Open Access Article
Open Access Article3D-Printed microfluidic device for protein purification in batch chromatography
Taieb
Habib
 a,
Chantal
Brämer
a,
Christopher
Heuer
a,
Chantal
Brämer
a,
Christopher
Heuer
 a,
Jan
Ebbecke
a,
Sascha
Beutel
a,
Jan
Ebbecke
a,
Sascha
Beutel
 a and
Janina
Bahnemann
*ab
a and
Janina
Bahnemann
*ab
aInstitute of Technical Chemistry, Leibniz University Hannover, Callinstraße 5, 30167 Hannover, Germany. E-mail: jbahnemann@iftc.uni.hannover.de
bCell Culture Technology, Technical Faculty, Bielefeld University, Universitätsstraße 25, 33625 Bielefeld, Germany
First published on 27th January 2022
Abstract
Modern 3D printers enable not only rapid prototyping, but also high-precision printing—microfluidic devices with channel diameters of just a few micrometres can now be readily assembled using this technology. Such devices offer a myriad of benefits (including miniaturization) that significantly reduce sample and buffer volumes and lead to lower process costs. Although such microfluidic devices are already widely used in the field of biotechnology, there is a lack of research regarding the potential of miniaturization by 3D-printed devices in lab-scale chromatography. In this study, the efficacy of a 3D-printed microfluidic device which provides a substantially lower dead-volume compared to established chromatography systems is demonstrated for batch purification applications. Furthermore, this device enables straightforward integration of various components (such as microfluidic valves and chromatographic units) in an unprecedentedly flexible fashion. Initial proof-of-concept experiments demonstrate successful gradient elution with bovine serum albumin (BSA), and the purification of a pharmaceutically relevant IgG monoclonal antibody (mAb).
1. Introduction
Cost and efficiency considerations in research laboratories often require lengthy and complex upfront planning for experimental design. Simply testing different experimental setups is, unfortunately, a highly uneconomical approach for many reasons: systems can be difficult to set up and dismantle, the increased consumption of materials typically leads to high costs and a great burden on the environment, etc.1–5 Especially in the case of custom-made microfluidic devices, production can take considerable time and effort6–8—and to make matters worse, the optimal experimental design is not always readily apparent at the outset; indeed, it may only become evident after multiple experimental iterations have already been attempted. Improved techniques for the fast and cheap production of various prototypes are therefore vitally needed in the modern research laboratory setting.6In recent years, high-resolution additive manufacturing techniques (referred to collectively herein as 3D printing) have emerged and facilitated the rapid prototyping of miniaturized and microfluidic systems in biotechnology.9,10 These techniques enable the production of multiple complex three-dimensional structures in a single step, by loading computer-aided design (CAD) files directly into the printer's software. The resulting devices can be quickly adjusted to changing experimental requirements, simply by modifying the design in the CAD software. Furthermore, 3D printing is highly automated and requires no special equipment or facilities. 3D printing thus represents a vital alternative to much-less-flexible standard techniques, such as soft-lithography (i.e., PDMS-based systems), which often require cleanroom procedures and the fabrication of master molds.8–13
One common 3D printing technique is termed multijet printing, which uses a photocurable resin which solidifies under UV light.14,15 This resin is applied through a printhead consisting of an array of nozzles which act collectively to create the desired 3D object (layer by layer). A wax-like support material (which is later removed in a post-processing step) is also initially printed alongside the resin, in order to facilitate complex overhanging structures.14,16 Post-processing typically involves heating the object, followed by incubation in a warm oil bath with ultrasonic agitation.14–17 Since this is the only step in the production process that requires manual labour, 3D printing provides tremendous advantages over traditional multistep soft photolithography with PDMS.12,18 Biocompatible resins for multijet printing are also widely available, facilitating applications of this technology in the field of biotechnology.19–22
In chromatography applications (and related liquid handling), one critical challenge is posed by the requirement of long tubing causing high dead volumes, which can render processes uneconomical.23 In addition, this challenge is often compounded by the need for various connectors and numerous different tubes to connect the chromatography system to its periphery (e.g., buffer solutions, pumps, fraction collectors, detectors).
Through 3D printing, complex microfluidic channels with custom-made connectors can be easily designed and tested, and the whole system can also be miniaturized (thereby reducing fluid consumption and associated costs). Additionally, microfluidic structures—such as mixer units—can be readily integrated and thereby provide the means to mix fluids in small dimensions and consequently decrease dead volumes.24
In this study a 3D-printed microfluidic device for batch chromatography in a fully automated fashion is presented combining these important advantages of microfluidics. By replacing conventional tubings and directly mounting the valves needed for an automated purification, system miniaturization is achieved. In addition, the device can be readily modified (rapid prototyping) to changing experimental requirements.
2. Experimental
2.1 Materials
BSA was purchased from Sigma-Aldrich (now Merck KGaA, Darmstadt, Germany). All other chemicals were supplied by Carl-Roth (Karlsruhe, Germany). The supernatant containing the mAb IgG was obtained from cell cultivation with Chinese hamster ovary (CHO) cells.2.2 Design of a four-valve device for a single chromatographic unit
To facilitate batch chromatography, we wanted the device to be capable of automating the various processes of washing and equilibrating the entire system; sample application; elution with subsequent fraction collection; and, finally, cleaning the system. In order to perform the batch chromatography, four inlets (1. sample, 2. wash buffer, 3. elution buffer, and 4. cleaning-in-place (CIP) buffer), four microfluidic valves, and two outlets (waste and elution fraction) are required. The valves are 3/2 solenoid valves with three ports and two switching positions. These ports are COM (common) port, which is always operated, NO (normally open) port, which is operated in the default state, and NC (normally closed) port, which is operated when activated. Additionally, the device should integrate a mixer to generate a gradient for stepwise elution. A 4-channel peristaltic pump is used to introduce the four different fluids into the device through their respective inlets. A schematic overview of the system and its various components is provided in Fig. 1.The sample is loaded onto the device via the first inlet. The channel of the first inlet then leads to valve V2, which is responsible for adding sample or buffer to the chromatographic unit. The buffers are pumped to the device through inlets two through four, with the wash buffer applied through the second inlet, the elution buffer applied through the third inlet, and the CIP buffer applied through the fourth inlet. The channels for the wash and elution buffers actually converge within the device, however, such that the buffers can be mixed by means of an integrated mixer. The channels of the buffers all flow into valve V1, which leads to either V2 (and thus to the chromatographic unit), or to V3 (which bypasses the chromatographic unit and leads to the waste). The chromatographic unit and the subsequent spectrophotometer are connected via tubings which lead to valve V4. This last valve is used to either direct the liquid to V3 (and thus into the waste), or to direct it into the sample collector.
2.3 Fabrication of microfluidic devices by 3D printing
All 3D models were designed using the 3D-CAD-program SOLIDWORKS (Dassault Systèmes SolidWorks Corporation). The multijet printer ProJet® MJP 2500 Plus (3D Systems, Rock Hill, SC, USA) was then used to produce the microfluidic devices. The polyacrylate photocurable resin VisiJet® M2R-CL (3D Systems, Rock Hill, SC, USA) was used to print the actually device, and hydroxylated wax (VisiJet® M2 SUP, 3D Systems, Rock Hill, SC, USA) acted as support material.19 The printer was operated in HD mode, with a nominal resolution of 800 × 900 × 790 dpi and a layer resolution of 32 μm.After printing, the device underwent several post-processing steps according to Siller et al.: first, the printed parts are cooled at −18 °C for 5 minutes, followed by incubation in a hot water vapor bath, a hot ultrasonic oil bath and lastly in a hot ultrasonic water bath with detergent.20
2.4 Experimental setup
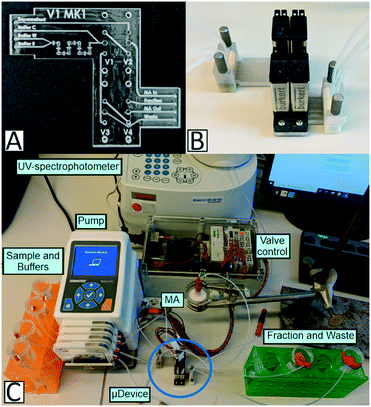
A peristaltic pump (Reglo ICC, Ismatec, Wertheim, Germany) was used to pump the buffers and supernatant solutions into the microfluidic device with a flow rate of 5 mL min−1. To connect the tubes (BOLA PTFE tubing, Bohlender GmbH, Grünsfeld, Germany; inner diameter of 0.8 mm, outer diameter of 1.6 mm) with the device, a microfluidic connector (4-way linear connector, the Dolomite Centre Ltd, Royston, England) with a 4 mm top interface (top interface 4-way 4 mm, the Dolomite Centre Ltd, Royston, England) and featuring a female to male luer adapter and a flangeless fitting (IDEX Health & Science, LLC, Middleboro, USA) was attached to the device. Four flangeless microfluidic valves (Whisper Valve Type 6724; Bürkert GmbH & Co. KG) were directly attached, via screws, into the integrated threads of the 3D-printed microfluidic device.The functionality tests of the device were conducted with the aid of a blue dye solution. Here, the device was directly connected to a UV-spectrophotometer (GENESYS 10S UV-vis) and the absorbance at 430 nm was measured. Initially, the device was washed with water. Then, the dye solution and water were pumped together. Afterwards, either the dye solution or water were pumped through the device.
For the batch chromatography experiments, the respective membrane adsorber (MA) as the chromatographic unit was connected to the device via tubes, and a UV-spectrophotometer was used for protein detection. Since BSA is negatively charged at neutral pH, an anion exchange (AEX) MA (Sartobind®Q75, Sartorius AG, Göttingen, Germany) was used. In the case of mAb IgG, a Sartobind® Protein A (Sartorius AG, Göttingen, Germany) MA was used, which utilizes the interaction of Protein A with immunoglobulins.25,26 The setup was run in a bind-and-elute mode, where the target protein first binds to the MA and can then be eluted by an appropriate elution buffer.27,28 The detection was performed in a UV flow-through cuvette, with an optical path length of 2 mm (Hellma GmbH & Co. KG) that was directly positioned in the spectrophotometer.
In the case of BSA, a 1 g L−1 solution in a 20 mM KH2PO4 buffer (pH 7) was used. This buffer also served as the equilibration and wash buffer for protein purification. Elution was performed stepwise by generating a gradient with the wash buffer and a 20 mM KH2PO4 buffer containing 1 M NaCl (pH 7). The two buffers were simultaneously pumped through their respective inlets—albeit, at different flow rates. Initially, the elution and wash buffer were pumped for two minutes with a flow rate of 0.5 mL min−1 and 4.5 mL min−1, respectively, and then mixed by the HC mixer. As a result, the buffer mixture has a NaCl concentration of 0.1 mol L−1. This was followed by three more minutes of elution with a NaCl concentration of 0.2 mol L−1 and then for a further three minutes with a concentration of 0.3 mol L−1. Finally, the elution was performed for five minutes with the pure elution buffer, and thus with a NaCl concentration of 1 mol L−1.
For the mAb experiments, the supernatant was also diluted in wash buffer—but to a concentration of 0.2 g L−1. 1× PBS buffer with a pH of 7.4 was used as wash buffer. Contrary to the purification of BSA, a 0.1 M citrate buffer with 0.15 M NaCl and a pH of 3.5 was used for elution. Furthermore, the elution was performed solely with the elution buffer without a gradient. In the mAb experiments, several elution fractions were collected and subsequently analysed via high-pressure liquid chromatography (HPLC). Additionally, a cleaning in place (CIP) step was performed with 1 M NaCl and 50 mM NaOH buffer, followed by regeneration of the MA. In the CIP step, impurities such as mAb fragments and aggregates or unspecifically bound host cell proteins (HCPs) are removed from the membrane. Initially, the system is flushed with the buffers. In this process, the tubing of the peristaltic pump is rinsed with the respective buffer and the device with valves, chromatographic unit, UV unit and the remaining tubing is rinsed with wash buffer. This consumes 470 μL of elution and CIP buffer and 1828 μL of wash buffer. Afterwards, the MA is equilibrated with 6 column volumes (CV) wash buffer. Subsequently, 2 CV of the diluted supernatant is loaded onto the MA. The MA is then washed with 4 CV of wash buffer. Thereupon, the elution is conducted with 5 CV of elution buffer, followed by 2.5 CV of CIP buffer.
The components of the system were controlled and automated by a program based on python™.
2.5 Analysis of elution fractions via HPLC
A Chromaster HPLC system (VWR International, Radnor, PA, USA) with a size exclusion chromatography (SEC) column (Yarra™ 3 μm SEC-3000, Torrance, CA, USA) was used to perform the actual substantive analysis. For the analysis, a buffer with 0.1 M NaPO4 (NaH2PO4 and Na2HPO4 mixed) with 0.1 M Na2SO4 at pH 6.6 was used. The analysis was performed at a flow rate of 1 mL min−1 and with a total run time of 20 min. The temperature of the column oven and the autosampler were 25 °C and 10 °C, respectively, and the injection volume was 5 μl. Samples were filtered through a 0.22 μm cellulose acetate membrane that was obtained from Sartorius Stedim Biotech GmbH (Göttingen, Germany). For the calibration of the system, purified mAb from Sartorius Stedim Biotech GmbH was used.The recovery rate of the batch purification was calculated using the ratio of the mAb amount before purification and the mAb amount in the elution fractions determined by HPLC.
3. Results and discussion
3.1 Construction and assembly of the 3D-printed microfluidic device
This device was designed to be capable of fully automating all steps in batch purification, as well as significantly reducing the dead volume in the process by replacing conventional tubing and using microfluidic connectors. Fig. 2 illustrates this 3D-printed device and its components, derived from the chromatography scheme in Fig. 1. The inlets (Fig. 2A) that are used to introduce buffer solutions contain microfluidic ball check valves that minimize the backflow from these converging channels. These ball check valves consist of a free-flowing sphere within the channel, a constriction upstream of the sphere, and a column as a barrier downstream of the sphere to allow fluids to flow only unilaterally (Fig. 2B). Additionally, this device also integrated a microfluidic HC mixer (Fig. 2C). HC mixers split and recombine the fluids (SAR principle), and contain H-shaped structures which further increase mixing efficiency.24,29 The channels within the device were designed to be 1 mm in both height and width. To optimize the flow behaviour of fluids and minimize dead volume within the channels, the edges of the channels were rounded off. In order to mount the microfluidic valves (Whisper Valve Type 6724; Bürkert GmbH & Co. KG) on the device, M2 threads were directly 3D-printed into the final device (Fig. 2D). | ||
| Fig. 2 CAD-Drawing of the novel 3D-printed microfluidic device for batch chromatography. (A) 3D-Printed microfluidic device with an inlet and an outlet connector as means to interface with the periphery of the chromatographic system via polytetrafluoroethylene (PTFE) tubings. The channels within the device are 1 mm in height. The device has three distinct components in order to perform the batch purification; a HC mixer for, a ball check valve and integrated 3D-printed M2 threads for directly mounting the four 3/2 valves onto the device. (B) The ball check valve consists of a free-flowing sphere in the channels, a tailback placed before the sphere with an opening smaller than the diameter of the sphere and a pillar. The ball check valve allows fluids to only flow in one direction. (C) Integrated HC mixer, which enables rapid mixing with low dead-volumes. The mixer consists of seven identical mixing units, which thoroughly mix two fluids at laminar flow rates.23 (D) Principal mechanism of the 3/2 valves. When deactivated, fluids can only flow between the NO and the COM ports. By activating the valves, the fluids can flow between the NC and the COM ports. | ||
In general, the channels within the device were designed to be 1 mm in both height and width. To optimize the flow behaviour of fluids and minimize dead volume within the channels, the edges of the channels were rounded off.
The dead volume of all channels in the device was 291.7 μl. In addition, the internal volume of the microfluidic valves was 38 μl each, summed up to a total of 152 μl. Together with the connections for the buffers, and to and from the MA and UV-spectrophotometer, the maximum dead volume of the whole system (see Fig. 3) in operation was 1828 μl.
3.2 Functionality test of the 3D-printed device
One of the key features of the fabricated microfluidic device is the integrated HC mixer. Integrated micromixers—such as the HC mixer—enable rapid and efficient mixing of fluids in a microfluidic environment, thereby reducing the dead volume and final size of the system.24,30,31 Microfluidic mixers already found application in liquid chromatography and facilitated thorough mixing of two fluids with low internal volumes and short mixing distances for channels with heights of about 100 μm.30,31 Enders et al. presented a comprehensive comparison of four 3D-printed microfluidic mixers and demonstrated the advantages of the HC-mixer, a split-and-recombine micromixer, for channel dimensions and flowrates typical for 3D-printed devices.24 The HC mixer design is used in the microfluidic device developed here due to its ease of reproducibility and integrability.In an initial experiment, the mixer efficiency was examined with the aid of a blue dye solution. For this purpose, the dye solution and water were introduced separately and together through inlets 3 and 4, respectively, and subsequently mixed by the micromixer. By using a spectrophotometer, the absorbance of the mixture was measured at 430 nm as shown in Fig. 4. The intensity of the absorbance signal at 430 nm directly correlates to the concentration of the blue dye solution, and thus can be used to evaluate the mixing efficiency. The dye solution showed an absorbance maximum of 0.2 AU, while water and the completely mixed solution should have an absorbance of 0.1 AU. In addition, this experiment served to assess the setup for both its tightness and stability.
Thorough mixing could be observed after 30 seconds. Furthermore, when the dye solution was solely pumped, an absorbance of 0.2 AU could be observed. When water was pumped, no significant absorbance was measured. This established that the device was capable of rapidly mixing two liquids, and thus was suitable for generating a gradient. Furthermore, no leakages were observed under these flow rates—which suggested that the device could be used for batch chromatography.
3.3 Purification of a model protein in batch mode
In a subsequent proof-of-concept experiment, the batch purification of the model protein BSA was performed using the 3D-printed device. A BSA solution (1 g L−1) was introduced through inlet 1, and was thereby loaded onto the AEX MA. This was followed by a spectrophotometer that detected the absorbance of the sample at 280 nm, in order to analyse the sample for proteins. Subsequently, the sample was directed via valve 4 to valve 3 and pumped to the waste. Afterwards, the MA was washed by pumping the wash buffer through inlet 3 and activating valve 2. For the gradient elution, wash buffer and elution buffer were pumped through inlet 3 and 4, respectively. The two buffers were mixed in the HC mixer, and then subsequently pumped to the MA. However, the flowrates of the respective pump channels were adjusted in order to achieve the desired NaCl concentration.The chromatogram (Fig. 5) revealed that no proteins were detectable during the sample application and subsequent washing off in the MA. This suggests that the MA was able to specifically bind the BSA. While a NaCl concentration of 0.1 mol L−1 did not lead to a significant elution of BSA, a concentration of 0.2 mol L−1 NaCl resulted in a clear peak in the chromatogram. Since BSA of technical purity was used, impurities were observed at higher NaCl concentrations.
In total, 90.4% of the model protein BSA could be purified with a NaCl concentration of 0.2 mol L−1. This coincides with the results of previous works by Brämer et al.32,33 Additionally, a gradient elution was successfully performed via means of the integrated microfluidic HC mixer.
3.4 Purification of a mAb
The purification of the monoclonal antibody was performed with diluted supernatant from a CHO cell cultivation containing 0.2 g L−1 mAb—and, as with the purification of BSA, the purification of the monoclonal antibody was monitored by the spectrophotometer (see Fig. 6). A Sartobind® Protein A MA was used for capturing the mAb.Drawing on the works of Brämer et al., the elution of the mAb was performed using a citrate buffer with a pH of 3.5 and 0.15 M NaCl.32,33 The optimized buffer increased the solubility of the mAb, and also enhanced elution of the MA. Contrary to the purification of BSA, no gradient elution was performed. After elution, a CIP step followed by the regeneration of the MA were performed.
Since the sample containing the mAb also contained a variety of HCP (which are unable to bind to the MA), a peak could be observed in the chromatogram during the sample application. However, the mAb specifically binds to the MA, and could thus be isolated from the supernatant. A small fraction of the flowthrough was collected for HPLC analysis, and the rest was discarded via valve 3. After the sample application, the MA was washed. The elution of the mAb was then performed for 4 min and collected via valve 4. The peak for the mAb could be observed after 1 min; at that point, the elution was collected in three fractions, which were analysed via HPLC. Finally, a small CIP peak was also observed, which was collected for further HPLC analysis.
Through the analysis of the samples via HPLC, the recovery rate was calculated to be 87.4%. In the elution peak, a high antibody purity of 90.2% could be observed. These results are comparable with those found in literature (between 85–95% for recovery and 90–98% for purity) for batch purifications of mAb.32–34 Notably, the operational time and buffer consumption were drastically reduced—by >50% and >30%, respectively—in comparison to established systems.35–37 This underscores the substantial benefits that can be derived from a microfluidic system.
Conclusions
In this study, a 3D-printed microfluidic device was designed and tested for the automated purification of proteins in batch. Compared to other established manufacturing methods (such as soft photolithography with PDMS), 3D printing did, in fact, enable the easy integration of microfluidic valves via 3D-printed threads and chromatography units with a subsequent spectrophotometer for monitoring via microfluidic connectors. With the aid of a dye solution, the system was shown to be capable of thoroughly and evenly mixing fluids within a few seconds.In a first proof-of-concept experiment, a MA with an AEX membrane was used to purify BSA. By means of the integrated HC mixer, a stepwise gradient elution was successfully achieved.
Finally, a mAb was successfully purified with a Protein A MA from a supernatant solution obtained from a CHO cell cultivation. Within 12 minutes, 30 mL of sample could be purified—a temporal reduction of nearly 50% compared with established processes.32–34 In addition, the low buffer consumption facilitated the screening of optimal buffers.
While this device was used for the automated batch purification of proteins, the authors firmly believe that the principle of a 3D-printed microfluidic device as a means to miniaturize conventional tubings in chromatographic systems can find application in larger and more complex systems in the analytical scale. Continuous chromatography systems such as the periodic counter-current chromatography (PCCC) heavily rely on the complex circuitry facilitated by a multitude of valves. Although these systems ensure higher utilization of the chromatographic units, they are difficult to operate due to their complexity, and they have high dead volumes. Through 3D-printing, the device can be easily upscaled in its complexity and may therefore be used in continuous protein purification. Furthermore, the system can be simply connected to continuous cultivation systems. However, the microfluidic nature of the device limits its use as a system to small-scale purification of target proteins—which can be particularly useful in the field of bioprocess optimization where small-scale bioreactors (such as Ambr® systems) are used. Due to its fabrication via rapid prototyping, the presented device also offers the potential for rapid and cost-effective design adaptation and direct connection to various bioreactors. This underlines the capabilities of 3D printing as a tool for custom chromatography systems and the significance of the miniaturised 3D-printed device presented in this work.
Author contributions
Conceptualization, J. B., S. B., T. H. and C. B.; methodology, T. H. and C. B.; investigation, T. H. and J. E.; data curation, T. H.; writing-original draft preparation, T. H.; writing-review and editing, T. H., C. H. and J. B.; visualization, T. H.; supervision, J. B.; project administration, J. B.; funding acquisition, J. B.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support of the German Research Foundation (DFG) via the Emmy Noether Programme (346772917).Notes and references
- R. M. Izzo, Waste minimization and pollution prevention in university laboratories, Chem. Health Saf., 2000, 7, 29–33 CrossRef.
- G. Bistulfi, Reduce, reuse and recycle lab waste, Nature, 2013, 502, 170 CrossRef CAS PubMed.
- M. A. Urbina, A. J. R. Watts and E. E. Reardon, Labs should cut plastic waste too, Nature, 2015, 528, 479 CrossRef CAS PubMed.
- S. Webb, Going Greener, BioTechniques, 2016, 60, 224–228 CrossRef.
- A. Sawyer, The unsustainable lab, BioTechniques, 2019, 66, 5–7 CrossRef CAS PubMed.
- S. O. Onuh and Y. Y. Yusuf, Rapid prototyping technology applications and benefits for rapid product development, J. Intell. Manuf., 1999, 10, 301–311 CrossRef.
- J. C. McDonald, D. C. Duffy, J. R. Anderson, D. T. Chiu, H. Wu, O. J. Schueller and G. M. Whitesides, Fabrication of microfluidic systems in poly(dimethylsiloxane), Electrophoresis, 2000, 21, 27–40 CrossRef CAS PubMed.
- D. C. Duffy, J. C. McDonald, O. J. Schueller and G. M. Whitesides, Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane), Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- B. C. Gross, J. L. Erkal, S. Y. Lockwood, C. Chen and D. M. Spence, Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences, Anal. Chem., 2014, 86, 3240–3253 CrossRef CAS PubMed.
- P. F. O'Neill, A. Ben Azouz, M. Vázquez, J. Liu, S. Marczak, Z. Slouka, H. C. Chang, D. Diamond and D. Brabazon, Advances in three-dimensional rapid prototyping of microfluidic devices for biological applications, Biomicrofluidics, 2014, 8, 52112 CrossRef PubMed.
- Y. Xia and G. M. Whitesides, Soft Lithography, Angew. Chem., Int. Ed., 1998, 37, 550–575 CrossRef CAS PubMed.
- N. Bhattacharjee, A. Urrios, S. Kang and A. Folch, The upcoming 3D-printing revolution in microfluidics, Lab Chip, 2016, 16, 1720–1742 RSC.
- A. Bonyár, H. Sántha, B. Ring, M. Varga, J. Gábor Kovács and G. Harsányi, 3D Rapid Prototyping Technology (RPT) as a powerful tool in microfluidic development, Procedia Eng., 2010, 5, 291–294 CrossRef.
- R. Walczak and K. Adamski, Inkjet 3D printing of microfluidic structures—on the selection of the printer towards printing your own microfluidic chips, J. Micromech. Microeng., 2015, 25, 85013 CrossRef.
- E. M. Sachs, J. S. Haggerty, M. J. Cima and P. A. Williams, US Pat., 5204055A, 1993 Search PubMed.
- R. Walczak, K. Adamski, A. Pokrzywnicka and W. Kubicki, Inkjet 3D Printing – Studies on Applicability for Lab-on-a-chip Technique, Procedia Eng., 2016, 168, 1362–1365 CrossRef.
- R. Walczak, Inkjet 3D printing – towards new micromachining tool for MEMS fabrication, 2018 Search PubMed.
- A. Naderi, N. Bhattacharjee and A. Folch, Digital Manufacturing for Microfluidics, Annu. Rev. Biomed. Eng., 2019, 21, 325–364 CrossRef CAS PubMed.
- 3d Systems Inc, VisiJet M2R-CL (MJP), https://de.3dsystems.com/materials/visijet-m2r-cl-mjp, (accessed 10.2021).
- I. G. Siller, A. Enders, T. Steinwedel, N.-M. Epping, M. Kirsch, A. Lavrentieva, T. Scheper and J. Bahnemann, Real-Time Live-Cell Imaging Technology Enables High-Throughput Screening to Verify in Vitro Biocompatibility of 3D Printed Materials, Materials, 2019, 12, 2125 CrossRef CAS PubMed.
- M. Wan, S. Liu, Y. Qu Da Huang, Y. Hu, Q. Su, W. Zheng, X. Dong, H. Zhang, Y. Wei and W. Zhou, Biocompatible heterogeneous bone incorporated with polymeric biocomposites for human bone repair by 3D printing technology, J. Appl. Polym. Sci., 2021, 138, 50114 CrossRef CAS.
- C. Heuer, J.-A. Preuß, T. Habib, A. Enders and J. Bahnemann, 3D printing in biotechnology—An insight into miniaturized and microfluidic systems for applications from cell culture to bioanalytics, Eng. Life Sci., 2021 DOI:10.1002/elsc.202100081.
- V. John, Hinshaw, What Is “Dead” Volume and Why Should Chromatographers Worry About It?, LCGC North Am., 2015, 33, 850–855 Search PubMed.
- A. Enders, I. G. Siller, K. Urmann, M. R. Hoffmann and J. Bahnemann, 3D Printed Microfluidic Mixers-A Comparative Study on Mixing Unit Performances, Small, 2019, 15, e1804326 Search PubMed.
- K. Jensen, A normally occurring Staphylococcus antibody in human serum, APMIS, 1958, 44, 421–428 Search PubMed.
- T. Löfkvist and J. Sjöquist, Chemical and serological analysis of antigen preparations from staphylococcus aureus, Acta Pathol. Microbiol. Scand., 1962, 56, 295–304 CrossRef.
- Sartorius AG, Sartobind® Q 75, https://www.sartorius.com/shop/ww/de/eur/produkte-labor-membranchromatographie/sartobind-q-75/p/93IEXQ42DB-12--V, (accessed 10.2021).
- Sartorius AG, Sartobind® Protein A, https://www.sartorius.com/shop/ww/de/eur/anwendungen-labor-chromatographie/sartobind-protein-a/p/93PRAP06HB-12--A, (accessed 10.2021).
- V. Viktorov, M. R. Mahmud and C. Visconte, Design and characterization of a new H-C passive micromixer up to Reynolds number 100, Chem. Eng. Res. Des., 2016, 108, 152–163 CrossRef CAS.
- N.-T. Nguyen, M. Hejazian, C. Ooi and N. Kashaninejad, Recent Advances and Future Perspectives on Microfluidic Liquid Handling, Micromachines, 2017, 8, 186 CrossRef.
- M. A. Ianovska, P. P. M. F. A. Mulder and E. Verpoorte, Development of small-volume, microfluidic chaotic mixers for future application in two-dimensional liquid chromatography, RSC Adv., 2017, 7, 9090–9099 RSC.
- C. Brämer, S. Schreiber, T. Scheper and S. Beutel, Continuous purification of Candida antarctica lipase B using 3-membrane adsorber periodic counter-current chromatography, Eng. Life Sci., 2018, 18, 414–424 CrossRef PubMed.
- C. Brämer, F. Lammers, T. Scheper and S. Beutel, Development and Testing of a 4-Columns Periodic Counter-Current Chromatography System Based on Membrane Adsorbers, Separations, 2019, 6, 55 CrossRef.
- M. Y. Arica, E. Yalçin and G. Bayramoğlu, Preparation and characterisation of surfaces properties of poly(hydroxyethylmethacrylate-co-methacrylolyamido-histidine) membranes: application for purification of human immunoglobulin G, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 807, 315–325 CrossRef CAS PubMed.
- E. Mahajan, A. George and B. Wolk, Improving affinity chromatography resin efficiency using semi-continuous chromatography, J. Chromatogr. A, 2012, 1227, 154–162 CrossRef CAS PubMed.
- J. Pollock, G. Bolton, J. Coffman, S. V. Ho, D. G. Bracewell and S. S. Farid, Optimising the design and operation of semi-continuous affinity chromatography for clinical and commercial manufacture, J. Chromatogr. A, 2013, 1284, 17–27 CrossRef CAS PubMed.
- D. Baur, M. Angarita, T. Müller-Späth, F. Steinebach and M. Morbidelli, Comparison of batch and continuous multi-column protein A capture processes by optimal design, Biotechnol. J., 2016, 11, 920–931 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |