Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Real-time monitoring of epithelial barrier function by impedance spectroscopy in a microfluidic platform†
João
Fernandes
 a,
Nikita
Karra
a,
Nikita
Karra
 a,
Joel
Bowring
a,
Joel
Bowring
 a,
Riccardo
Reale
a,
Riccardo
Reale
 a,
Jonathan
James
b,
Cornelia
Blume
a,
Jonathan
James
b,
Cornelia
Blume
 bcd,
Theresa J.
Pell
bcd,
Theresa J.
Pell
 e,
Wendy C.
Rowan
e,
Wendy C.
Rowan
 e,
Donna E.
Davies
e,
Donna E.
Davies
 bcd,
Emily J.
Swindle
bcd,
Emily J.
Swindle
 bcd and
Hywel
Morgan
bcd and
Hywel
Morgan
 *ac
*ac
aElectronics and Computer Science, Faculty of Physical Sciences and Engineering, University of Southampton, UK. E-mail: hm@ecs.soton.ac.uk
bClinical and Experimental Sciences, Faculty of Medicine, University of Southampton, UK
cInstitute for Life Sciences, University of Southampton, UK
dNIHR Southampton Biomedical Research Centre, University Hospital Southampton, UK
eNovel Human Genetics Research Unit, GlaxoSmithKline R&D, Stevenage, Hertfordshire, UK
First published on 22nd April 2022
Abstract
A multichannel microfluidic platform for real-time monitoring of epithelial barrier integrity by electrical impedance has been developed. Growth and polarization of human epithelial cells from the airway or gastrointestinal tract was continuously monitored over 5 days in 8 parallel, individually perfused microfluidic chips. Electrical impedance data were continuously recorded to monitor cell barrier formation using a low-cost bespoke impedance analyser. Data was analysed using an electric circuit model to extract the equivalent transepithelial electrical resistance and epithelial cell layer capacitance. The cell barrier integrity steadily increased overtime, achieving an average resistance of 418 ± 121 Ω cm2 (airway cells) or 207 ± 59 Ω cm2 (gastrointestinal cells) by day 5. The utility of the polarized airway epithelial barrier was demonstrated using a 24 hour challenge with double stranded RNA to mimic viral infection. This caused a rapid decrease in barrier integrity in association with disruption of tight junctions, whereas simultaneous treatment with a corticosteroid reduced this effect. The platform is able to measure barrier integrity in real-time and is scalable, thus has the potential to be used for drug development and testing.
1 Introduction
In multicellular organisms, physical barriers comprised of epithelial or endothelial cells are essential for tissue compartmentalization and performance of specialised functions.1 Epithelial barriers (e.g., the skin, airway and gut) protect tissues from the external environment whereas endothelial barriers separate the blood and tissue compartments. Both barrier cell types elaborate a range of cell–cell adhesion complexes that link with the cytoskeleton to form a robust sheet-like structure that functions as a selective permeability barrier. Key to this function are tight junctions that form between adjacent cells by intercellular transmembrane proteins and intracellular proteins that link to the actin cytoskeleton.2 These junctions form around the entire perimeter of each cell, forming a continuous belt-like structure to control its barrier (or “gate”) properties, thereby regulating paracellular passage of ions, water, and various macromolecules across the epithelial layer. In epithelial cells, tight junctions are concentrated near the apical surface above the adherens junctions (zonula adherens) where they also prevent lateral diffusion and intermixing of molecules in the apical membrane with those in the lateral membrane. This “fence” function maintains cell polarity, another key property for normal epithelial barrier function.Tight junctions are multimolecular structures which all utilise three types of adhesive transmembrane proteins. These are the MARVEL domain proteins, occludin and tricellulin, the junctional adhesion molecules (JAMs) and the claudins. Claudin domain proteins regulate the permeability selectivity of specific tissue barriers via pattern of expression interactions with other claudin family members;3 this provides the potential to customise their interactions and give rise to the wide range of permselective barriers found in the human micro-environment.4 The transmembrane proteins of the tight junction link to underlying plaque proteins, such as zona occludin-1 (ZO-1), that in turn associate with the cytoskeleton. These tight junction complexes regulate not only paracellular solute and water flux but also control diverse processes including gene transcription, tumour suppression, cell proliferation, and cell polarity.5 Given their multifunctional roles, it is perhaps not surprising that epithelial barrier function is dysregulated in a range of diseases including asthma, COPD,6–10 inflammatory bowel disease and skin diseases such as psoriasis and eczema.11 Consequently, therapeutic approaches that restore normal barrier function are being actively researched.12
Investigations of epithelial barrier integrity typically use in vitro models in which cells are grown in standard well plates or on suspended nanoporous supports (Transwell® inserts) where the development and integrity of the epithelial barrier is monitored using transepithelial electrical resistance (TER); the resistance correlates to the number and function of the tight junctions.13,14 Although TER measurements are easy to perform in Transwell® inserts using Ag/AgCl “chopstick” electrodes, damage to the epithelial layer caused by handling and fluctuations in temperature (removing cells from the incubator) need to be avoided, as these can affect the measurements.10 Unfortunately, this method does not allow continuous analysis of the cells during formation of the epithelial barrier or modulation following stimulation. Furthermore, the lack of interstitial flow in static Transwell® models does not recapitulate the in vivo condition.15
Over recent years, organ-on-chip (OoC) models have been developed to address the need for more representative in vitro models. These are biomimetic devices that aim to recreate complex organ-level physiological functions, clinically relevant disease phenotypes and pharmacological responses that arise from the structural and functional integration of multiple tissues or interfaces.15 Several devices have been fabricated to recapitulate epithelial cell barriers in vitro models such as skin,16–18 gut19 and lungs.15,20–25 These in vitro models are capable of recreating key in vivo conditions such as interstitial flow, which supplies metabolites to cells and clears metabolic waste in addition to inducing shear stress on cell cultures (due to liquid flow and pressure differentials), and mechanical stimulation, to mimic breathing motions within alveolus on chip models.26
Electrical impedance spectroscopy (EIS) has been used in OoC devices to analyse barrier integrity.27–30 EIS provides information not only about the epithelial barrier resistance but also epithelial barrier capacitance which correlates with the cell membrane surface area, offering information about cell stacking or development of complex membrane folds such as microvilli.27 While EIS allows continuous data collection of cell barrier parameters,27,28,31–34 typically these systems require electrodes on both sides of the barrier (i.e., a trans configuration), necessitating the use of submerged conditions to ensure electrical connection to the electrodes. For example, Alexander et al.,35 cultured fibroblasts and EpiDermTM reconstructed human epidermis in an OoC device at an air–liquid interface (ALI), for each TER measurement, the apical layer was temporarily submerged in PBS using an automated pumping system. To solve these issues, we have previously presented a device with the measuring electrodes placed beneath the cell layer (cis configuration), allowing continuous monitoring of barrier function at baseline and in response to challenge (Sun et al.,36). This method was utilised in a lung-on-chip device presented by Mermoud et al.,37 incorporating mechanical stretch and demonstrated frequency-dependent changes in impedance resulting from the detergent permeabilization of the epithelial monolayers.
In this paper we present a barrier on a chip model composed of an 8-channel microfluidic system capable of culturing different types of epithelial cells with integrated real-time impedance capability. Electrical impedance measurements were made using integrated micro-electrodes placed within the basolateral compartment of each chip (directly beneath the cell layer), providing real-time electrical data without perturbation of the cells. Impedance data were fitted to an equivalent circuit model to obtain the TER and epithelial cell layer capacitance. We have demonstrated the establishment of an electrically tight epithelial barrier, in addition to changes in barrier function using a viral mimetic (double stranded RNA (dsRNA)) and reversal of this effect using a corticosteroid, providing proof-of-concept for the system as a drug discovery platform.
1.1 System design
A diagram of the barrier-on-chip system is shown in Fig. 1. A disposable microfluidic chip serves as a simple interchangeable consumable item that interfaces with a microfluidic manifold in a ‘plug and play’ format (Fig. 1A). The microfluidic chip is secured with magnets, providing an easy, rapid assembly suitable for a non-specialist user. Medium is pumped through individual chips using a syringe pump and a bubble trap, with waste/cellular secretions collected in a tube at the outlet (Fig. 1B). The compact platform accommodates 8 microfluidic chips in parallel, with two systems capable of fitting onto a standard incubator shelf. Syringe pumps used for perfusion of cell culture media are kept outside the incubator (and refrigerated, if required), with the connecting tubing introduced through the door of the incubator.Details of the microfluidic chip are shown in Fig. 2. It is fabricated from glass and PMMA and is similar in size to a conventional Transwell® and has a cell culture surface area of 0.2 cm2. The upper (apical) chamber is separated from the lower (basolateral) chamber by a semi-permeable membrane, with a pore diameter of 0.4 μm and pore density of 108 pores per cm2, onto which epithelial cells are grown (Fig. 2B). The base of the chip contains inlet and outlet holes which mate with a microfluidic manifold (Fig. 1B). The support membrane is bonded to a spacer piece approximately 275 μm thick made from self-adhesive tape and PMMA, creating a microfluidic channel through which media flows at a rate of 30 μL h−1. The shape of the microfluidic channel ensures equal flow velocity across the entire cell barrier.15
The cells are grown on the apical side of the membrane (Fig. 2A) in a submerged environment (100 μL volume). The chamber is sealed with a thin gas permeable membrane (PDMS), creating a sterile environment for cell culture and is easily accessible to introduce compounds onto the apical side of the cell culture when required (via puncturing). The chip interfaces fluidically with the manifold via O-rings and snaps into place using location features and two pairs of magnets.
Each chip has two pairs of electrodes (Fig. 2B) that are used to perform periodical EIS measurements. One pair of electrodes, denominated ‘media electrodes’ (Fig. 2B), is upstream of the cell electrodes; these measure the electrical properties of the medium, and monitor the system (consistent fluid flow, stability and bubble formation). The electrical parameters of the media measured with these electrodes are used in the circuit model. The second pair of electrodes located beneath the cells (‘cell electrodes’) measures the electrical properties of the epithelial barrier. The electrical current path between these electrodes is dominated by the basolateral medium, but current also flows through the cell layer. The magnitude of this current depends on the integrity of the cell barrier. This arrangement also allows the ‘TER’ to be measured at the air–liquid interface as well as the liquid–liquid interface.
In the case of ALI cultures, the presence of air rather than medium on the apical side leads to a high value of apical resistance that could potentially mask the epithelial barrier resistance and capacitance. In practice there is always a layer of fluid or mucus (typically 10 μL) above the cells when grown at ALI.38 Previous studies by Reale et al.,39 demonstrated the reduction in sensitivity at ALI. Polarized 16HBE14o- cells were grown in Transwell® supports and the volume of apical media reduced to 10 μL before EIS measurement using coplanar electrodes beneath the cell culture. The data is summarized in ESI† Fig. S1 and demonstrates changes in cell barrier can be followed, albeit with reduced sensitivity.
The electrodes in each microfluidic chip are connected to a PCB header board (Fig. 1A) through wires. The PCB routes signals to and from the chips to a custom-made impedance spectrometer based on the AD5933 network analyser IC (Analog devices) controlled with a Raspberry PI expansion board. A custom script written in Python controls the analyser, which can measure the impedance of up to 32 chips in parallel (4 systems). A graphical user interface enables the system to be controlled remotely via WiFi, and a screenshot of this display is shown in ESI† Fig. S2. Impedance is measured at 100 mVp–p, between 1 kHz to 100 kHz. This frequency window is sufficient for circuit analysis and characterization of cell barrier resistance and capacitance. In a typical experiment impedance frequency sweeps were recorded automatically at intervals of 17 minutes. Liquid was perfused under the cell layer continuously at a rate of 30 μL per hour.
To minimise toxicity to the cells metal oxide electrodes were used (rather than Ag/AgCl). Specifically, ruthenium oxide which is biocompatible and provides a large effective surface area, similar to platinum black but much more stable minimising electrode polarisation issues.40
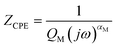 | (1) |
If the impedance is a pure capacitance the CPE exponent (αM) is equal to 1, and a pure resistance is equal to 0. For most metal (or metal oxide) electrodes the exponent varies between 0.5 and 0.9. The magnitude of the impedance is defined by the parameter QM. For the low frequency (103 to 105 Hz) range used in these experiments the media impedance approximates to the resistance, and so the measured impedance of the ‘media electrodes’ is simply the sum of the CPE and medium resistance, seen in eqn (2). Although there are two ZCPE in the model, one for each electrode, for simplicity these are represented as a single value in the equation.
| ZMedia Electrode = ZCPE + RM | (2) |
The cell chamber consists of two compartments separated by the high porosity membrane. Each compartment, basolateral and apical, has a medium resistance of RB and RA respectively. The cell chamber electrodes provide the resistance of the nanoporous support membrane and cell layer, together with the epithelial cell layer capacitance. All other parameters in this equivalent circuit can be fixed and determined by the media electrodes. The resistance of the nano porous support (RSM) was modelled following Hall et al.,41 where the pores were modelled as cylindrical channels with a fixed conductivity defined by their geometry together with an access resistance. This gives a total pore resistance for the membranes of approximately 13.3 Ω, much smaller than the resistance of other components in the electrical circuit model. Given this is in series with the apical resistance (RA) the two were assigned a single variable. The cell barrier impedance (ZCB) was modelled as the parallel combination of a resistor (RCB) and a capacitor (CCB) which account for the ohmic resistance of the tight junctions and the capacitive behaviour of the cell membrane respectively.36 Thus, the cell electrode circuit model is defined by eqn (3). A more detailed list of equations is provided in ESI† detailed equation list.
 | (3) |
The electrical parameters were determined from the equivalent circuit using the following algorithm:
1. CPE (QM and αM) and medium resistance (RSM) determined from the ‘media electrodes’ by fitting to eqn (2).
2. Impedance data from the “cell electrodes” was fitted to eqn (3) using the 3 parameters determined from step 1. To optimise the fit, these parameters were assigned upper and lower boundaries of ±5%.
3. Values for the cell layer resistance and capacitance were determined by curve fitting.
4. Cell layer resistance was multiplied by the nanoporous support membrane surface area to give a TER equivalent (Ω cm2).
The impedance data are related to the physical properties of the system (conductivity and permittivity) through the electrode chamber geometry. These geometric constants were determined through numerical simulation to be 1.9 × 10−3 m−1, 4.1 × 10−2 m−1 and 1.5 × 10−3 m−1 for the media electrode, cell electrodes apical and cell electrodes basolateral compartment respectively. For the media electrode, cell electrodes apical and cell electrodes basolateral compartment respectively. For the fit, the CPE exponent (which depends on electrode surface roughness) was assumed to be identical for both sets of electrodes. The CPE magnitude was scaled by the difference in electrode area (factor of 2.3). Apical and basolateral cell media resistance for the cell electrode were determined from the fit for the media electrodes, with the appropriate conversions from differences in geometry (see ESI† detailed equation list). An upper and lower boundary of 5% was applied to the value of apical resistance, since this is affected by the volume of liquid held in the apical compartment, which can vary between chips or over the course of the experiment. A full list of variables and applied boundaries is given in ESI† Table S1.
2 Experimental
Electrodes were fabricated using standard photolithography on 1.1 mm thick glass wafer following the method described by Mingels et al.40 Electrodes were made of 20 nm titanium, 200 nm platinum, 20 nm titanium and 120 nm ruthenium oxide. Following metal deposition, chips were annealed at 420 °C overnight. The microfluidic chip consisted of 3 layers of PMMA either solvent bonded or assembled with 50 μm double-sided adhesive tape (3M). The nanoporous support membrane (Inpore Track Etched Membrane) was adhered to the PMMA chamber with tape, then fixed to the channel layer in a similar manner. The PMMA layers were either machined using a laser cutter or a CNC micro milling machine. Inlet and outlet holes in the glass chips were made with powder blasting. The microfluidic chip was secured onto the manifolds using magnets (10 × 5 × 3 mm) and O-rings. Manifolds were micro-milled from Polyetheretherketone (PEEK). Syringes were connected to the bubble traps (Darwin Microfluidics) with PTFE tubing.Prior to cell culture, the microfluidic platform was decontaminated by flowing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 sodium hypochlorite solution throughout the system and submerging the apical compartment for 20 minutes. Subsequently the system was copiously rinsed with sterile DI water. Syringes primed with culture media were then attached to the assembly and 100 μL of cell culture media added into the apical compartment of each chip. The system was placed inside a humidified incubator at 37 °C and 5% CO2 with media pumped at a flow rate of 30 μL per hour overnight to ensure fluidic and electrical stability before introducing cells.
50 sodium hypochlorite solution throughout the system and submerging the apical compartment for 20 minutes. Subsequently the system was copiously rinsed with sterile DI water. Syringes primed with culture media were then attached to the assembly and 100 μL of cell culture media added into the apical compartment of each chip. The system was placed inside a humidified incubator at 37 °C and 5% CO2 with media pumped at a flow rate of 30 μL per hour overnight to ensure fluidic and electrical stability before introducing cells.
2.1 Cell preparation
The human colorectal carcinoma cell line (Caco-2) and human bronchial epithelial cell line (16HBE14o-) were maintained in Dulbecco's modified Eagle medium (DMEM) or minimal essential medium (MEM) respectively supplemented with 1× Glutamax, 10% foetal bovine serum (FBS), and 1% penicillin/streptomycin. The DMEM was additionally supplemented with 1% sodium pyruvate and 1% non-essential amino acid solution. All cell culture flasks were coated with 30 μg mL−1 collagen (Advanced Biomatrix). For experiments cells were incubated with HBSS without Ca2+ and Mg2+ for 10 minutes at 37 °C, prior to a 5 minute incubation with 1× trypsin/EDTA solution. Following cell detachment, trypsin was neutralised with complete DMEM or MEM (depending on the cell line) and the cell suspension centrifuged at 300 × g for 5 minutes at 20 °C. After discarding the cell-free supernatant, the cell pellet was resuspended in complete medium and counted. Cells were seeded onto collagen coated (30 μg mL−1, 30 min, 37 °C) microfluidic chips at a density of 3 × 105 cells per cm2 in 100 μL of DMEM or 7.5 × 105 cells per cm2 in 100 μL of MEM for the Caco-2 and 16HBE14o- cells respectively. The microfluidic chip was left undisturbed in the incubator at 37 °C for 20 hours (Caco-2) or 1 hour (16HBE14o-) to allow cells to adhere to the support membrane. After which, media was perfused through each chip at a flow rate of 30 μL per hour. Additionally, cells were seeded onto collagen coated Transwells® inserts, inside a 24 well-plate with 200 and 500 μL of complete MEM or DMEM in the apical and basolateral compartments respectively. Medium was changed every 2 days and TEER measurements were recorded daily using an EVOM Epithelial Voltohmmeter.2.2 Reagent preparation
Two different challenge experiments were performed. The first experiment involved apically treating cells with Triton X-100 (TX-100, final 1% solution v/v). The second experiment involved apical stimulation with a synthetic analogue of dsRNA (to mimic viral infection) polyinosinic:polycytidylic acid (Poly I:C, final concentration 25 μg mL−1) (Invivogen) or a corticosteroid, fluticasone propionate (final concentration 100 nM) alone or together in complete MEM media. Complete MEM media was administered apically as a control.2.3 Immunofluorescent staining
Membranes containing cells were washed with 1× PBS and fixed in 4% PFA for approximately 20 minutes at room temperature, then washed and stored in PBS. Membranes were removed from the chips, transferred to a glass slide, and submerged in blocking buffer (2% BSA, 0.1% Tween-20 in PBS). Permeabilization buffer (0.1% TX-100 in PBS) was then applied to the samples for 15 minutes at room temperature followed by blocking buffer for 1 hour at room temperature. Samples were stained with Acti-stain555-phalloidin (70 nM, Cytoskeleton, PHDH1-A) and AlexaFluor®488-conjugated mouse anti-human occludin antibody (2.5 μg mL−1, Life Technologies, Clone OC-3F10) in blocking buffer overnight at 4 °C in a humidified chamber. Samples were then washed in wash buffer (0.1% Tween-20 in PBS) three times, followed by an incubation with the nuclear stain 4′,6-diamidino-2-phenylindole (DAPI, 2 μg mL−1 in dH2O, ThermoFisher) for 15 minutes in the dark. Samples were rinsed with wash buffer followed by dH2O prior to cover slips being mounted onto slides with Mowiol (Sigma). Samples were aligned (Leica DMI 6000 inverted fluorescence microscope) and images were captured at 63× in xyz mode using confocal microscopy (Leica TCS-SP8 laser scanning microscope with the LAS X software (Leica)) using laser wavelengths of 405 nm (DAPI), 561 nm (Actin) and 488 nm (Occludin).3 Results and discussion
To assess variability between microfluidic chips, MEM cell culture medium was first perfused through all 8 basolateral channels for 12 hours, with 100 μL of media in the apical chamber. Impedance data were collected continuously, with the mean value of the medium resistance and electrode CPE determined from the circuit model. This is summarised in Table 1, which shows that the culture medium resistance and CPE exponent have low standard deviations, with the values of cell medium resistance close to the expected values derived from the numerically calculated geometric cell constants (see below). However, the magnitude of the CPE was more variable, which may be due to variations in the electrode area arising from the manual assembly of the chip and variability in the metal oxide of the electrodes. The culture medium conductivity was measured with a conductivity meter (YSI 3200) as 1.46 S m−1. Based on the calculated geometrical cell constants of the chip and electrodes (see ESI† Geometrical cell constants), using this value of conductivity the cell media resistance should be approximately 390 Ω and 467 Ω for the media and cell electrode chambers respectively.| Culture medium resistance (Ω) | Media electrode | Cell electrode | ||||
|---|---|---|---|---|---|---|
| CPE magnitude | CPE exponent | Apical resistance (Ω) | CPE magnitude | CPE exponent | ||
| Average | 376 | 4.2 × 10−5 | 0.6 | 467 | 9.2 × 10−5 | 0.64 |
| Standard deviation | 14.0 (3.7%) | 1.19 × 10−5 (28.3%) | 0.04 (6.7%) | 18.8 (4.0%) | 3.1 × 10−5 (34%) | 0.04 (6.3%) |
These values are similar to the experimental values (Table 1). Impedance measurements were made for chips with and without a nanoporous support membrane. At 1 kHz, the resistance differed by approximately 28 Ω, a value close to the calculated value for the resistance of the membrane, and negligible compared to the medium resistance.
3.1 Cell culture
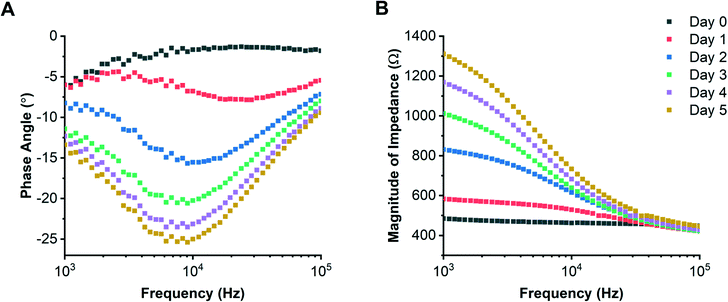
The Caco-2 colorectal epithelial cell line19,42–46 was grown on individual chips for 5 days with media flowing beneath the support membrane, and EIS data acquired every 17 minutes. Examples of daily impedance spectra (phase angle and magnitude) are shown in Fig. 4. The circuit model was first used to determine the electrode CPE and medium resistance, then using eqn (3) the epithelial barrier resistance and capacitance. Values for the medium resistance and electrode CPE are given in the ESI.†R-Squared values were calculated for the modelled data; see ESI† Fig. S4 and S5. Examples of equivalent TER and cell layer capacitance (determined from each impedance spectrum) are shown in Fig. 5.The cell layer resistance and capacitance increased over the course of 5 days, reaching an average of 207 ± 59 Ω cm2 and 1.97 ± 0.13 μF cm−2 respectively. The perturbation seen at day 1 for Caco-2 cells coincides with initiation of basolateral perfusion, since these cells required 24 hours to fully adhere unlike the 16HBE14o- cells which only required 1 hour. Epithelial cell barrier capacitance increased over time and was similar to values reported by Helm et al.,34 (approximately 2 μF cm−2 after 5 days of culture with flow). This increase has previously been attributed to formation of microvilli.34 For comparison, Caco-2 cells were also grown in standard Transwell® inserts where they developed a TER of 1258 ± 40 Ω cm2 over 5 days of culture. The difference in TER observed between the microfluidic system and static Transwell® cultures has been reported previously,34 where cells cultured in a fluidic system have a lower TER: 200 Ω cm2 compared with 600 Ω cm2 for no flow. Azizgolshani et al.,47 also showed that the TER of Caco-2 cells grown with flow was approximately 200 Ω cm2 after 11 days of culture. Our data shows a lower TER with flow, consistent with previous publications.
To compare the response of epithelial cells from a different tissue, bronchial epithelial cells (16HBE14o-) were cultured for 5 days with flow. Example daily impedance spectra (phase angle and impedance magnitude) are shown in Fig. 6; values of electrode CPE and medium resistance are given in the ESI† along with R-squared values (see ESI† Fig. S6 and S7) For this cell type, cell layer resistance and capacitance reached an average of 418 ± 121 Ω cm2 and 0.13 ± 0.02 μF cm−2 after 5 days of culture. The epithelial cell layer capacitance (Fig. 7A) stabilised quickly to approximately 0.13 ± 0.02 μF cm−2 (day 0.5) after initial cell seeding (day 0), probably because the cells were seeded at a high density (unlike the Caco-2 cells which were seeded at a lower density). The 16HBE14o- cell layer capacitance is smaller than reported for other cell layers which are typically in the range of 1–4 μF cm−2.32,36,48 Caco-2 cells have been shown to develop microvilli, which would lead to a higher capacitance27,34 but the 16HBE14o- cells stack on top of one another (see ESI† Fig. S8), which would result in a lower value of cell layer capacitance.
The cell layer resistance increased throughout the experiment, reaching an average peak value of 418 ± 121 Ω cm2 on day 5 (Fig. 7B). This barrier resistance was almost double the value obtained with the Caco-2 cells, highlighting the ability of the device to demonstrate cell type-related differences in barrier permeability. The same bronchial epithelial cells were also grown (in parallel) on commercial Transwell® inserts. This gave a day 5 average TER of 803 ± 34 Ω cm2, consistent with reported values for cells grown without flow (between 330–2500 Ω cm2 (ref. 10, 15 and 49)). These data indicate that the TER in the microfluidic device with flow are lower than no-flow, similar to the findings with the Caco-2 cell line. This is supported by previous studies with polarized 16HBE14o- cells where a lower TER was measured in cells on Transwell® inserts with flow (560 Ω cm2)50 and in cells perfused in Ussing chambers (TER of 105 Ω cm2).51 It is likely that these differences under flow have a biological explanation. For example, tight junctions can participate in the regulation of the mechanical properties of epithelial monolayers and are finely tuned by the contractility of the actomyosin cytoskeleton,52 a highly dynamic structure that is sensitive to shear stress.53 Actomyosin contractility is required both to allow claudins fibrils to operate correctly, and to reinforce the actin cytoskeleton to prevent leaks.52
3.2 Apical challenge
The 16HBE14o- cells were grown for 5 days with a flow rate of 30 μL per hour. After formation of the epithelial barrier, cells were then apically exposed to 1% TX-100.36 This caused an immediate disruption of the epithelial barrier, evidenced by the sharp drop in the normalised TER from 100% to 0%, as compared with control chips without TX-100 (Fig. 8A). To extend the barrier challenge into a more physiologically relevant context, viral infection was simulated by challenging cells with a dsRNA viral analogue Poly I:C (25 μg mL−1). dsRNA is produced during replication of RNA viruses and is a pathogen-associated molecular pattern detectable by pattern recognition receptors within infected cells. It triggers an anti-viral response including inflammation, disassembly of tight junctional complexes and actin cytoskeleton reorganization without cell apoptosis. Reorganization leads to an increase in barrier permeability, leading to a decrease in TER.8 Within the microfluidic chip, addition of medium to the apical compartment caused an initial perturbation in the cell barrier irrespective of whether the medium contained dsRNA, or corticosteroid (Fig. 8B). This may be due to small mechanical perturbations and/or temperature fluctuations during introduction of the challenge even though the cells remain in the incubator. This effect was not visible for the TX-100 experiment, as the incubator was open for less time during stimulation, minimizing the temperature drop. When the challenge medium also contained dsRNA, the TER dropped quickly, reaching a minimum 8 hours after challenge. Addition of a corticosteroid (fluticasone propionate) in combination with dsRNA reduced the severity of the drop in TER in the initial hours after introducing the stimulus, which then stabilised to approximately 25% of the initial value. This is consistent with Transwell® data (Fig. 8C), where the addition of the corticosteroid mitigates the barrier disrupting effect of dsRNA.54Immunofluorescence staining supported the EIS findings. Compared to control cells (Fig. 9A) which showed pericellular staining for occludin at cell junctions and a well organised actin cytoskeleton, cells treated with dsRNA (Fig. 9B) exhibited occludin reorganisation to the cell–cell junctions, although the cell layer was still intact, as evidenced by the presence of regular DAPI stained nuclei and an actin cytoskeleton. Treatment of cells with dsRNA in the presence of a corticosteroid (Fig. 9D) decreased the impact of dsRNA on the integrity of the epithelial cell barrier, with an improvement in the junctional staining for occludin compared with dsRNA alone. These findings are similar to those of Heijink et al.,54 where fluticasone (500 μg) was able to limit barrier disruption following dsRNA (12.5 μg mL−1) addition to 16HBE14o- cells grown on Transwell supports.
4 Conclusions
We have described a novel multi-channel microfluidic platform with integrated impedance measuring capability for real-time analysis of barrier function by TER. The platform combines plug and play fluidic and electrical functionality suitable for multidisciplinary use (discovery research, target discovery, drug testing, toxicological testing etc.) generating a more physiological relevant environment compared to conventional static cultures.Conventional TER measurements are usually obtained using chop-stick electrodes in a transformation which requires a submerged apical compartment which is unsuitable for ALI cultures. Furthermore, periodic removal from the incubator disrupts the physiological environment and is problematic for temperature sensitive cells.10 Impedance spectroscopy with fixed cis orientation electrodes minimises user-error and significantly reduces these issues. The system has individual perfusion to deliver media to and remove waste from cells at a physiological flow rate synonymous with interstitial flow. This is particularly beneficial for down-stream analysis of mediators, providing the opportunity for collection of samples with a temporal resolution if required in the future.
The system uses 2 pairs of electrodes to measure the medium and cell layer impedance separately. A simple circuit model extracts the cell barrier electrical properties from the impedance data to give an equivalent TER enabling comparison with conventional static cultures. Real-time impedance measurements and calculated TER were utilised to analyse the growth and polarisation of Caco-2 and 16HBE14o- cells over 5 days with a continuous microfluidic flow (30 μL per hour), demonstrating an increase in TER over time. Cells challenged with compounds after 5 days of growth were monitored continuously over time. Dissolution of the cell membrane by addition of Triton-X-100 led to the immediate disruption of barrier integrity while the addition of dsRNA (Poly I:C 25 μg mL−1) caused a time-dependent decrease in barrier integrity as a result of tight junction disassembly, where the response was modulated by a corticosteroid (fluticasone (100 nM)), consistent with previous findings.54–56
The development of a barrier on a chip platform with a tuneable microfluidic flow providing individual perfusion along with the ability to measure barrier integrity in real-time through integrated impedance electrodes provides a distinct advantage over other platforms. This platform is scalable and easy to use with a simple chip to manifold interface. It has the potential to be used in drug development and testing, where the influence of compounds can be analysed in a temporal fashion, facilitating the delivery of new treatments for disease. The platform can analyse eight separate cultures in parallel and can be scaled with 32 chips in a single incubator, enabling testing of several different compounds simultaneously. Future work requires optimisation in the fabrication methodology to decrease chip-to-chip variability, and studies of both submerged and ALI cell barriers with human primary cells.
Data accessibility
All data supporting this study is openly available from the University of Southampton repository at https://doi.org/10.5258/SOTON/D2138.Author contributions
Author contributions are as follows: João Fernandes was involved in the conceptualization, data curation, formal analysis, investigation, methodology, resources, software, validation and writing – original draft; Nikita Karra was involved in the conceptualization, data curation, formal analysis, investigation, methodology, resources, validation and writing – original draft; Joel Bowring was involved in resources and software, Riccardo Reale was involved in conceptualization; Jonathan James was involved in resources and methodology; Cornelia Blume was involved in supervision and resources; Theresa J. Pell was involved in funding acquisition and project administration; Wendy C. Rowan was involved in funding acquisition and project administration; Donna E. Davies was involved in supervision, resources and writing – review and editing; Emily J. Swindle was involved in conceptualization, supervision, funding acquisition, project administration, resources and writing – review and editing; Hywel Morgan was involved in conceptualization, supervision, funding acquisition, project administration, resources and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the EPSRC, GlaxoSmithKline, IUK (102522) and the Asthma Allergy and Inflammation Research charity. We would like to thank Katie Chamberlain for her assistance with fabrication of glass chips, Mark Long, Anthony Gardener and Jamie Stone for their technical assistance with micro milling and 3D printing and David Johnstone for his support with confocal microscopy. 16HBBE14o- cells were gifted from Prof. D.C. Gruenert, San Francisco, USA. Caco-2 cells were gifted by Prof. P. Calder and provided by his PhD student Luke Dirkin, University of Southampton, UK.Notes and references
- A. M. Marchiando, W. V. Graham and J. R. Turner, Annu. Rev. Pathol.: Mech. Dis., 2010, 5, 119–144 CrossRef CAS PubMed.
- S. Tsukita, M. Furuse and M. Itoh, Nat. Rev. Mol. Cell Biol., 2001, 2, 285–293 CrossRef CAS PubMed.
- G. Krause, L. Winkler, S. L. Mueller, R. F. Haseloff, J. Piontek and I. E. Blasig, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 631–645 CrossRef CAS PubMed.
- M. K. Findley and M. Koval, IUBMB Life, 2009, 61, 431–437 CrossRef CAS PubMed.
- E. E. Schneeberger and R. D. Lynch, Am. J. Physiol., 2004, 286, C1213–C1228 CrossRef CAS PubMed.
- C. Xiao, S. M. Puddicombe, S. Field, J. Haywood, V. Broughton-Head, I. Puxeddu, H. M. Haitchi, E. Vernon-Wilson, D. Sammut, N. Bedke, C. Cremin, J. Sones, R. Djukanović, P. H. Howarth, J. E. Collins, S. T. Holgate, P. Monk and D. E. Davies, J. Allergy Clin. Immunol., 2011, 128, 549–556.e12 CrossRef CAS PubMed.
- K. Sweerus, M. Lachowicz-Scroggins, E. Gordon, M. LaFemina, X. Huang, M. Parikh, C. Kanegai, J. V. Fahy and J. A. Frank, J. Allergy Clin. Immunol., 2017, 139, 72–81.e1 CrossRef CAS PubMed.
- F. Rezaee, N. Meednu, J. A. Emo, B. Saatian, T. J. Chapman, N. G. Naydenov, A. De Benedetto, L. A. Beck, A. I. Ivanov and S. N. Georas, J. Allergy Clin. Immunol., 2011, 128, 1216 CrossRef CAS PubMed.
- Y. Soini, Respir. Res., 2011, 12, 70–70 CrossRef CAS PubMed.
- P. J. Callaghan, B. Ferrick, E. Rybakovsky, S. Thomas and J. M. Mullin, Biosci. Rep., 2020, 40, 1–16 CrossRef PubMed.
- L. S. Poritz, L. R. Harris, A. A. Kelly and W. A. Koltun, Dig. Dis. Sci., 2011, 56, 2802–2809 CrossRef CAS PubMed.
- F. M. Carlier, C. de Fays and C. Pilette, Front. Physiol., 2021, 12, 1–27 Search PubMed.
- D. W. Powell, Am. J. Physiol., 1981, 4, 275–288 Search PubMed.
- B. Srinivasan, A. R. Kolli, M. B. Esch, H. E. Abaci, M. L. Shuler and J. J. Hickman, J. Lab. Autom., 2015, 20, 107–126 CrossRef CAS PubMed.
- C. Blume, R. Reale, M. Held, T. M. Millar, J. E. Collins, D. E. Davies, H. Morgan and E. J. Swindle, PLoS One, 2015, 10, 1–13 CrossRef PubMed.
- R. S. Tavares, T. Phuong-Tao, I. Maschmeyer, S. S. Maria-Engler, M. Schäfer-Korting, A. Winter, C. Zoschke, R. Lauster, U. Marx and L. R. Gaspar, Int. J. Pharm., 2020, 589, 119788 CrossRef CAS PubMed.
- G. Sriram, M. Alberti, Y. Dancik, B. Wu, R. Wu, Z. Feng, S. Ramasamy, P. L. Bigliardi, M. Bigliardi-Qi and Z. Wang, Mater. Today, 2018, 21, 326–340 CrossRef CAS.
- M. Wufuer, G. H. Lee, W. Hur, B. Jeon, B. J. Kim, T. H. Choi and S. H. Lee, Sci. Rep., 2016, 6, 1–12 CrossRef PubMed.
- N. Ashammakhi, R. Nasiri, N. R. de Barros, P. Tebon, J. Thakor, M. Goudie, A. Shamloo, M. G. Martin and A. Khademhosseni, Gut-on-a-chip: Current progress and future opportunities, Biomaterials, 2020, 255, 120196 CrossRef CAS PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- A. M. Ghaemmaghami, M. J. Hancock, H. Harrington, H. Kaji and A. Khademhosseini, Drug Discovery Today, 2012, 17, 173–181 CrossRef CAS PubMed.
- T. Nguyen, S. H. Jung, M. S. Lee, T. E. Park, S. K. Ahn and J. H. Kang, Lab Chip, 2019, 19, 3706–3713 RSC.
- J. W. Song, J. Paek, K. T. Park, J. Seo and D. Huh, Biomicrofluidics, 2018, 12, 042211 CrossRef PubMed.
- M. Humayun, C.-W. Chow and E. W. K. Young, Lab Chip, 2018, 18, 1298–1309 RSC.
- A. Doryab, G. Amoabediny and A. Salehi-Najafabadi, Biotechnol. Adv., 2016, 34, 588–596 CrossRef CAS PubMed.
- N. Karra, E. Swindle and H. Morgan, Organs-on-a-Chip, 2019, 1, 100002 CrossRef.
- T. Gerasimenko, S. Nikulin, G. Zakharova, A. Poloznikov, V. Petrov, A. Baranova and A. Tonevitsky, Front. Bioeng. Biotechnol., 2020, 7, 474 CrossRef PubMed.
- E. Ferrari, C. Palma, S. Vesentini, P. Occhetta and M. Rasponi, Biosensors, 2020, 10, 110 CrossRef CAS PubMed.
- T. Sun and H. Morgan, Microfluid. Nanofluid., 2010, 8, 423–443 CrossRef CAS.
- T. Sun, S. Tsuda, K. P. Zauner and H. Morgan, Biosens. Bioelectron., 2010, 25, 1109–1115 CrossRef CAS PubMed.
- Y. B. Arlk, M. W. Van Der Helm, M. Odijk, L. I. Segerink, R. Passier, A. Van Den Berg and A. D. Van Der Meer, Biomicrofluidics, 2018, 12, 042218 CrossRef PubMed.
- M. Odijk, A. D. Van Der Meer, D. Levner, H. J. Kim, M. W. Van Der Helm, L. I. Segerink, J. P. Frimat, G. A. Hamilton, D. E. Ingber and A. Van Den Berg, Lab Chip, 2015, 15, 745–752 RSC.
- N. Zhang, F. Stauffer, B. R. Simona, F. Zhang, Z. M. Zhang, N. P. Huang and J. Vörös, Biosens. Bioelectron., 2018, 112, 149–155 CrossRef CAS PubMed.
- M. W. van der Helm, O. Y. F. Henry, A. Bein, T. Hamkins-Indik, M. J. Cronce, W. D. Leineweber, M. Odijk, A. D. van der Meer, L. I. Segerink, D. E. Ingber, J. C. T. Eijkel, M. J. Cronce, M. Odijk, A. Bein, W. D. Leineweber, A. D. van der Meer, A. van den Berg, M. W. van der Helm, D. E. Ingber, T. Hamkins-Indik, L. I. Segerink and O. Y. F. Henry, Lab Chip, 2019, 19, 452–463 RSC.
- F. A. Alexander, S. Eggert and J. Wiest, Genes, 2018, 9, 114 CrossRef PubMed.
- T. Sun, E. J. Swindle, J. E. Collins, J. A. Holloway, D. E. Davies, H. Morgan, E. Davies, H. Morgan, D. E. Davies and H. Morgan, Lab Chip, 2010, 10, 1611–1617 RSC.
- Y. Mermoud, M. Felder, J. D. Stucki, A. O. Stucki and O. T. Guenat, Sens. Actuators, B, 2018, 255, 3647–3653 CrossRef CAS.
- P. R. Harvey, R. Tarran, S. Garoff and M. M. Myerburg, Am. J. Respir. Cell Mol. Biol., 2011, 45, 592–599 CrossRef CAS PubMed.
- R. Reale, Ph.D. thesis, University of Southampton, 2017 Search PubMed.
- R. Mingels, S. Kalsi, Y. Cheong and H. Morgan, Sens. Actuators, B, 2019, 297, 126779 CrossRef CAS.
- J. E. Hall, Access resistance of a small circular pore, J. Gen. Physiol., 1975, 66, 531–532 CrossRef CAS PubMed.
- K. Verhoeckx, P. Cotter, I. López-Expósito, C. Kleiveland, T. Lea, A. Mackie, T. Requena, D. Swiatecka and H. Wichers, The impact of food bioactives on health: In vitro and Ex Vivo models, Springer, 2015, pp. 1–327 Search PubMed.
- L. Wang, J. Wu, J. Chen, W. Dou, Q. Zhao, J. Han, J. Liu, W. Su, A. Li, P. Liu, Z. An, C. Xu and Y. Sun, Talanta, 2021, 226, 122097 CrossRef CAS PubMed.
- G. Preksha, R. Yesheswini and C. V. Srikanth, Indian J. Pathol. Microbiol., 2021, 64, S52–S57 CrossRef PubMed.
- S. E. Blutt, J. R. Broughman, W. Zou, X. L. Zeng, U. C. Karandikar, J. In, N. C. Zachos, O. Kovbasnjuk, M. Donowitz and M. K. Estes, Exp. Biol. Med., 2017, 242, 1633–1642 CrossRef CAS PubMed.
- D. Marrero, F. Pujol-Vila, D. Vera, G. Gabriel, X. Illa, A. Elizalde-Torrent, M. Alvarez and R. Villa, Biosens. Bioelectron., 2021, 181, 113156 CrossRef CAS PubMed.
- H. Azizgolshani, J. R. Coppeta, E. M. Vedula, E. E. Marr, B. P. Cain, R. J. Luu, M. P. Lech, S. H. Kann, T. J. Mulhern, V. Tandon, K. Tan, N. J. Haroutunian, P. Keegan, M. Rogers, A. L. Gard, K. B. Baldwin, J. C. de Souza, B. C. Hoefler, S. S. Bale, L. B. Kratchman, A. Zorn, A. Patterson, E. S. Kim, T. A. Petrie, E. L. Wiellette, C. Williams, B. C. Isenberg and J. L. Charest, Lab Chip, 2021, 21, 1454–1474 RSC.
- G. Linz, S. Djeljadini, L. Steinbeck, G. Köse, F. Kiessling and M. Wessling, Biosens. Bioelectron., 2020, 165, 112345 CrossRef CAS PubMed.
- D. Papazian, P. A. Würtzen and S. W. K. Hansen, Int. Arch. Allergy Immunol., 2016, 170, 1–21 CrossRef CAS PubMed.
- C. Blume, R. Reale, M. Held, M. Loxham, T. M. Millar, J. E. Collins, E. J. Swindle, H. Morgan and D. E. Davies, Immun., Inflammation Dis., 2017, 5, 45–56 CrossRef CAS PubMed.
- A. L. Cozens, M. J. Yezzi, K. Kunzelmann, T. Ohrui, L. Chin, K. Eng, W. E. Finkbeiner, J. H. Widdicombe and D. C. Gruenert, Am. J. Respir. Cell Mol. Biol., 1994, 10, 38–47 CrossRef CAS PubMed.
- S. Citi, Biophys. Rev., 2019, 11, 783–793 CrossRef CAS PubMed.
- N. M. Cronin and K. A. DeMali, Biology, 2021, 11, 52 CrossRef PubMed.
- I. H. Heijink, M. R. Jonker, M. de Vries, A. J. van Oosterhout, E. Telenga, N. H. ten Hacken, D. S. Postma and M. van den Berge, Respir. Res., 2016, 17, 1–13 CrossRef PubMed.
- C. Blume, J. David, R. E. Bell, J. R. Laver, R. C. Read, G. C. Clark, D. E. Davies and E. J. Swindle, Pathogens, 2016, 5, E53 CrossRef PubMed.
- A. Singanayagam, N. Glanville, N. Bartlett and S. Johnston, Lancet, 2015, 385, S88 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1lc01046h |
| This journal is © The Royal Society of Chemistry 2022 |