiokostas, W. Ding, G. Shahane and M. Orsi, Effects of lipid composition on membrane permeation, Soft Matter, 2018, 14(42), 8496–8508 RSC
.
- J. De Gier, J. G. Mandersloot and L. L. M. Van Deenen, Lipid composition and permeability of liposomes, Biochim. Biophys. Acta, Biomembr., 1968, 150(4), 666–675 CrossRef CAS
.
- B. Schlicht and M. Zagnoni, Droplet-interface-bilayer assays in microfluidic passive networks, Sci. Rep., 2015, 5, 1–8 Search PubMed
.
- M. S. Friddin, G. Bolognesi, Y. Elani, N. J. Brooks, R. V. Law and J. M. Seddon,
et al., Optically assembled droplet interface bilayer (OptiDIB) networks from cell-sized microdroplets, Soft Matter, 2016, 12(37), 7731–7734 RSC
.
- F. G. Downs, D. J. Lunn, M. J. Booth, J. B. Sauer, W. J. Ramsay and R. G. Klemperer,
et al., Multi-responsive hydrogel structures from patterned droplet networks, Nat. Chem., 2020, 12(4), 363–371 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc01155c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |