Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selective bacterial separation of critical metals: towards a sustainable method for recycling lithium ion batteries†
Virginia
Echavarri-Bravo
 ae,
Houari
Amari‡
be,
Jennifer
Hartley
ae,
Houari
Amari‡
be,
Jennifer
Hartley
 ce,
Giovanni
Maddalena
ce,
Giovanni
Maddalena
 ae,
Caroline
Kirk
d,
Maarten W.
Tuijtel§
ae,
Caroline
Kirk
d,
Maarten W.
Tuijtel§
 a,
Nigel D.
Browning
befg and
Louise E.
Horsfall
a,
Nigel D.
Browning
befg and
Louise E.
Horsfall
 *ae
*ae
aSchool of Biological Sciences, University of Edinburgh, Edinburgh, EH9 3FF, UK. E-mail: Louise.Horsfall@ed.ac.uk
bDepartment of Mechanical, Materials and Aerospace Engineering, University of Liverpool, Liverpool, L69 3GQ, UK
cSchool of Chemistry, University of Leicester, Leicester, LE1 7RH, UK
dSchool of Chemistry, University of Edinburgh, Edinburgh, EH9 3FJ, UK
eFaraday Institution (ReLiB project), Quad One, Harwell Science and Innovation Campus, Didcot, UK
fSivananthan Laboratories, 590 Territorial Drive, Bolingbrook, IL 60440, USA
gPhysical and Computational Sciences Directorate, Pacific Northwest National Laboratory, P.O. Box 999, Richland, WA 99352, USA
First published on 17th October 2022
Abstract
The large scale recycling of lithium ion batteries (LIBs) is essential to satisfy global demands for the raw materials required to implement this technology as part of a clean energy strategy. However, despite what is rapidly becoming a critical need, an efficient and sustainable recycling process for LIBs has yet to be developed. Biological reactions occur with great selectivity under mild conditions, offering new avenues for the implementation of more environmentally sustainable processes. Here, we demonstrate a sequential process employing two bacterial species to recover Mn, Co and Ni, from vehicular LIBs through the biosynthesis of metallic nanoparticles, whilst Li remains within the leachate. Moreover the feasibility of Mn recovery from polymetallic solutions was demonstrated at semi-pilot scale in a 30 L bioreactor. Additionally, to provide insight into the biological process occurring, we investigated selectivity between Co and Ni using proteomics to identify the biological response and confirm the potential of a bio-based method to separate these two essential metals. Our approach determines the principles and first steps of a practical bio-separation and recovery system, underlining the relevance of harnessing biological specificity for recycling and up-cycling critical materials.
Introduction
It is well established that one of the most important measures to slow climate change is a reduction in CO2 emissions. Road transportation is highly dependent on carbon-based fuels and responsible for 20% of CO2 emissions worldwide.1 Therefore, there is mounting pressure to move towards transportation alternatives with lower carbon footprints, leading to many national governments incentivising the transition to electric vehicles (EV) for mobility. These pressures and incentives are resulting in an increasing demand for lithium ion batteries (LIBs),2 currently the best technological solution to power EV based on energy density.3 Life cycle assessment of LIBs shows that the availability of raw materials needed to fulfil the demand for EV LIBs by 2050 is estimated to be ‘very critical’ for both lithium and cobalt, and ‘critical’ for nickel.4,5 Thus, the development of efficient technologies to enable selective recovery and recycling of the components and materials present in spent LIBs is vital for minimising risks in the supply chain and reducing the waste burden.6 Moreover, moving to a circular economy for LIBs would reduce reliance on the current sources of raw materials associated with human rights abuses and decrease mining activities reported to negatively impact upon human and environmental health.7,8Adaptive and flexible recycling solutions are needed to address the wide variety of continuously evolving cathode chemistries in a highly competitive market. Current recycling methods are multi-step processes, often starting with physical separation of the various battery parts, or shredding and comminution, followed by a combination of other physical, hydrometallurgical and/or pyrometallurgical processes.3,6,7 Pyrolysis involves the calcination (>400 °C) of the battery and although it is a mature technology it has been decreasing in popularity for EV LIB recycling. It results in high CO2 and toxic emissions, and the significant loss of value to the materials streams including Li and Al.7 Hydrometallurgical methods involve the dissolution of battery parts, often using sulfuric acid or hydroxides,9 with other secondary treatments (e.g. thermal, sonication) that result in the production of battery leachates with varying concentrations of metals dependent on the chemistry of the cathode. Development of more efficient recycling options continues at all stages of the recycling loop, with new technologies investigated, such as high powered ultrasound with increasing potential for the rapid delamination of electrodes and to facilitate direct recycling of the cathode.10 Nevertheless, hydrometallurgy is always being applied to some extent and polymetallic solutions that require metal separation and refining are generated.
As such, this too is an evolving area of study and novel solutions have been reported such as ferro-chemistry-based approaches and bio-electrochemical reduction to separate Li from Co.11,12 However when other elements are also present in the cathode, such as Mn and Ni (e.g. LiNixCoyMnzO2, NCM) the downstream recovery and separation of metals contained in the battery leachates is commonly achieved by solvent-exchange (SX) and chemical precipitation. The energy inputs and the use of hazardous chemicals are then major limiting factors for achieving a cost effective and sustainable process.3,7 SX methods using organophosphorus solvents are the most frequent approach to selectively separate and purify metals, with D2EHPA being the most common method for Mn separation and Cyanex®272 for Co and Ni. However SX involves the use of toxic solvents to treat aqueous polymetallic streams13 and Mn removal using SX is not cost-efficient unless subsidised by the recovery of Co and Ni.14 Other limitations of SX include the treatment of heterogeneous feedstocks that may hinder the control of the process; the potential environmental impacts of organic solvents if the reuse rates are low, and the wastewater generated.14 There are other chemical alternatives to SX which may first involve the selective leaching of Li using organic acids (e.g. tartaric, formic, oxalic) followed by precipitation as Li2CO3 from the addition of NaOH and Na2CO3.15,16 The remaining metals in the cathode once dissolved in inexpensive acids (e.g. H2SO4) are precipitated as hydroxides17 or carbonates15 to be used as precursor material for new active electrodes. The high requirements of alkaline compounds (e.g. NaOH, NH4OH, Na2CO3) with an elevated environmental footprint is one of the major drawbacks of the chemical precipitation approach.18 The energy inputs of the chemical reactions vary within a wide range, as solvent extraction takes place ∼25 °C, while chemical precipitation as hydroxides or carbonates may require 40–65 °C15–17 or above (80 °C) to shorten reaction times, enhance metal specificity and evaporate the solvent. Multiple combinations of optimised parameters are reported in the literature dependent upon the initial concentration of metals, leaching process, target efficiency and purity.19 Nowadays the development of greener methods that enable the selective separation of metals from battery leachates and allows for their return to use is a major challenge for the industry.3
Here the incorporation of biological methods into the process (Scheme 1) may provide the key, as bioprocessing occurs at relatively low temperatures (≤30 °C), in aqueous solutions and does not involve the use of hazardous solvents or compounds such as NaOH to raise the pH due to other alkaline compounds (e.g. carbonates and ammonia) produced by the bacteria.20 In this study, bacteria were used for metal bio-recycling, as they grow quickly and are engineered more easily in comparison to other microorganisms, should the process require such optimisation. We examined the selective separation and recovery of the most relevant metals (Co, Li, Mn and Ni) present in EV LIB (NMC) leachates prepared with strong mineral acids using two bacterial species, Shewanella oneidensis MR-1 and Desulfovibrio alaskensis G20. These bacteria reportedly precipitate dissolved Mn and Ni ions in the form of Mn oxide21 and Ni sulfide nanoparticles (NPs)22 respectively and additionally, during the course of this work, we confirmed the synthesis of Co NPs by D. alaskensis G20. The biological mechanisms responsible for metal removal such as biosorption, bioprecipitation and bioreduction are ubiquitous but, in the main, are metal and bacterial species-specific.23 Metal oxido-reduction reactions resulting in nanoparticle synthesis are usually associated with the production of metal reducing and metal binding compounds such as enzymes and non-enzymatic proteins.20 Thus we interrogated the proteome of D. alaskensis G20 to gain insight into the biological pathways responsible for the precipitation of Co and Ni. It is crucial to recycle both metals as they are currently essential in the majority of EV LIB chemistries; Co is a critical element and there is an increasing demand for Ni due the rapid adoption of high-Ni cathodes.24,25
Results and discussion
Bioprecipitation of nickel, manganese, and cobalt complexes
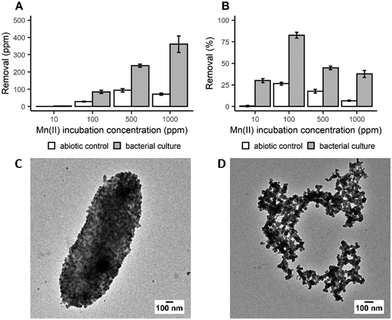
In order to determine the efficiency of Mn bioprecipitation by S. oneidensis MR-1, experiments using single-metal solutions were carried out at 20 °C, over a period of 20 h. Manganese recovery was investigated in systems (pH 7–8.5) containing 10 to 1000 ppm of dissolved Mn2+ ions based upon the composition of real EV LIBs leachates. It was observed that while total Mn recovery increased with increasing initial Mn2+ concentration (Fig. 1A), the highest recovery efficiency was observed from the 100 ppm solution, with 83% of the dissolved Mn2+ recovered (Fig. 1B). The low recoveries at 10 ppm suggests that there is a minimum dissolved Mn2+ concentration threshold for recovering Mn efficiently. However, not all of this Mn recovery is associated with bacterial activity, as Mn precipitates were also detected in the abiotic control (27% recovery at 100 ppm incubation concentration). Nevertheless, taking incubation times (20 h) and cell densities into account, the removal rate obtained in this study is encouraging compared to previous work with Shewanella putrefaciens which required longer incubation times, up to 10 days, to achieve 80% removal at an incubation concentration of 125 ppm.26 Analysis by X-ray powder diffraction (XRPD) identified the white precipitate obtained at the end of the incubation period as MnCO3 (ESI, Fig. S1A†). Transmission electron microscopy (TEM) imaging confirmed that this biogenic MnCO3 was in the form of nanoparticles both coating the bacterial cells (Fig. 1C) and was also detached from cell biomass (Fig. 1D). This latter material has the greater potential for resynthesis into LIB electrode active materials.27,28 It is very likely that Mn2+ precipitated as MnCO3 due to carbonate species produced during bacterial growth (ESI, Fig. S2A†). The synthesis of carbonates increases the pH of the culture during bacterial growth (ESI, Fig. S2B†) supporting Mn biomineralisation and bioprecipitation. S. oneidensis does not produce urease, the enzyme known for mediating fungal synthesis of MnCO328 and the molecular mechanisms responsible for the synthesis of carbonate by this bacterium are still unknown29 and deserve further investigation.Following the work with Mn we extended our investigations with S. oneidensis MR-1 to precipitate other relevant metals, Li, Co and Ni, without success. This confirmed this bacterium was a suitable candidate for the selective recovery of Mn from LIBs leachates.
With a similar batch process approach, we demonstrated that D. alaskensis G20 was able to remove both Co2+ and Ni2+ from the dissolved fraction with an efficiency above 70% at 10 ppm of metal ions. The net mass recovery of Ni varied little across the different incubation concentrations (Fig. 2A). While the removal of Co did increase with higher incubation concentrations, the removal efficiency for both metals was most efficient at lower concentrations (Fig. 2B).
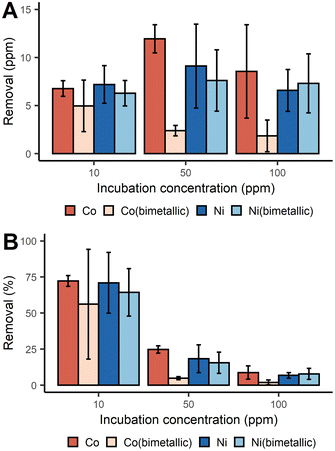 | ||
| Fig. 2 Removal of Co2+ and Ni2+ with D. alaskensis G20 expressed as mean ± SD (n = 3) measured as (A) ppm and (B) percent. | ||
Unlike the Mn systems, there was no Co or Ni precipitation observed in either of the abiotic control solutions. As vehicular LIBs leachates can often contain both Co and Ni, the removal of Co and Ni from bimetallic solutions was also studied. It was found that in general the presence of Ni2+ significantly decreased the removal of Co2+ at all three incubation concentrations tested (Fig. 2A). Significant differences in the removal of Co were found between the single and bimetallic treatments (two-way ANOVA, p-value <0.001). The differing removal profiles of these metals underpins the existence of different biological molecules and mechanisms responsible for Co2+ and Ni2+ precipitation which were investigated with a proteomics study (ESI, proteomics analysis, Fig. S3†). For instance the abundance of the UPF0173 metal-dependent hydrolases (Dde_0151) and MJ0042 family finger-like proteins (Dde_0116) increased after 2 h incubation with 100 ppm Co2+ compared to the control treatment, but decreased in the bimetallic treatment. These two protein families bind Zn2+, but can also bind Co2+ without significant loss of functionality.30,31 To reduce the risk of mis-metallation with Ni, or perhaps to remedy such, it is understandable that a decrease in their abundance is observed and this might then also explain why Co removal from the dissolved fraction dropped when Ni2+ was present at concentrations ≥50 ppm.
Characterisation of Co and Ni nanoparticles
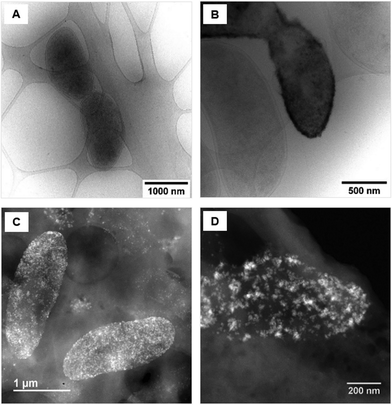
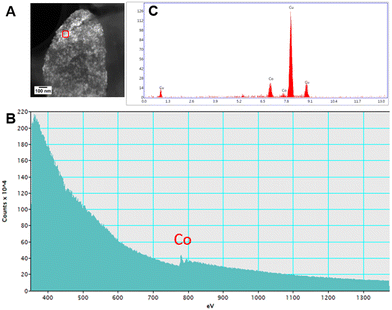
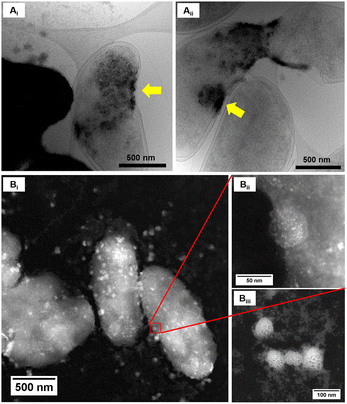
Upon examination of the bacterial cells post-treatment using cryo-electron microscopy (cryo-EM), areas of high-density were observed in the bacterial envelope of cells incubated with 50 ppm Co2+ (Fig. 3B) compared to cells in the control treatment (Fig. 3A).High resolution scanning transmission electron microscopy (STEM) imaging also showed the presence of high-density areas covering the surface of bacterial cells incubated with Co2+ (Fig. 3C) and Ni2+ (Fig. 3D) at 10 ppm, depicting the formation of metallic nanoparticles. To our delight the characterisation revealed zero-valent Co NPs attached to the cells (Fig. 4A), the first time, to the best of our knowledge, that biosynthesis of such nanoparticles of this critical metal have been reported. The absence of oxygen in the electron energy loss spectroscopy (EELS) (Fig. 4B) and sulfur in the EDXS (Fig. 4C) spectra rules out the possibility that the NPs are sulfides. The synthesis of biogenic zero-valent Co NPs by the bacterium Geobacter sulfurreducens had been previously speculated but not demonstrated.32 The biological mechanisms involved in the formation of zero-valent Co NPs are unknown and deserve further investigation that has commenced with the proteomics work presented in this study. From this we hypothesise that certain proteins, such as quinone-interacting membrane-bound oxidoreductases (ESI, Fig. S3A and B,† Dde_1113), and other redox active elements (e.g. cytochromes, flavins33,34) could be involved in the reduction of Co2+ to Co0 during the anaerobic metabolism, under negative oxidation–reduction potential conditions. Other metal-binding proteins in the membrane (ESI, Fig. S3,† Dde_0155, Dde_2208, Dde_3518) may play a role in the nucleation of Co resulting in nanoparticle formation and stabilisation. Cryo-EM images show that Co nanoparticles are synthesised in the bacterial envelope eventually compromising the integrity of the membrane (Fig. 5A). The bacterial envelope under the STEM presented different degrees of degradation depending on the fixative used, ethanol 50% v/v was less aggressive than acetone 50% v/v. The images obtained from samples of D. alaskensis G20 incubated with Co salts and fixed with acetone showed mesoporous nanostructures of a diameter ∼50 nm (Fig. 5B and 6A). EELS analysis confirmed the presence of Co in these mesoporous nanoparticles. The increased concentration of metal-binding proteins during incubation with Co in different locations of the bacterial envelope (ESI, Fig. S3†) such as Dde_2670 in the inner membrane and Dde_0155, Dde_2208, Dde_1113, and Dde_3518 in the periplasm, support the hypothesis that these Co-based nanostructures are formed due to biological processes. We could not confirm the oxidation state of Co in this instance because elemental edges associated to C, Ca and O were also present in the EELS spectra (Fig. 6B and C). The presence of Ca could be attributed to accumulation as a result of the direct electron transfer from cytochromes and hydrogenases35 or alternatively may be associated to the degraded bacterial envelope surrounding the NPs.36 In addition to the biological mechanisms of metal bioprecipitation, the formation of nanocrystalline cobalt and nickel sulfides must be considered due to the presence of biogenic hydrogen sulfide (H2S).37 Whilst the cells were thoroughly washed and resuspended in nutrient-free buffer prior to the beginning of the metal ion removal experiments, biogenic H2S was produced ([H2S] <160 μmol L−1) (ESI, Fig. S4†) during maintenance of the bacterial cell steady-state.38 The formation of nanoparticles made of Co and S (ESI, Fig. S5A†), and Ni and S (ESI, Fig. S5B†) on the surface of the bacterial envelope was confirmed by STEM/EDXS. Precipitation of Co and Ni as nanocrystalline metal sulfides by the activity of sulfate reducing bacteria (SRB) has been reported previously and is attributed to the presence of biogenic H2S.37 However, the presence of sulfur in the nanoparticles could also be due to the presence of cysteinyl ligands in relevant proteins binding iron-sulfur clusters. Some proteins binding 4Fe–4S were significantly more abundant after incubation with Ni2+ (ESI, Fig. S3B and C,† Dde_0718, Dde_2176, Dde_1830, Dde_2943). There is a possibility that the iron sites within these metalloproteins have been replaced by Ni2+ and/or Co2+, as observed in rubredoxins-related studies,39 and thus end up serving as an anchor and metal nucleation site for the nanoparticles synthesised as observed on the bacterial outer membrane.40 Metal removal by H2S is relatively efficient and in the bacterial cell-free supernatant was above 70% due to the presence of dissolved H2S (24.5 mmol L−1), however it exhibits no selectivity for Co or Ni (ESI, Fig. S6†).41
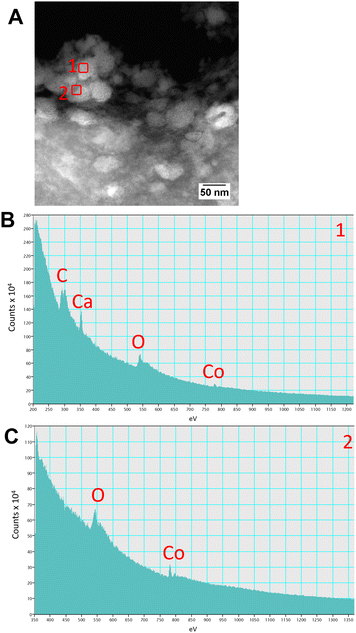 | ||
| Fig. 6 Characterisation of the mesoporous nanostructures using (A) STEM and (B and C) EELS spectra obtained from regions (A) 1 and 2 highlighted with red squares. | ||
Selective bioprecipitation of dissolved Mn from vehicular LIBs leachates
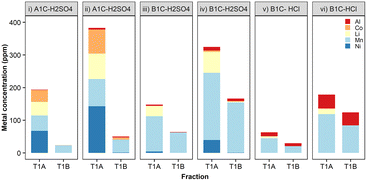
To investigate metal removal/precipitation from vehicular battery leachates, two commercial LIB cathodes (Nissan Leaf) were leached with two different mineral acids, H2SO4 or HCl, both widely used in hydrometallurgy.42 These cathode materials contained either Li, Ni, Mn and Co in a ratio of 50% nickel, 30% manganese, and 20% cobalt (NMC-532), or a mixture of approximately 70% lithium manganese oxide spinel (LMO) with 25% lithium nickel manganese cobalt oxide,43 referred to hereafter as A1C and B1C, respectively. The concentration of metals (Al, Co, Mn, Li and Ni) in these LIB leachates was dependent upon the origin of the cathode material, solvent used and leaching temperature (ESI, Table S1†). Prolonged leaching times, from 5 to 30 minutes in 0.1 M H2SO4 at 50 °C doubled the concentration of Co, Li and Mn, and increased Al and Ni concentrations even further. The final dissolved metal composition of the leachates was very different according to the battery type, cathode chemistry and acid used. The leaching data for these cathodes in H2SO4 and HCl at 20 °C is available in ESI, Fig. S7.†The specificity for dissolved Mn and the removal rates achieved by S. oneidensis MR-1 with vehicular leachates were in agreement with the results we obtained in the previous experiments using metal salts, i.e. higher concentrations of metal ions resulted in greater total metal precipitates. The capability exhibited by this bacterium for the selective precipitation of Mn2+ out of the mixed metals contained in the crude leachates is extremely relevant for establishing the principles of a bio-separation process (Fig. 7, ESI Table S2†). The removal efficiency of Mn peaked at 75% (154 ppm) from leachates of the cathode material B1C dissolved in 0.1 M H2SO4 for 30 minutes at 50 °C (Fig. 7iv). The removal of the other metals was low, generally well below 5% total precipitated mass, except for Co when present at concentrations above 70 ppm (Fig. 7ii) and Al contained in HCl leachates (Fig. 7v and vi). Fortunately, a recent advance using selective hydrometallurgical methods combined with high-intensity ultrasonication can now be employed to provide leachates with a lower concentration of Al through delamination of the active materials from the foil current collectors.10
Two bacterial treatment bioprocessing approach
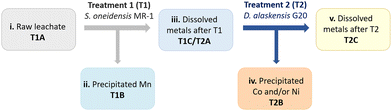
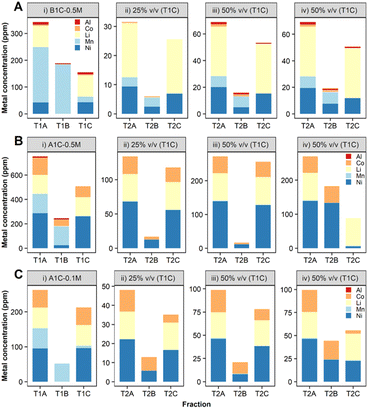
In order to ensure a complete and selective bio-recycling process, a two-bacterial bioprocessing approach was developed by using S. oneidensis MR-1 to selectively precipitate out Mn (treatment 1, Scheme 2), followed by treatment of the stripped solution using D. alaskensis G20 to precipitate Ni and/or Co (treatment 2, Scheme 2).To enable the use of leachates with higher acid content (0.5 M H2SO4), a carbonate–bicarbonate buffer was added to S. oneidensis MR-1 cell culture. Small-scale (2 ml) experiments confirmed this buffer would aid the biological removal of Mn from leachates prepared in 0.5 M H2SO4 (ESI, Fig. S8†). The results obtained were very encouraging with the B1C leachate (Fig. 8A(i)-T1B, ESI Table S3†) as the precipitation of Mn was enhanced to 89% and the dissolved Mn present in the downstream fraction (T1C) was reduced to just 10 ppm (mass balance ESI Fig. S9†). A higher (97%) Mn removal was achieved from the A1C leachate, however the precipitated Mn exhibited lower purity (63% total precipitated metal mass) due to co-precipitation of Al (4%), Co (22%) and Ni (10%) (Fig. 8B(i)-T1B, ESI Table S4,† mass balance Fig. S10†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) in agreement with the results obtained during the bimetallic experiments at 50–100 ppm. The release of extracellular proteins with high Ni-affinity as a mechanism of bacterial stress response caused by this metal may also explain differences between Co and Ni removals.44 Our proteomics study showed proteins that might be involved in metal reduction processes, such as oxidoreductases,45,46 which were significantly more abundant after 20 h incubation with 10 ppm of Ni2+ compared to the treatment with Co2+ (ESI, Fig. S3C†) confirming a distinctive cellular response depending on the metal. Some of these oxidoreductases, such as the FAD/NAD (P)-binding domain protein (Dde_1381) and the FAD-dependent pyridine nucleotide-disulfide oxidoreductase (Dde_2176), are classified within the xenobiotics biodegradation and metabolism pathways and might be responsible for reducing Ni2+ into a less toxic form. Since Ni was present at toxic concentrations for D. alaskensis G20 in the A1C 0.5 M H2SO4 leachate, the removal of Ni and Co from a leachate (A1C, 0.1 M H2SO4) containing lower concentrations of both metals was investigated (Ni
3) in agreement with the results obtained during the bimetallic experiments at 50–100 ppm. The release of extracellular proteins with high Ni-affinity as a mechanism of bacterial stress response caused by this metal may also explain differences between Co and Ni removals.44 Our proteomics study showed proteins that might be involved in metal reduction processes, such as oxidoreductases,45,46 which were significantly more abundant after 20 h incubation with 10 ppm of Ni2+ compared to the treatment with Co2+ (ESI, Fig. S3C†) confirming a distinctive cellular response depending on the metal. Some of these oxidoreductases, such as the FAD/NAD (P)-binding domain protein (Dde_1381) and the FAD-dependent pyridine nucleotide-disulfide oxidoreductase (Dde_2176), are classified within the xenobiotics biodegradation and metabolism pathways and might be responsible for reducing Ni2+ into a less toxic form. Since Ni was present at toxic concentrations for D. alaskensis G20 in the A1C 0.5 M H2SO4 leachate, the removal of Ni and Co from a leachate (A1C, 0.1 M H2SO4) containing lower concentrations of both metals was investigated (Ni![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Co ratio of 2
Co ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). This time D. alaskensis G20 showed preference for precipitating Co, and by increasing the volume of leachate from 25 to 50% v/v, the removal increased by 70% for Co, 12.2 ppm, and 40% for Ni, 7.9 ppm (Fig. 8C, ESI Table S5†). The removal yields of Co and Ni presented here are still low as experiments have been developed in a single batch process in 2 ml volumes as a first step to show the fundamentals of the approach and to provide benchmark data for process optimisation. The Li that remains in the dissolved fraction at pH > 8, with similarities in terms of composition and concentration to some lithium salar brines,47 could then be recovered by chemical precipitation as Li2CO3.
1). This time D. alaskensis G20 showed preference for precipitating Co, and by increasing the volume of leachate from 25 to 50% v/v, the removal increased by 70% for Co, 12.2 ppm, and 40% for Ni, 7.9 ppm (Fig. 8C, ESI Table S5†). The removal yields of Co and Ni presented here are still low as experiments have been developed in a single batch process in 2 ml volumes as a first step to show the fundamentals of the approach and to provide benchmark data for process optimisation. The Li that remains in the dissolved fraction at pH > 8, with similarities in terms of composition and concentration to some lithium salar brines,47 could then be recovered by chemical precipitation as Li2CO3.
Conclusion
The need for more efficient and greener methods for recycling metals contained in LIBs is driving research to consider the application of less conventional methodologies. For decades microorganisms have been used extensively in the areas of metal bioremediation and bioleaching however their uses for metal bio-separation are still in the early stages. Our work with two different bacterial strains shows the potential for the separation and recovery of all the relevant metals contained in LIBs. First S. oneidensis MR-1 precipitates dissolved Mn followed by the use of D. alaskensis G20 for the recovery of Co and/or Ni, leaving Li in the downstream leachate as could be found in a lithium salt brine. The results presented here show the principles for establishing a bio-based technology and will be used as the benchmark to define areas of research needed for enhancing removal yields and improving metal selectivity. To date there is a wide range of synthetic biology tools available for manipulation of S. oneidensis MR-1 that could be applied for improving the removal efficiency of Mn2+. The removal of Co and Ni was higher with whole bacterial culture treatment however no selectivity for either metal was observed. In contrast, the utilisation of bacterial cell-only treatment offers greater advantages such as better control of the physicochemical properties of the nanoparticles produced and the potential for enhancing the specificity for Co or Ni by using D. alaskensis G20 engineered strains with their design informed by the proteomics analysis provided herein. Biological approaches are ideally suited for implementation alongside existing technologies and could form part of battery recycling processes to provide a sustainable incentive for industry and as a solution to fit EV LIB technology within a circular economy. The implementation of life cycle assessments (LCAs) will ultimately be the tool to compare the sustainability of the methodologies for recycling end of life LIBs.48Another exciting finding achieved in this work was the synthesis of novel nanoparticles. The identification of biogenic zero-valent Co NPs shows the potential of biology for producing unique nanoparticles with perhaps novel physicochemical properties. This finding provides us with new insight into the nanoparticle synthesis pathways of D. alaskensis G20 and deserves further investigation due to the relevance of bio-based Ni and Co nanoparticles as electrocatalysts in Hydrogen Evolution Reactions (HER).49,50
Experimental
Bacterial cultures preparation
S. oneidensis MR-1 was cultured aerobically in Luria Bertani (LB) media without NaCl, LB no salts (LBNS).51 Overnight cultures (5 ml) were used to inoculate larger volumes (100–200 ml) at 200 rpm and 20 °C until stationary phase (OD600 ∼ 6). Cultures of D. alaskensis G20 were grown in Postgate Media C (PGMC) as described elsewhere,22 washed with MOPS buffer (pH 7.5, 10 mM) and re-suspended in fresh MOPS buffer (OD600 = 1).Measurement of dissolved carbonate
Bacterial cultures of S. oneidensis MR-1 were collected at different growth levels (monitored by OD600) and centrifuged for 20 min at 4500 rpm, 20 °C. Afterwards supernantants were filter-sterilised (∅ < 0.2 μm) and stored at 4 °C until analysis. The carbonate analysis of the bacterial supernatant was performed by coulometric titation using a CM 5012 CO2 coulometer (UIC Inc., Joliet, IL, USA) and CM 5130 acidification unit. The coulometer measured the inorganic carbon mass (MIC) which was converted to the equivalent mass of CO32− (1 μg C = 5 μg of CO32−, under the assumption that most of the inorganic carbon is in a bicarbonate/carbonate form) using eqn (1) where V (ml) is the volume of the sample:| [CO32−] = (MIC × 5)/V | (1) |
Measurement of dissolved H2S
The concentration of H2S was measured in fully grown cultures of D. alaskensis G20 and cells resuspended in MOPS in the absence of metal using a H2S microsensor following manufacturer's guidelines (UNISENSE).Bioprecipitation of nickel, manganese, and cobalt complexes
Stock metal solutions were made in ultrapure water to a concentration of 10 g L−1 using Co·Cl2·6H2O, LiCl, MnSO4·H2O and NiCl2·6H2O as a source of Co2+, Li+, Mn2+ and Ni2+ ions respectively. Bacterial cell cultures and abiotic controls, fresh media and buffers, were incubated with metal solutions in 15 ml falcon tubes at a final volume of 2 ml, pH was not adjusted. S. oneidensis MR-1 treatments took place aerobically (20 °C and 200 rpm), whereas D. alaskensis G20 treatments took place in an anaerobic atmosphere (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) % CO2, 10
% CO2, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) % H2 in N2 atmosphere, static, 30 °C).
% H2 in N2 atmosphere, static, 30 °C).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C), and 100 μl of supernatant (dissolved fraction) acidified with 900 μl of 20% HNO3 (v/v). The acidified samples were digested for 5 h at 80 °C and diluted in ultrapure water prior to inductively coupled plasma optical emission spectrometry (ICP-OES) analysis on an Optima 8300 (PerkinElmer).51 Metal removal (%) from the dissolved fraction was calculated as the difference between the total concentration (Tc) of the metal added and the concentration of metal that remained in the dissolved fraction (Dc) as depicted in eqn (2):
000g and 4 °C), and 100 μl of supernatant (dissolved fraction) acidified with 900 μl of 20% HNO3 (v/v). The acidified samples were digested for 5 h at 80 °C and diluted in ultrapure water prior to inductively coupled plasma optical emission spectrometry (ICP-OES) analysis on an Optima 8300 (PerkinElmer).51 Metal removal (%) from the dissolved fraction was calculated as the difference between the total concentration (Tc) of the metal added and the concentration of metal that remained in the dissolved fraction (Dc) as depicted in eqn (2):| Metal removal (%) = ((Tc − Dc)/Tc) × 100 | (2) |
Differences between Co and Ni precipitation across different conditions were analysed with ANOVA.
Selective bioprecipitation of dissolved Mn from vehicular LIBs leachates
The LIB leachates used were prepared with two different cathode materials: A1C (NMC-532) and B1C (70% lithium manganese oxide spinel (LMO) with 25% lithium nickel manganese cobalt oxide) obtained from uncycled electric vehicle batteries. The cathode materials were dissolved in H2SO4 (0.1 M) at 50 °C over a period of 5 to 30 minutes with no agitation. Leachates of B1C cathode were also produced in HCl (0.1 M) at 20 °C for up to 5 h with no agitation. The bioprecipitation of metals contained in vehicular LIBs leachates was investigated following the same methodology used to investigate metal removal from single-metal solutions. Cultures of S. oneidensis MR-1 were incubated with 10% v/v of raw leachates (2 ml final volume) prepared in H2SO4 (0.1 M) and HCl (0.1M) for 20 h at 20 °C and 200 rpm. The concentration of metals was analysed by ICP-OES.Nanoparticle characterisation
Two bacterial-treatment bioprocessing approach
The different stages of the bioprocess approach are summarised graphically in Scheme 2 showing the two-bacterial treatments and the resulting fractions involved. The first treatment (T1) consisted of processing the raw leachate mixed with S. oneidensis MR-1 to precipitate mainly Mn (T1B). Afterwards the fraction containing the remaining dissolved metals (T1C) was further processed with D. alaskensis G20 (treatment 2, T2) to precipitate Ni and/or Co. The study was developed with leachates prepared with A1C and B1C cathode material delaminated with H2SO4 (0.5 M, 20 min at 50 °C). Carbonate–bicarbonate buffer (final concentration 91 mM) was added to the S. oneidensis MR-1 cell suspension just before the addition of the acidic leachate (10% v/v). Once treated with S. oneidensis MR-1 the precipitated metals (T1B) were separated from the metals in the dissolved fraction (T1C) by centrifugation (10 min at 4500 rpm). Fraction T1C was filter-sterilised (∅ < 0.2 μm) before incubation with D. alaskensis G20 cells (cell-only treatment, OD600 = 2, resuspended in MOPS buffer, pH 7.5) for 20 h at different concentrations of the pretreated leachate (T1C), by volume, 25% and 50%, labelled as T2A. The metal removal using the whole bacterial culture of D. alaskensis G20 (cells and extracellular matrix) was investigated with 50% v/v of leachate. Removal of dissolved metal was calculated as the difference between the total concentration of metal and the remaining concentration in the supernatant after centrifugation as described previously (eqn (2)) using ICP-OES.Author contributions
L. E. H. conceived and directed the project, V. E. B. planned and conducted experimental work, with support from G. M. H. A. and N. D. B. were responsible for STEM, EDXS and EELS analysis. Automotive LIB leachates were supplied by J. H. X-ray powder diffraction analysis was undertaken by C. K. and CryoEM by M. W. T. All authors contributed to discussions and manuscript writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Lisa Imrie and the EdinOmics facility for proteomics data and analysis, to Steve Mitchell at the Biology Scanning EM Facility (BioSem) for his help with the TEM (Wellcome Trust Multi User Equipment Grant (WT104915MA)), to Dr Gavin Sim (carbonate analysis, School of Geosciences, University of Edinburgh), Dr Lorna Eades for her support with the ICP-OES (School of Chemistry, University of Edinburgh) and Dr Neil Renault and Dr Shiwen Zhuang (IBioIC) (IBioIC, scale-up work). Shewanella oneidensis MR-1 was generously provided by Prof. Jeff Gralnick at Minnesota University.This research was supported by the Faraday Institution (grant codes FIRG005 and FIRG027, project website https://relib.org.uk) and EPSRC Fellowship (EP/N026519/1). CryoEM data was acquired in the cryoEM facility in School of Biological Sciences at the University of Edinburgh, which was set up with funding from the Wellcome Trust (087658/Z/08/Z) and SULSA. MWT is supported by Wellcome Centre for Cell Biology (203149).
References
- G. Santos, Transp. Policy, 2017, 59, 71–74 CrossRef.
- N. Rietmann and T. Lieven, J. Cleaner Prod., 2019, 206, 66–75 CrossRef.
- E. Fan, L. Li, Z. Wang, J. Lin, Y. Huang, Y. Yao, R. Chen and F. Wu, Chem. Rev., 2020, 120, 7020–7063 CrossRef CAS PubMed.
- M. Weil, S. Ziemann and J. Peters, The Issue of Metal Resources in Li-Ion Batteries for Electric Vehicles, Springer International Publishing, Cham, 2018 Search PubMed.
- B. K. Sovacool, S. H. Ali, M. Bazilian, B. Radley, B. Nemery, J. Okatz and D. Mulvaney, Science, 2020, 367, 30–33 CrossRef CAS PubMed.
- T. Or, S. W. D. Gourley, K. Kaliyappan, A. Yu and Z. Chen, Carbon Energy, 2020, 2, 6–43 CrossRef CAS.
- G. Harper, R. Sommerville, E. Kendrick, L. Driscoll, P. Slater, R. Stolkin, A. Walton, P. Christensen, O. Heidrich, S. Lambert, A. Abbott, K. Ryder, L. Gaines and P. Anderson, Nature, 2019, 575, 75–86 CrossRef CAS PubMed.
- W. Mrozik, M. A. Rajaeifar, O. Heidrich and P. Christensen, Energy Environ. Sci., 2021, 14, 6099–6121 RSC.
- O. Velázquez-Martínez, J. Valio, A. Santasalo-Aarnio, M. Reuter and R. Serna-Guerrero, Batteries, 2019, 5, 68 CrossRef.
- C. Lei, I. Aldous, J. M. Hartley, D. L. Thompson, S. Scott, R. Hanson, P. A. Anderson, E. Kendrick, R. Sommerville, K. S. Ryder and A. P. Abbott, Green Chem., 2021, 23, 4710–4715 RSC.
- Y. Jiang, X. Chen, S. Yan, S. Li and T. Zhou, Chem. Eng. J., 2021, 426, 131637 CrossRef CAS.
- T. Huang, T. Junjun, W. Liu, D. Song, L.-X. Yin and S. Zhang, Waste Manage., 2021, 126, 377–387 CrossRef CAS PubMed.
- G. Alvial-Hein, H. Mahandra and A. Ghahreman, J. Cleaner Prod., 2021, 297, 126592 CrossRef CAS.
- L. Brückner, J. Frank and T. Elwert, Metals, 2020, 10, 1107 CrossRef.
- X. Chen, D. Kang, J. Li, T. Zhou and H. Ma, J. Hazard. Mater., 2020, 389, 121887 CrossRef CAS PubMed.
- W. Gao, X. Zhang, X. Zheng, X. Lin, H. Cao, Y. Zhang and Z. Sun, Environ. Sci. Technol., 2017, 51, 1662–1669 CrossRef CAS PubMed.
- H. Zou, E. Gratz, D. Apelian and Y. Wang, Green Chem., 2013, 15, 1183–1191 RSC.
- M. Abdelbaky, L. Schwich, E. Crenna, J. R. Peeters, R. Hischier, B. Friedrich and W. Dewulf, Procedia CIRP, 2021, 98, 97–102 CrossRef.
- S. Windisch-Kern, E. Gerold, T. Nigl, A. Jandric, M. Altendorfer, B. Rutrecht, S. Scherhaufer, H. Raupenstrauch, R. Pomberger, H. Antrekowitsch and F. Part, Waste Manage., 2022, 138, 125–139 CrossRef CAS PubMed.
- Y. Choi and S. Y. Lee, Nat. Rev. Chem., 2020, 4, 638–656 CrossRef CAS.
- M. H. Wright, S. M. Farooqui, A. R. White and A. C. Greene, Appl. Environ. Microbiol., 2016, 82, 5402–5409 CrossRef CAS PubMed.
- M. J. Capeness, M. C. Edmundson and L. E. Horsfall, New Biotechnol., 2015, 32, 727–731 CrossRef CAS PubMed.
- M. Sethurajan and S. Gaydardzhiev, Resour., Conserv. Recycl., 2021, 165, 105225 CrossRef CAS.
- J. Baars, T. Domenech, R. Bleischwitz, H. E. Melin and O. Heidrich, Nat. Sustain., 2021, 4, 71–79 CrossRef.
- M. Li and J. Lu, Science, 2020, 367, 979–980 CrossRef CAS PubMed.
- N. Chubar, T. Visser, C. Avramut and H. de Waard, Geochim. Cosmochim. Acta, 2013, 100, 232–250 CrossRef CAS.
- L. Xiao, S. Wang, Y. Wang, W. Meng, B. Deng, D. Qu, Z. Xie and J. Liu, ACS Appl. Mater. Interfaces, 2016, 8, 25369–25378 CrossRef CAS PubMed.
- Q. Li, D. Liu, Z. Jia, L. Csetenyi and G. M. Gadd, Curr. Biol., 2016, 26, 950–955 CrossRef CAS PubMed.
- J. E. Johnson, P. Savalia, R. Davis, B. D. Kocar, S. M. Webb, K. H. Nealson and W. W. Fischer, Environ. Sci. Technol., 2016, 50, 4248–4258 CrossRef CAS PubMed.
- W. M. B. L. Vallee, Methods Enzymol., 1993, 226, 52–71 Search PubMed.
- B. Bennett, in Metals in Biology. Biological Magnetic Resonance, ed. G. Hanson and L. Berliner, Springer New York, New York, NY, 2010, vol. 29, pp. 345–370 Search PubMed.
- H. Dulay, M. Tabares, K. Kashefi and G. Reguera, Front. Microbiol., 2020, 11, 2992 Search PubMed.
- H. Yu, X. Yan, W. Weng, S. Xu, G. Xu, T. Gu, X. Guan, S. Liu, P. Chen, Y. Wu, F. Xiao, C. Wang, L. Shu, B. Wu, D. Qiu, Z. He and Q. Yan, J. Hazard. Mater., 2022, 426, 127795 CrossRef CAS PubMed.
- K. L. Keller, B. J. Rapp-Giles, E. S. Semkiw, I. Porat, S. D. Brown and J. D. Wall, Appl. Environ. Microbiol., 2014, 80, 855–868 CrossRef PubMed.
- F. Goulhen, A. Gloter, F. Guyot and M. Bruschi, Appl. Microbiol. Biotechnol., 2006, 71, 892–897 CrossRef CAS PubMed.
- C. F. Chang, H. Shuman and A. P. Somlyo, J. Bacteriol., 1986, 167, 935–939 CrossRef CAS PubMed.
- J. Sitte, K. Pollok, F. Langenhorst and K. Küsel, Geomicrobiol. J., 2013, 30, 36–47 CrossRef CAS.
- C. P. Kempes, P. M. van Bodegom, D. Wolpert, E. Libby, J. Amend and T. Hoehler, Front. Microbiol., 2017, 8, 31 Search PubMed.
- I. Moura, M. Teixeira, J. J. G. Moura and J. LeGall, J. Inorg. Biochem., 1991, 44, 127–139 CrossRef CAS PubMed.
- C. H. Luo, V. Shanmugam and C. S. Yeh, NPG Asia Mater., 2015, 7, e209 CrossRef CAS.
- G. Calvert, A. H. Kaksonen, K. Y. Cheng, J. Van Yken, B. Chang and N. J. Boxall, Minerals, 2019, 9, 563 CrossRef CAS.
- D. L. Thompson, J. M. Hartley, S. M. Lambert, M. Shiref, G. D. J. Harper, E. Kendrick, P. Anderson, K. S. Ryder, L. Gaines and A. P. Abbott, Green Chem., 2020, 22, 7585–7603 RSC.
- J. S. Terreblanche, D. L. Thompson, I. M. Aldous, J. Hartley, A. P. Abbott and K. S. Ryder, J. Phys. Chem. C, 2020, 124, 14622–14631 CrossRef CAS.
- Y. Liu, A. Serrano, V. Wyman, E. Marcellin, G. Southam, J. Vaughan and D. Villa-Gomez, J. Hazard. Mater., 2021, 402, 123506 CrossRef CAS PubMed.
- A. E. Otwell, R. W. Sherwood, S. Zhang, O. D. Nelson, Z. Li, H. Lin, S. J. Callister and R. E. Richardson, Environ. Microbiol., 2015, 17, 1977–1990 CrossRef CAS PubMed.
- M. Arenas-Salinas, J. I. Vargas-Pérez, W. Morales, C. Pinto, P. Muñoz-Díaz, F. A. Cornejo, B. Pugin, J. M. Sandoval, W. A. Díaz-Vásquez, C. Muñoz-Villagrán, F. Rodríguez-Rojas, E. H. Morales, C. C. Vásquez and F. A. Arenas, Front. Microbiol., 2016, 7, 1160 Search PubMed.
- R. L. López Steinmetz and S. Salvi, Earth-Sci. Rev., 2021, 217, 103615 CrossRef.
- F. Arshad, J. Lin, N. Manurkar, E. Fan, A. Ahmad, M.-N. Tariq, F. Wu, R. Chen and L. Li, Resour., Conserv. Recycl., 2022, 180, 106164 CrossRef CAS.
- Q. Wang, R. Guo, Z. Wang, D. Shen, R. Yu, K. Luo, C. Wu and S. Gu, Fuel, 2021, 293, 120440 CrossRef CAS.
- G. Li, J. Yu, J. Jia, L. Yang, L. Zhao, W. Zhou and H. Liu, Adv. Funct. Mater., 2018, 28, 1801332 CrossRef.
- M. J. Capeness, V. Echavarri-Bravo and L. E. Horsfall, Front. Microbiol., 2019, 10, 997 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Proteomics data. See DOI: https://doi.org/10.1039/d2gc02450k |
| ‡ Present address: Leibniz Institute for Crystal Growth, Max-Born-Str. 2, 12489 Berlin, Germany. |
| § Present address: Department of Structural Biology, Max Planck Institute of Biophysics, Max-von-Laue Strasse 3, 60438 Frankfurt am Main, Germany. |
| This journal is © The Royal Society of Chemistry 2022 |