Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Oxidative cleavage of polysaccharides by a termite-derived superoxide dismutase boosts the degradation of biomass by glycoside hydrolases†
João Paulo L.
Franco Cairo
 abc,
Fernanda
Mandelli
d,
Robson
Tramontina
e,
David
Cannella‡
abc,
Fernanda
Mandelli
d,
Robson
Tramontina
e,
David
Cannella‡
 b,
Alessandro
Paradisi
b,
Alessandro
Paradisi
 c,
Luisa
Ciano§
c,
Luisa
Ciano§
 c,
Marcel R.
Ferreira
c,
Marcel R.
Ferreira
 f,
Marcelo V.
Liberato
e,
Lívia B.
Brenelli
bd,
Thiago A.
Gonçalves
a,
Gisele N.
Rodrigues
d,
Thabata M.
Alvarez
g,
Luciana S.
Mofatto
f,
Marcelo V.
Liberato
e,
Lívia B.
Brenelli
bd,
Thiago A.
Gonçalves
a,
Gisele N.
Rodrigues
d,
Thabata M.
Alvarez
g,
Luciana S.
Mofatto
 h,
Marcelo F.
Carazzolle
h,
José G. C.
Pradella
d,
Adriana F.
Paes Leme
i,
Ana M.
Costa-Leonardo
h,
Marcelo F.
Carazzolle
h,
José G. C.
Pradella
d,
Adriana F.
Paes Leme
i,
Ana M.
Costa-Leonardo
 j,
Mário
Oliveira-Neto
f,
André
Damasio
j,
Mário
Oliveira-Neto
f,
André
Damasio
 a,
Gideon J.
Davies
a,
Gideon J.
Davies
 c,
Claus
Felby
b,
Paul H.
Walton
c,
Claus
Felby
b,
Paul H.
Walton
 *c and
Fabio M.
Squina
*c and
Fabio M.
Squina
 *e
*e
aDepartment of Biochemistry and Tissue Biology, Institute of Biology, University of Campinas (UNICAMP), Campinas, São Paulo, Brazil
bDepartment of Geosciences and Natural Resource Management, Faculty of Science, University of Copenhagen, Rolighedsvej 23, DK-1958 Frederiksberg C, Denmark
cDepartment of Chemistry, University of York, York, YO10 5DD, UK. E-mail: paul.walton@york.ac.uk
dBrazilian Biorenewables National Laboratory, Brazilian Center for Research in Energy and Materials, Campinas, São Paulo, Brazil
ePrograma de Processos Tecnológicos e Ambientais da Universidade de Sorocaba (UNISO), Sorocaba, SP, Brazil. E-mail: fabio.squina@prof.uniso.br
fDepartamento de Física e Biofísica, Instituto de Biociências, Universidade Estadual Paulista, UNESP, Botucatu, São Paulo, Brasil
gPrograma de Mestrado e Doutorado em Biotecnologia Industrial, Universidade Positivo, Curitiba, PR, Brasil
hLaboratório de Genômica e Expressão, Departamento de Genética, Evolução e Bioagentes, Instituto de Biologia, Universidade de Campinas, UNICAMP, Campinas, São Paulo, Brasil
iLaboratório Nacional de Biociências (LNBio) do Centro Nacional de Pesquisa em Energia e Materiais (CNPEM), Campinas, São Paulo, Brasil
jLaboratório de Cupins, Departamento de Biologia Geral e Aplicada, Instituto de Biociências, Universidade Estadual Paulista, UNESP, Rio Claro, São Paulo, Brasil
First published on 12th May 2022
Abstract
Wood-feeding termites effectively degrade plant biomass through enzymatic degradation. Despite their high efficiencies, however, individual glycoside hydrolases isolated from termites and their symbionts exhibit anomalously low effectiveness in lignocellulose degradation, suggesting hereto unknown enzymatic activities in their digestome. Herein, we demonstrate that an ancient redox-active enzyme encoded by the lower termite Coptotermes gestroi, a Cu/Zn superoxide dismutase (CgSOD-1), plays a previously unknown role in plant biomass degradation. We show that CgSOD-1 transcripts and peptides are up-regulated in response to an increased level of lignocellulose recalcitrance and that CgSOD-1 localizes in the lumen of the fore- and midguts of C. gestroi together with termite main cellulase, CgEG-1-GH9. CgSOD-1 boosts the saccharification of polysaccharides by CgEG-1-GH9. We show that the boosting effect of CgSOD-1 involves an oxidative mechanism of action in which CgSOD-1 generates reactive oxygen species that subsequently cleave the polysaccharide. SOD-type enzymes constitute a new addition to the growing family of oxidases, ones which are up-regulated when exposed to recalcitrant polysaccharides, and that are used by Nature for biomass degradation.
1. Introduction
Lignocellulosic biomass (LB) is the main feedstock in the production of second-generation biofuels.1 Derived from the plant cell walls (PCWs), the carbohydrate portion of LB is composed mainly of the following polysaccharides: cellulose, (40–50% of the biomass total composition), diverse hemicelluloses (25–30%), and pectin (5–10%). Lignin comprises around 20–25% of LB depending on origin.2 The complex 3D structures of the biopolymers in LB hinder its use as a feedstock in biofuel production, since its sustainable breakdown into soluble saccharides is both chemically and mechanically difficult. As such, LB demands extensive pre-treatment steps to reduce both its physical and chemical recalcitrance before it can be processed into soluble sugars by enzymes.2 At the operational level of a biorefinery, this degradation is currently achieved through the addition of enzyme cocktails which contain a combination of glycoside hydrolases (GHs) and accessory enzymes such as carbohydrate esterases. This enzymatic saccharification of the plant material is a crucial, yet expensive, processing step during the production of second-generation biofuels.3Biomass-degrading organisms have evolved a variety of enzymes to overcome LB recalcitrance. However, despite the effectiveness of these organisms in utilizing LB, this efficiency is not yet fully replicated by industrial cocktails.4 Therefore, in the last decade, studies have concentrated on discovering what additional carbohydrate-active enzymes (CAZymes) can increase LB degradation efficiency when used in conjunction with other enzymes. A significant recent breakthrough in this regard was the discovery of Cu-containing lytic polysaccharide monooxygenases (LPMOs)5,6 belonging to the Auxiliary Activities (AAs) families AA9-AA11, AA13-AA17.7 These enzymes catalyze the oxidative cleavage of various polysaccharides (cellulose, hemicelluloses, pectin, starch and chitin), boosting significantly the activity of the enzymatic cocktails during biomass degradation both in Nature and industry.7 Also, as part of the endeavor to discover new enzymes, the exploration of Nature's biodiversity throughout ‘omics’ studies, has now been extended beyond bacteria and fungi,8–10 to look at more diverse organisms, for instance firebrats, beetles and termites.11–13
In this context, termites, particularly within the worker caste, are notably efficient decomposers of LB, degrading up to 90% of the ingested plant biomass.14 In what might be coined ‘the perfect biorefinery’ studies have shown how termites first reduce particle size in their mandibles before arrival of the masticated LB in the fore and midguts where endogenous cellulases, glucosidases and other CAZymes, such as esterases and laccases, liberate monosaccharides and oligosaccharides.15 Finally, the part-digested material moves to the hindgut where CAZymes from symbiotic microorganisms generate sugars that will be fermented to acetate and short-chain fatty acids that are the primary source of termite nutrition.16,17
Given their high efficiency, termites now present an opportunity for the discovery of new enzymes that could be used in biomass utilization.11,18 In this regard, many studies have focused on the lower termite Coptotermes gestroi,19–21 which was introduced in Brazil at the beginning of the last century and rapidly became a major urban pest.22 The especially high biomass-degrading capacity of C. gestroi has made it a rich source of enzymes, with some already used in biomass-to-bioproducts applications.23,24 Notwithstanding its potential however, studies on the use of C. gestroi and other termite-derived glycoside hydrolases, from both endogenous and symbiotic origins, have repeatedly and somewhat puzzlingly shown that the catalytic efficiencies of isolated enzymes on recalcitrant PCW are low.20,25–27 This observation shows that our understanding of the consortia of enzymes used by these organisms to degrade biomass is incomplete. Our current study thus seeks to determine the ‘missing’ enzymatic components (auxiliary activity, AA enzymes) in the digestive secretome of C. gestroi.
In terms of what is currently understood about AA enzymes in termites, it was recently in shown that two LPMOs belonging to the family AA15 are chitin-active enzymes in C. gestroi, but that these have no ostensible roles in lignocellulose degradation.28C. gestroi and other lower termites do secrete in their guts non-LPMO redox-active enzymes related to pro-oxidation, antioxidation and detoxification processes (dubbed PAD enzymes).18,19,27 These PADs include p450s, alcohol dehydrogenases, glutathione S-transferases, catalases, aldo-keto reductases (AKR) and copper-dependent superoxide dismutases (SOD).19,29,30 Yet, despite their prevalence, there is limited direct evidence about the role of PAD enzymes in lignocellulose degradation and detoxification of lignin-derived phenolic compounds in termite gut or other organisms.27,31
Among these potential new families of AA enzymes, those classified as Cu/Zn superoxide dismutases perhaps are the most promising since previous studies have shown that the SOD group displays strikingly high expression levels in termites that had been fed with lignocellulose19,27 or in bacteria and fungi grown on plant biomass.8,29,32 Indeed, a patent claims that Cu/Zn SOD from termite can increase the saccharification of lignocelluloses when mixed with termite endogenous and symbiotic GHs.33
SOD enzymes are widely distributed. They catalyze the dismutation of the superoxide anion radical (O2˙−) into oxygen and hydrogen peroxide.34 SODs are metalloenzymes, divided into four families depending on the metals found at the active site: Cu/Zn, Ni, Mn, or Fe.34 One of the most studied is the Cu/Zn SOD group, carrying in their catalytic site a copper ion (Cu2+) and a zinc ion (Zn2+) linked by an imidazolate bridge. The redox-active Cu directly interacts with the substrates in the following pair of reactions:
| Cu2+-SOD + O2˙− → Cu+-SOD + O2 | (1) |
| Cu+-SOD + O2˙− + 2H+ → Cu2+-SOD + H2O2 | (2) |
Beyond their principal role of superoxide scavenging, SODs are also known to produce hydroxyl radicals (˙OH) when exposed to high concentrations of hydrogen peroxide.35–37 For the purposes of the study described herein, the ˙OH radical is able to cleave glucose-based polysaccharides, generating several oxidized ion products as recently reported by Boulos and Nyström.38 Fenton chemistry (Fe2+ + H2O2 → Fe3+ + ˙OH + OH−) is similarly utilized by brown-rot fungi to depolymerize lignocellulosic biomass.39
Here we report the isolation of a Cu/Zn SOD from C. gestroi. We show that during termite feeding experiments on sugarcane bagasse, CgSOD-1 transcripts and peptides are up-regulated in response to the increased level of lignocellulose recalcitrance. We further demonstrate that CgSOD-1 localizes in the fore- and midgut of C. gestroi together with its major cellulase. Furthermore, this SOD works in synergy with endogenous termite GHs for polysaccharide breakdown, although this boosting effect is reduced/disappears when other biomass-degrading enzymes are added, revealing the complex relationship between enzyme components and the efficiency of biomass degradation. We show that this SOD degrades polysaccharides through an oxidative mode of action, from which it boosts the action of glycosidic hydrolases in an analogous manner to that of LPMO enzymes. Thus CgSOD-1 adds a new member to the group of ‘Auxiliary Activity’ enzymes used by Nature in biomass utilization.
2. Results
Lignocellulose recalcitrance influences Cu/Zn SOD expression in C. gestroi
Recently, the expression of PAD enzymes was reported to be up-regulated when termites were fed on lignocelluloses.18,19,27 The expression of nine putative genes encoding for Cu/Zn SODs, identified in the genome of C. gestroi,19 was evaluated by transcriptomics and proteomics after 21 days of a feeding experiment. Termite workers were fed either raw sugarcane bagasse (SCB), pretreated phosphoric acid sugarcane bagasse (PASCB) or pretreated sodium chlorite/HCl delignified sugarcane bagasse (DELSCB) as sole carbon sources (Fig. 1a). The highest abundance of SOD transcripts was observed in SCB-fed animals, as shown in the heat map chart (Fig. 1b). According to the dendrogram analysis (Fig. 1c), the gene expression of CGSOD-1, CGSOD-8 and CGSOD-9 clustered together, and these transcripts presented the highest number of normalized Reads Per Kilobase Millions (RPKMs) among all Cu/Zn SODs. The same three genes presented a near doubling in up-regulation in SCB compared to both PASCB and DELSCB (log2 of fold change <−0.5 and False Discovery Rate (FDR) <0.05) (Table S1†). Similarly, the expression of two main C. gestroi's cellulases from family GH9 (CGEG1 and CGEG2) as well as the two main β-glucosidases GH1 (CGBG1 and CGBG2) was also monitored, revealing that these enzymes are also highly up-regulated in response to SCB compared to the other feeding conditions (Table S1†).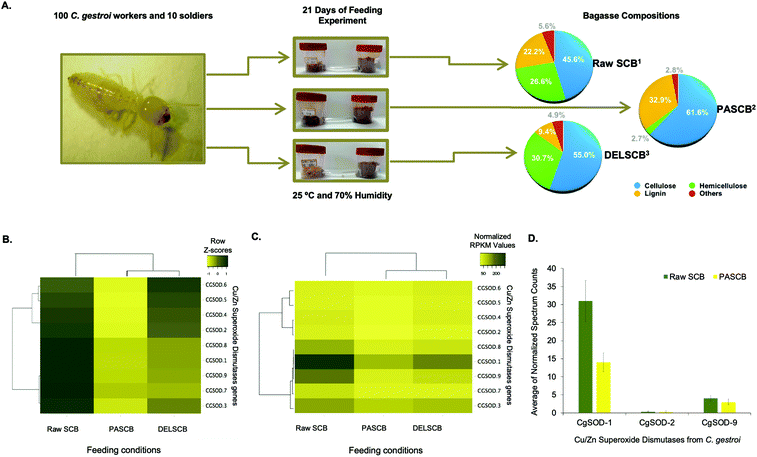 | ||
| Fig. 1 Feeding assay scheme and plots from the differential expression of superoxide dismutase in workers of C. gestroi fed on different sugarcane bagasse. (A) Schematic view of feeding assays using 100 C. gestroi workers and 10 soldiers. (1) The compositional analysis of Raw Sugarcane Bagasse (SCB) was: 45.6% cellulose, 26.6% hemicellulose, and 22.2% lignin.40(2) Pre-treated Phosphoric Acid Sugarcane Bagasse (PASCB) has 61.7% cellulose, 2.8% hemicellulose, and 33.0% lignin41 and (3) Delignified Sugarcane Bagasse (DELSCB) has 55% cellulose, 30.7% hemicellulose, and 9.4% lignin.42 (B) Nine Cu/Zn SOD genes were identified previously in the genome of C. gestroi. Heat map analysis of the Z-score for the complete-linkage hierarchical clustering based on Pearson distance measure, showing the differential gene expression pattern of Cu/Zn SODs in Raw SCB, PASCB and DELSCB diets. (C) Heat map analysis showing the number of normalized RPKMs for each diet. The normalized RPKM was represented by the average of RPKMs among the replicates. Colour from light yellow to dark yellow indicates low to high gene transcription levels. (D) The normalized peptide spectrum counts for three endogenous Cu/Zn SOD enzymes from C. gestroi between raw SCB and PASCB feeding conditions. | ||
Following on from the transcriptomics, proteomics similarly identified three Cu/Zn SODs (Fig. 1d), including two differentially expressed in the RNA-Seq: CgSOD-1 and CgSOD-9. The proteome revealed that CgSOD-1 was differentially expressed, resulting in levels ∼2× higher in SCB-fed termites than PASCB-fed individuals (p-value: 0.0032). CgSOD-1 also exhibited the highest number of spectrum counts (and total peptides), among the three Cu/Zn SODs identified (Table S2†). The two major C. gestroi cellulases (CgEG-1 and CgEG-2) and β-glucosidases (CgBG-1 and CgBG-2) were likewise identified in the proteomics analysis. Collectively, our data support that CgSOD-1 and cellulases are overproduced in SCB-feeding experiments compared to other conditions (Table S2†).
Since SCB was not pretreated, it represented the material with the highest recalcitrance level (i.e., lower porosity and higher crosslinking between the polysaccharides and lignin).40 PASCB has 90% less hemicellulose than SCB as well as partially modified lignin, as a result of the pre-treatment process.41 Likewise, DELSCB presents instead a lower lignin content (about 65% less) than SCB and a comparable amount of hemicellulose, albeit the latter is modified as a result of the pre-treatment step.42 Thus, the up-regulation of SOD transcript and proteins during feeding, especially for CgSOD-1, correlates with the degree of LB in this substrate.
CGSOD-1 encodes a canonical Cu/Zn superoxide dismutase
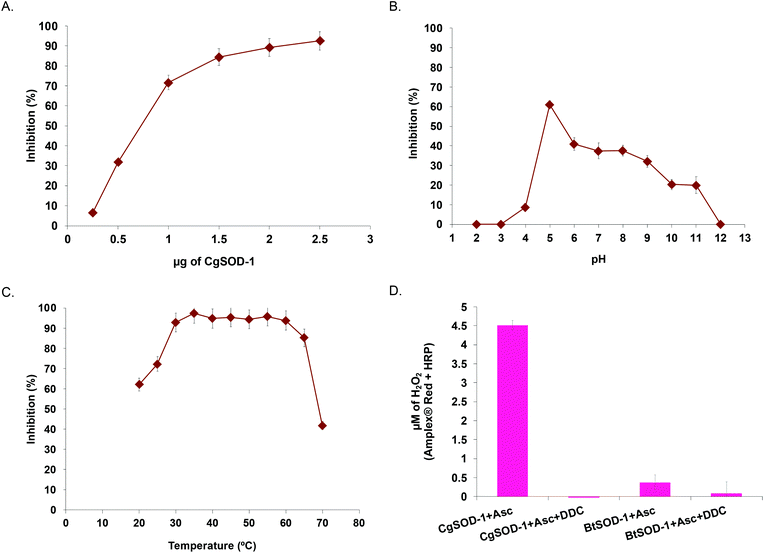
To evaluate whether CGSOD-1 encodes a functional superoxide dismutase, the gene was amplified from a C. gestroi cDNA library and heterologously expressed in E. coli (Fig. S1a†). CgSOD-1 production was confirmed by LC-MS/MS (Fig. S1B and C†) and its initial activity was assayed using the pyrogallol method (inhibition of autoxidation), presenting an IC50 of 1.17 μg (4.7 μg mL−1) (Fig. 2a). This is a high scavenger activity when compared with bacterial (1.6 mg mL−1)43 and fungal (1.3 mg mL−1)44 SODs. In addition, CgSOD-1 was active for superoxide dismutation from pH 4.0 to 11.0, with the optimum activity around pH 5.0 (Fig. 2b). The optimum temperature activity ranged from 30 °C to 60 °C, retaining ∼80% of the activity from 30 °C to 65 °C, (Fig. 2c and S2a,† respectively). The CgSOD-1 secondary folding (Fig. S2b†) and thermal stability were confirmed by Far-UV Circular Dichroism (CD), giving a melting temperature of 66(1) °C (R2 = 0.9996) (Fig. S2c†). CgSOD-1 produced around 4.5 μmol L−1 of H2O2 (after 30 min reaction at 30 °C), using ascorbic acid as electron/superoxide donor (Fig. 2d) and activity was lost in the presence of the Cu chelator diethyldithiocarbamate (DDC). CgSOD-1 displayed higher activity than the prototypical bovine Cu/Zn BtSOD-1 from Bos taurus, (Fig. 2d).Given the high sequence similarity of CgSOD-1 to previous Cu/Zn SODs (around 75%), a three-dimensional (3D) structure of CgSOD-1 was generated using SWISS-MODEL (QMEAN of 1.08) and the crystal structure PDB: 3L9Y from Bombyx mori as a template. The CgSOD-1 was assembled as a homodimer (subunits A and B) in the 3D model, showing a typical β-barrel folding, with eight β-sheets and several loops. The copper and zinc metals ions were also modelled in the active site (Fig. 3a), according to their analogous positions in other Cu/Zn SODs. The electrostatic surface potentials for CgSOD-1 and BtSOD-1 were also calculated and, as for other SODs, a positively charged pocket of amino acids was found around the Cu ion for both proteins (Fig. 3b and c). Conversely, CgSOD-1 has an overall negative net charge around the Zn ion, while BtSOD-1 is positively and neutrally charged in the same region. A detailed comparison between the active site of CgSOD-1 and BtSOD-1 (PDB: 1E9Q) was performed, which confirmed highly conserved positions of amino acids around the metals (Fig. 3D), including the copper-His63-zinc imidazolate bridge essential for the enzymatic mechanism of Cu/Zn SODs. According to small-angle X-ray scattering (SAXS) experiments (Fig. S3a–d†), CgSOD-1 shows high flexibility with an estimated molecular mass in solution of 33.2 kDa, indicating the predominance of homodimers. The SAXS ‘ab initio’ model was also built and the three-dimensional dummy atom model (DAM) of CgSOD-1 was determined from the SAXS curves (Fig. 3e) (Table S3†), confirming the homodimer configuration as well as validating the molecular modelling prediction.
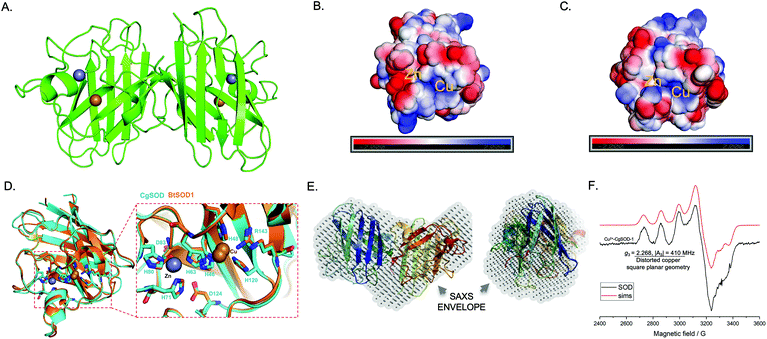 | ||
| Fig. 3 Structural and spectroscopic hallmarks for CgSOD-1. (A) Cartoon representation of the homology model for CgSOD-1 as a homodimer, showing the Zn (grey sphere) and Cu (golden sphere) metals in the catalytic pocket. The model was generated by SWISS-MODEL server, using the crystal structure PDB: 3L9Y from Bombyx mori as a template. The electrostatic potential surface at pH 6.0 of CgSOD-1 (B) and BtSOD-1 (C) shows the positively charged region (blue) present on both protein surfaces around the Cu ion. Around the Zn ion, a neutral (white) and positively charged surface (blue) is shown for BtSOD-1 and negatively charged (red) for CgSOD-1. (D) The active site of CgSOD-1 (blue) superposed with equivalent residues from the commercial Cu/Zn-SOD BtSOD-1 (orange) (1E9Q pdb) from Bos Taurus. All the residues that have contact with the metals are conserved in CgSOD-1. (E) SAXS analysis showing the superposition of ‘ab initio’ dummy atom model (DAM) (in grey) for CgSOD-1 envelope and the crystal structure BmCu-ZnSOD (3L9Y). The left model is rotated 90° along the x-axis from the right model. (F) Continuous-wave X-band EPR spectrum (black line) at 9.3 GHz and 160 K with simulation (red line) for CgSOD-1. The spectrum shows a typical type 2 copper (Cu2+) at the active site for CgSOD-1. | ||
X-band Electron Paramagnetic Resonance (EPR) spectroscopy studies were conducted to determine the electronic state of the active site copper from CgSOD-1. The frozen solution spectrum of the protein (Fig. 3f) is consistent with what was previously reported in the literature for other Cu/Zn SODs.45,46 The spin Hamiltonian parameters derived from the simulations of the continuous wave (cw) EPR spectrum of CgSOD-1 are indicative of a type 2 copper site according to the Peisach-Blumberg classification,47 with a certain degree of distortion from square planar geometry (g3 = 2.268, |A3| = 410 MHz) (Table S4†). The overall spectral envelope is characterized by a SOMO with mostly d(x2 − y2) character with small amounts of d(z2) mixing, that would arise from a distorted square planar structure.
CgSOD-1 is co-located with the termite main endo-cellulase and the site of H2O2 production in vivo
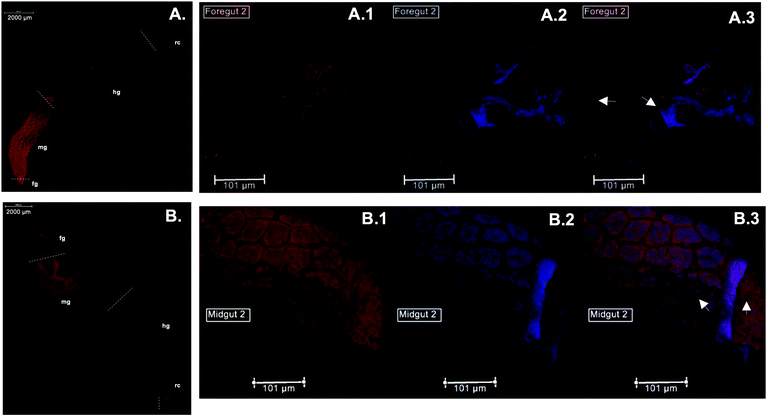
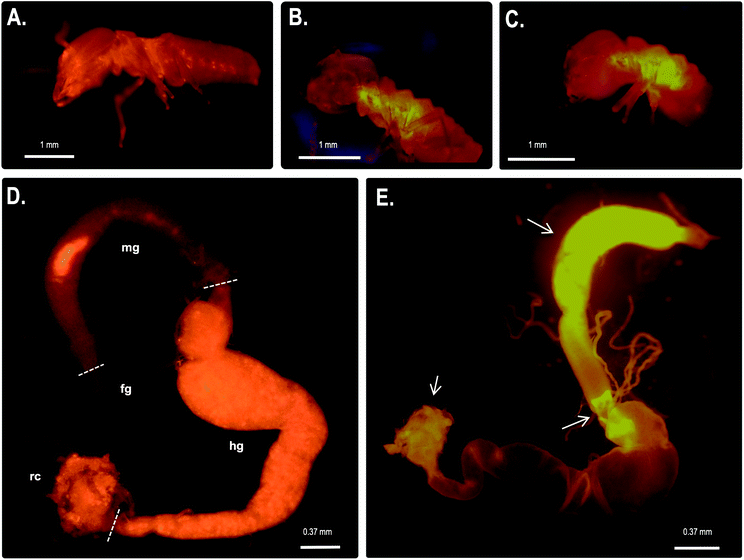
Immunolocalization assays using anti-CgSOD-1 and anti-CgEG-1 (endo-β-1,4-glucosidase 1 from Coptotermes gestroi) antibodies were performed on dissected C. gestroi guts. Here, the localization of CgEG-1 can be considered as a positive control because it is expected to locate in the foregut and midgut regions48 (Fig. 4a). CgEG-1 was found in the lumen of the gut region (Fig. 4 a.1 and a.3, controls in Fig. S4a, b and c†). For CgSOD-1, the gut tissue similarly shows a strong signal in the midgut, as well as in the foregut, mainly in the pro-ventricle part. A low fluorescence signal was also observed for CgSOD-1 in the rectum, as well as around the enteric valve (Fig. 4b), whilst no signal was detected at the hindgut. The expanded view from the midgut segment shows that CgSOD-1 is outside of the gut cells, confirming its presence in the lumen of the midgut (Fig. 4 b.1 and b.3).We also performed fluorescence microscopy to evaluate H2O2 production in vivo using the Amplex® Red reagent. As shown in Fig. 5b and c, a strong signal was detected in the thorax and abdomen of C. gestroi, demonstrating the formation of hydrogen peroxide inside the gut, further confirmed after gut dissection and Amplex® Red labelling. The strong CgSOD-1 immunofluorescence, in the fore- and midguts therefore correlates strongly with H2O2 production in vivo, Fig. 5f.
CgSOD-1 boosts the activity of termite glycoside hydrolases, promoting substrate oxidation through the generation of reactive oxygen species
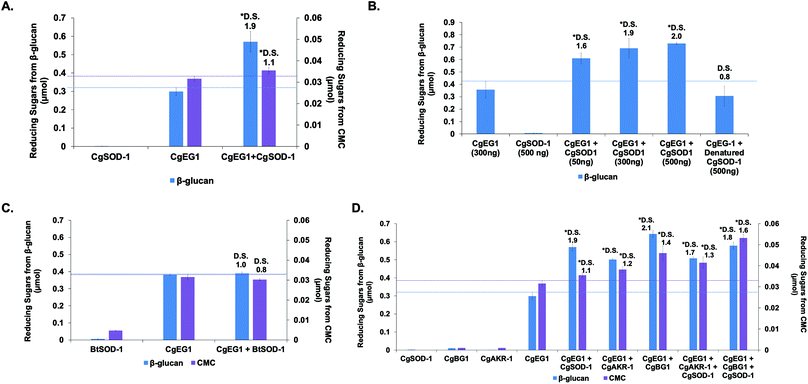
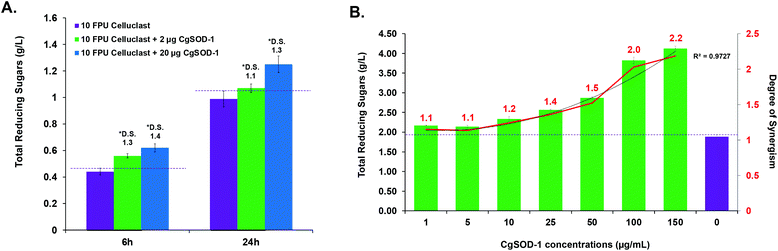
Previous reports from our group and the literature have shown that the cellulase CgEG-1-GH9 works in synergy for the saccharification of β-glucan and carboxymethylcellulose (CMC) with the main termite β-glucosidase, CgBG-1-GH1, and also with another PAD enzyme, the aldo-keto reductase 1 from C. gestroi (CgAKR-1).20,27,31CgSOD-1 localizes in the fore- and mid-guts of C. gestroi together with CgEG-1-GH9, and therefore we investigated whether CgSOD-1 could similarly boost the activity of CgEG-1 on model substrates. The degree of synergism (DS) among the enzymes was calculated as abc/(a + b + c) as described previously,49 where abc is the result in μmol of sugar released by the enzymes together and a, b and c are the results of each enzyme alone. DS ≥ 1.1 indicates a synergism effect for the combination of the enzymes.Furthermore, the increase of CgSOD-1 loads in the assays increased the reducing sugar amount released from β-glucan (Fig. 6b) from around 0.6 μmol to around 0.75 μmol, as well as the DS (1.6 to 2.0). As a control, the addition of denatured CgSOD-1 did not increase the saccharification yield of CgEG-1 (Fig. 6b), showing no DS as well as any putative synergistic effect of the free copper and zinc. Moreover, the ability of a different SOD, BtSOD-1, to work in synergy with CgEG-1 was also tested using β-glucan and CMC as substrates; however, no DS was found (Fig. 6c) in the tested conditions, and an inhibitory effect was found for CMC saccharification (DS of 0.8).
For CMC saccharification, the presence of CgSOD-1 together with CgEG-1 + CgAKR-1 (DS of 1.3) or CgEG-1 + CgBG-1 (DS of 1.6) increased the final sugar yields (0.042 and 0.055 μmol of reducing sugars respectively), when compared with the coupled enzymes (0.038 μmol for CgEG-1 + CgAKR-1 and 0.035 for CgEG-1 + CgBG-1) (Fig. 6d). Again, no inhibitory effect was found when compared with CgEG-1 alone (0.03 μmol for CMC).
Interestingly, our data point to a complex picture of the synergistic effects of biomass-degrading enzymes. In the first instance, CgSOD-1 significantly boosts the action of a single glycoside hydrolase. Somewhat counterintuitively however, no boosting and possibly even a slight inhibitory effect is seen when a third enzyme (CgBG-1 or CgAKR-1) is then added to the biomass-degrading system. While the ostensible contradiction brings into question whether CgSOD-1 is a natural boosting enzyme at all, we note that the effects of enzyme degradation by oxidative chemistry and the potential for inhibition of CgBG-1 by oxidized saccharides are factors which may be more evident in our laboratory-based system as compared to natural systems. For instance, both the temporal and relative concentrations of enzyme components are carefully controlled by organisms during biomass degradation—a feature absent in our current laboratory study. This constitutes an area for future study.
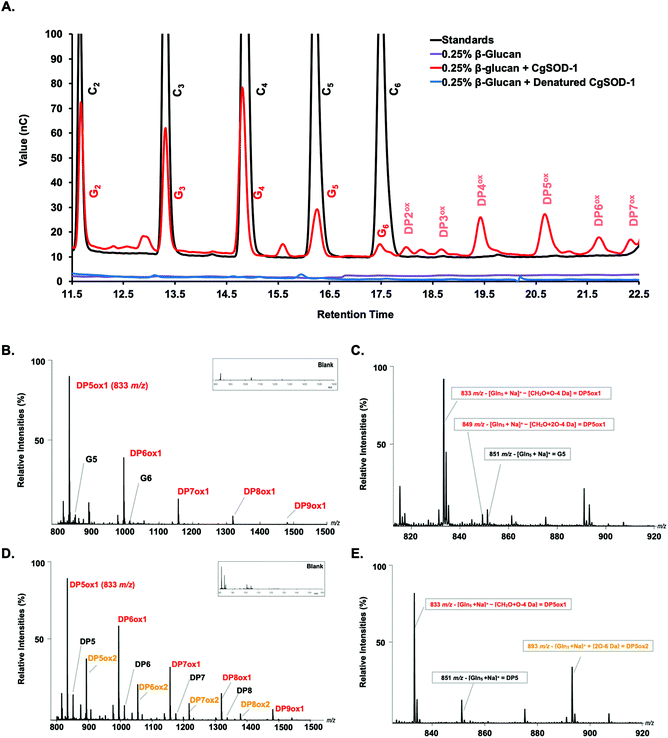
Notably, peaks were also detected in the region of the chromatogram where oxidized oligosaccharides elute with a degree of polymerization (DP) ranging from 2 to 7, including four minor signals at 18, 18.5, 21.5, and 22.5 minutes of retention and two major ones at 19.5 and 20.5 minutes (Fig. 7a). Peaks with similar retention times have previously been assigned to oxidized cello-oligosaccharide with DP2 and DP3 for the early peaks (18 and 18.5 min), and to DP4, DP5, DP6 and DP7 for the later peaks (19.5, 20.5, 21.5 and 22.5 min) (Fig. 7a).53
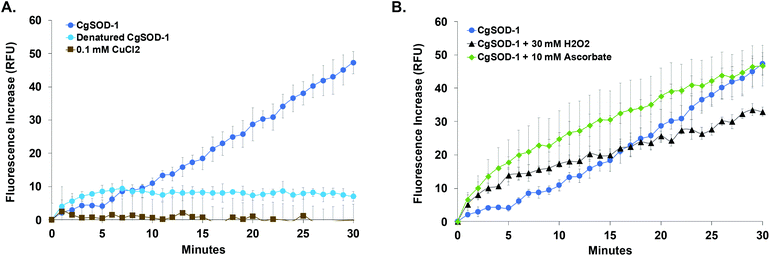
In this context, we note that the native glucose oligosaccharides eluted from the reaction could be also the result of on-column C4 oxidized sugar degradation due to the high pH of the chromatography method (known for C4 oxidized oligos from substrates oxidized by LPMOs, where the C4 oligos tend to be degraded to the native sugars one unit shorter).52 Incubation of denatured CgSOD-1 or 0.1 mmol L−1 CuCl2 with β-glucan and no signals arising from the polysaccharide degradation were detected (Fig. 7a and S5a†). Bovine Cu/Zn BtSOD-1 was also incubated following the reaction conditions described above; however, again the production of putative native or oxidized oligosaccharides was not observed (Fig. S5a†).
In further confirmation of the oxidative action of CgSOD-1, the degradation of β-glucan only by Celluclast® in the absence of CgSOD-1 was also investigated. The HPAEC-PAD chromatogram showed that glucose oligosaccharides released by Celluclast from β-glucan contained a mixture of β-1,4 and β-1,3 linkages ranging from G2 to G6, eluting in similar retention time (Fig. S5b†) to cello-oligosaccharides (only β-1,4) as previously reported.54,55 Interestingly, no oxidative products were identified from these reactions.
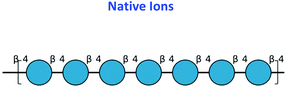
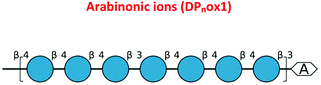
To validate further the enzyme-dependent production of oxidized sugars, β-glucan or Avicel were incubated with CgSOD-1 for 24 h and the reaction products analyzed by MALDI/ToF MS. Interestingly, the series of ion products, showing both native and oxidized gluco-oligosaccharides (Table 1), were comparable to previous reports of gluco-oligosaccharide oxidation through Fenton chemistry.38,56 For β-glucan, the acidic product ions were detected at m/z 833, 995, I1157, 1319 and 1481 (DPnox1), indicating an ion series with DP ranging from DP5 to DP9 (Glc5ox1 to Glc9ox1), separated by 162 Da, a hexose molecule (Fig. 7b). According to Simões et al.56 and Boulos and Nyström,38 the loss of 18 Da from sodiated non-oxidized native species ([Glcn + Na]+) is equivalent to a loss of a [CH2O + O − 4 Da] (Fig. 7c) from the reducing end glucose unit, which corresponds to the formation of arabinonic acid product due to oxidative cleavage of the C1–C2 bond combined with the formation of two keto groups along the oligosaccharide chain (Table 1).
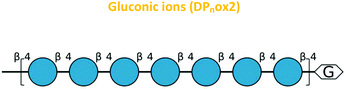
Another ion product from the arabinonic series with a loss of −2 Da was also identified at m/z 849 (Glc5 –CH2O + 2O − 4 Da) (Fig. 7c) (Table 1). Similar to the ion products derived from β-glucan oxidation (DPnox1), the Avicel degradation also exhibited another series of ion products at m/z 893, 1055, 1217, and 1379 (DPnox2) (Fig. 7d), which corresponds to a gain of 42 Da to [Glcn + Na]+ (Fig. 7e) (Table 1). These ion products were also separated by a hexose monomer (162 Da), suggesting an ion series with a degree of polymerization ranging from DP5 to DP8. The gain of 42 Da from the native ion is equivalent to the generation of gluconic acid (Glcn + 3O − 6 Da) in the reducing end glucose unit with a formation of three keto groups along the oligosaccharide (Fig. 7e) (Table 1).
Furthermore, reactions in the presence of 10 mmol L−1 ascorbic acid or 30 mmol L−1 hydrogen peroxide (in these concentrations, the ascorbic acid can act as a reducing agent/superoxide donor for SODs58 and hydrogen peroxide can stimulate the production of hydroxyl radicals by Cu/Zn SODs or MnSODs35–37) revealed that CgSOD-1, in the presence of both compounds, could initially increase the velocity of hydroxyl radical generation faster than CgSOD-1 alone (Fig. 8b). However, after 30 minutes of reaction, CgSOD-1 alone achieved equivalent or higher amounts of ˙OH (Fig. 8b), suggesting that the presence of these compounds at the tested concentrations and incubation time, may cause damage and inactivation of CgSOD-1.
CgSOD-1 increases the saccharification of sugarcane bagasse by commercial and laboratory fungal CAZymes preparations
The ability of CgSOD-1 to boost recalcitrant plant biomass saccharification by fungal lignocellulolytic CAZymes was also assessed. Two different Celluclast® loadings (5 and 10 FPU) and two different concentrations of CgSOD-1 were used on steam-exploded pretreated sugarcane bagasse (Fig. 9a and S6a†). After 6 hours of reaction, the assays containing 10 FPU Celluclast® exhibited DS of 1.3 and 1.4 for 2 and 20 μg of CgSOD1, respectively (Fig. 9a). After 24 hours, the reactions with 20 μg CgSOD-1 exhibited a DS of 1.3, reaching the highest reducing sugar release (1.2 g L−1) (Fig. 9a and Fig S6b†). Notably, and consistent with previous data, no synergistic effects were detected in the assays containing ®Celluclast with 20 μg of BtSOD-1 (Fig. S6c†). Similarly, DDC, a specific inhibitor for Cu/Zn SODs prevented the synergistic action of CgSOD-1 (Fig. S6d†).Supplementation of non-commercial Penicillium echinulatum hypercelluloytic secretome (free of preservatives) similarly exhibited an increase of saccharification yields of hydrothermally pretreated sugarcane bagasse in response to the CgSOD1 (Fig. 9b), reaching around 4 g L−1 of reducing sugar. In addition, the degree of synergy using 50 μg mL−1 of CgSOD-1 was 1.5, with 100 μg mL−1 enzyme was 2.0 and with 150 μg mL−1 enzyme was 2.2, indicating that the increase of CgSOD-1 concentration in the reaction increased the degree of synergism (R2 = 0.9727) and the sugar yields.
3. Discussion
In the last decade, reports on termite digestome have revealed the occurrence of CAZy and PAD enzymes18,19,27 but biochemical assays with classical cellulases from termites (GH9),20,26 as well as from their protist symbionts (GH5, GH7 and GH45),27,50 have demonstrated anomalously poor saccharification efficiencies. These findings led to the hypothesis described herein of whether auxiliary mechanisms, such as oxidative enzymes, are also involved in the termite digestome,18,19,31 through the production of ROS and subsequent oxidative cleavage of polysaccharide chains.31,32,37,59–61 In this context, PAD genes and enzymes are found at high concentrations in the worker caste of C. gestroi, commensurate with their roles in feeding the colony, rather than colony defense.19 Among these PAD enzymes, a highly expressed enzyme that annotates as a superoxide dismutase is of note, not least because SODs have previously been reported to be involved in lignin oxidation and production of Fenton reaction intermediates.36,37Our results show that the global expression of endogenous Cu/Zn SODs genes is upregulated in response to LB feeding. The plant biomass with the highest degree of recalcitrance tested in our experiments is consistent with previous reports on PAD gene upregulation in the lower termites Reticulitermes flavipes.27 and Coptotermes formosanus.62 We further show that CgSOD-1 and H2O2 production localise to the midgut and foregut of C. gestroi, a microoxic environment with an average pH of 6.0. This phenomenon is similar to that reported in the wood-feeding lower termites R. flavipes,63 and Coptotermes formosanus64 as well as for the litter-feeding Cornitermes cumulans, which also exhibit high levels of ROS in the fore- and mid-guts, allied with a high abundance of mRNA for CcSD (Cu/Zn SOD) and glutathione peroxidase (CcGPX).65–67
CgSOD-1 mediates an oxidative mechanism for the degradation of glucose polymers akin to that of the product profile exhibited by fungal and bacterial LPMOs5,6,68,69 and likewise acts in synergy with classical glycoside hydrolases. Notably, activity on β-glucan and CMC reported here is specific for CgSOD-1, no effects were observed for BtSOD-1, one of the most studied vertebrate Cu/Zn SODs.70,71 This model SOD has similar biochemical properties to CgSOD-1, such as optimal pH around 5.5–6 and optimal temperature 25–30 °C.70
Furthermore, although BtSOD-1 and CgSOD-1 share the same amino acids in the coordination sphere of Cu and Zn, there are several different residues around the active site. The electrostatic potential in both enzymes’ catalytic pockets was very similar around the copper metal (both positively charged). However, CgSOD-1 exhibits more negatively charged residues around the zinc metal, i.e., the substitution K68D. The shift from a positively charged protonated amino acid to a negatively charged deprotonated residue may explain the catalytic differences between these enzymes.
4. Conclusions
For the last two decades, scientists have reported that detritivore insect guts, such as caterpillars, beetles, and termites, produce considerable amounts of reactive oxygen species (ROS) involved in the degradation of plant-derived phenolic compounds and tannins.13,65,66 Recent studies have hypothesized that this ROS generation could enhance plant cell wall degradation by boosting the activity of endogenous cellulases.19,27,31 It is in this regard that the role Cu/Zn SODs discovered in our report now solves a long-running puzzle of how detritivore insect guts produce ROS in their guts and why endogenous termite hydrolases do not exhibit high catalytic performance on insoluble and recalcitrant polysaccharides. As such, CgSOD-1 can now be seen as an essential component of C. gestroi digestion physiology, working in synergy with other GH enzymes for plant biomass saccharification through the generation of redox species.Author contributions
JPLFC performed and/or conceived: RNA-Seq and proteomics experiments, cloning and protein expressions/purifications; enzymatic activities and saccharification experiments, conceived the immunolocalizations experiment and analyzed all data. FM performed protein expressions and purifications as well as enzymatic activities. RT performed saccharification assays and immunolocalization microscopies. DC conceived and performed the HPAEC experiments. AP performed Maldi-TOF MS experiments. LC performed EPR experiments and simulations. MRF collected SAXS data. MVL performed structural molecular modelling. LBB and TAG performed the HPAEC and HPLC experiments and performed saccharification assays. GNR and JGCP performed fungal cultivation, and fungal saccharification experiments. TMA performed protein expressions and purifications for SAXS analyses. LSM and MCF performed bioinformatic analysis. AFPL conceived the proteomics experiments. AMCL conceived termite feeding assays and provide the termites. MON conceived the SAXS experiments and analyzed the data. AD conceived cloning-expression and protein purifications as well as enzymatic activities. CF conceived the HPAEC analysis and analyzed the data. PHW conceived the EPR experiment and analyzed the data. FMS conceived and coordinated all the experiments. JPLFC, FM and DC drafted the manuscript with contributions from all authors. JPLFC, CF, GJD, PHW and FMS wrote and revised the paper.Conflicts of interest
The authors declare competing financial interests: Several of the authors have submitted patent (BR 10 2015 017256 7 A2) on the use of Cu/Zn SODs for biomass degradation and valorization.Acknowledgements
We would like to acknowledge Dr Celia Dietrich for the termite's supervision and care as well as professor Dr Ednildo Machado (UFRJ) for comments on the manuscript. This work was financially supported by fellowship and grants from FAPESP 2011/20977-2, 2015/06971-3, 2016/09950-0, 2017/11952-3 (JPLFC), 2014/04105-1 (MVL) and 2015/50590-4 (FMS), 2021/04254-3 and 2016/07926-4 (RT) 2013/03061-0 and 2015/07008-2 (LBB), and 2017/16089-1 (TAG). DC was financially supported by the Danish Council for Independent Research, project: “The lignin-cellulose electron shuttling mechanism”. We gratefully acknowledge the provision of time at the CNPEM facilities NGS/LNBR, LBI/LNBio, MAS/LNBio and SAXS2/LNLS, located in Campinas, Brazil. PHW, GJD and LC thank the UK Biotechnology and Biological Sciences Research Council (BB/L021633/1) for funding. GJD is supported by the Royal Society “Ken Murray” Research Professorship. AD and FMS thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for funding (304816/2017-5, 428527/2018-3 and 306279/2020-7). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.Notes and references
- L. Petridis and J. C. Smith, Nat. Rev. Chem., 2018, 2, 382–389 CrossRef CAS.
- M. S. Buckeridge and A. P. de Souza, BioEnergy Res., 2014, 7, 1065–1073 CrossRef CAS.
- H. Østby, L. D. Hansen, S. J. Horn, V. G. H. Eijsink and A. Várnai, J. Ind. Microbiol. Biotechnol., 2020, 47, 623–657 CrossRef PubMed.
- A. Zerva, C. Pentari, C. Ferousi, E. Nikolaivits, A. Karnaouri and E. Topakas, Bioresour. Technol., 2021, 342, 126058 CrossRef CAS PubMed.
- R. J. Quinlan, M. D. Sweeney, L. Lo Leggio, H. Otten, J.-C. N. Poulsen, K. S. Johansen, K. B. R. M. Krogh, C. I. Jorgensen, M. Tovborg, A. Anthonsen, T. Tryfona, C. P. Walter, P. Dupree, F. Xu, G. J. Davies and P. H. Walton, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15079–15084 CrossRef CAS PubMed.
- G. Vaaje-Kolstad, B. Westereng, S. J. Horn, Z. Liu, H. Zhai, M. Sorlie and V. G. H. Eijsink, Science, 2010, 330, 219–222 CrossRef CAS PubMed.
- T. M. Vandhana, J.-L. Reyre, D. Sushmaa, J.-G. Berrin, B. Bissaro and J. Madhuprakash, New Phytol., 2022, 233, 2380–2396 CrossRef CAS PubMed.
- G. M. M. Rashid, C. R. Taylor, Y. Liu, X. Zhang, D. Rea, V. Fülöp and T. D. H. Bugg, ACS Chem. Biol., 2015, 3, 150803112646000–150803112646000 Search PubMed.
- M. Couturier, S. Ladevèze, G. Sulzenbacher, L. Ciano, M. Fanuel, C. Moreau, A. Villares, B. Cathala, F. Chaspoul, K. E. Frandsen, A. Labourel, I. Herpoël-Gimbert, S. Grisel, M. Haon, N. Lenfant, H. Rogniaux, D. Ropartz, G. J. Davies, M.-N. Rosso, P. H. Walton, B. Henrissat and J.-G. Berrin, Nat. Chem. Biol., 2018, 14, 306–310 CrossRef CAS PubMed.
- C. Filiatrault-Chastel, D. Navarro, M. Haon, S. Grisel, I. Herpoël-Gimbert, D. Chevret, M. Fanuel, B. Henrissat, S. Heiss-Blanquet, A. Margeot and J.-G. Berrin, Biotechnol. Biofuels, 2019, 12, 55 CrossRef PubMed.
- G. Tokuda, in Advances in Insect Physiology, ed. R. Jurenka, Academic Press, 2019, vol. 57, pp. 97–136 Search PubMed.
- F. Sabbadin, G. R. Hemsworth, L. Ciano, B. Henrissat, P. Dupree, T. Tryfona, R. D. S. Marques, S. T. Sweeney, K. Besser, L. Elias, G. Pesante, Y. Li, A. A. Dowle, R. Bates, L. D. Gomez, R. Simister, G. J. Davies, P. H. Walton, N. C. Bruce and S. J. McQueen-Mason, Nat. Commun., 2018, 9, 756 CrossRef PubMed.
- J. A. Ceja-Navarro, U. Karaoz, M. Bill, Z. Hao, R. A. White, A. Arellano, L. Ramanculova, T. R. Filley, T. D. Berry, M. E. Conrad, M. Blackwell, C. D. Nicora, Y.-M. Kim, P. N. Reardon, M. S. Lipton, J. N. Adkins, J. Pett-Ridge and E. L. Brodie, Nat. Microbiol., 2019, 4, 864–875 CrossRef CAS PubMed.
- K. Katsumata, Z. Jin, K. Hori and K. Iiyama, J. Wood Sci., 2007, 53, 419–426 CrossRef CAS.
- A. Brune, Nat. Rev. Microbiol., 2014, 12, 168–180 CrossRef CAS PubMed.
- M. Calusinska, M. Marynowska, M. Bertucci, B. Untereiner, D. Klimek, X. Goux, D. Sillam-Dussès, P. Gawron, R. Halder, P. Wilmes, P. Ferrer, P. Gerin, Y. Roisin and P. Delfosse, Commun. Biol., 2020, 3, 1–12 CrossRef PubMed.
- Y. Nishimura, M. Otagiri, M. Yuki, M. Shimizu, J. Inoue, S. Moriya and M. Ohkuma, ISME J., 2020, 1–12 Search PubMed.
- A. Geng, Y. Cheng, Y. Wang, D. Zhu, Y. Le, J. Wu, R. Xie, J. S. Yuan and J. Sun, Biotechnol. Biofuels, 2018, 11, 24 CrossRef PubMed.
- J. P. L. Franco Cairo, M. F. Carazzolle, F. C. Leonardo, L. S. Mofatto, L. B. Brenelli, T. A. Gonçalves, C. A. Uchima, R. R. Domingues, T. M. Alvarez, R. Tramontina, R. O. Vidal, F. F. Costa, A. M. Costa-Leonardo, P. F. A. Leme, G. A. G. Pereira and F. M. Squina, Front. Microb., 2016, 7, 1518 Search PubMed.
- J. P. L. Franco Cairo, L. C. Oliveira, C. A. Uchima, T. M. Alvarez, A. P. S. Citadini, J. Cota, F. C. Leonardo, A. M. Costa-Leonardo, M. F. Carazzolle, F. F. Costa, G. A. G. Pereira and F. M. Squina, Insect Biochem. Mol. Biol., 2013, 43, 970–981 CrossRef CAS PubMed.
- J. P. L. Franco Cairo, F. C. Leonardo, T. M. Alvarez, D. A. Ribeiro, F. Büchli, A. M. Costa-Leonardo, M. F. Carazzolle, F. F. Costa, A. F. Paes Leme, G. A. Pereira and F. M. Squina, Biotechnol. Biofuels, 2011, 4, 50–50 CrossRef PubMed.
- A. M. Costa-Leonardo, R. C. Barsotti and C. R. R. Camargo-Dietrich, Sociobiology, 1999, 33, 339–356 Search PubMed.
- R. Tramontina, J. L. Galman, F. Parmeggiani, S. R. Derrington, T. D. H. Bugg, N. J. Turner, F. M. Squina and N. Dixon, Green Chem., 2020, 22, 144–152 RSC.
- R. Tramontina, L. B. Brenelli, A. Sousa, R. Alves, A. M. Zetty-Arenas, V. M. Nascimento, S. C. Rabelo, S. Freitas, R. Ruller and F. M. Squina, Enzyme Microb. Technol., 2020, 135, 109490 CrossRef CAS PubMed.
- J. Ni and G. Tokuda, Biotechnol. Adv., 2013, 31, 838–850 CrossRef CAS PubMed.
- M. E. Scharf, Z. J. Karl, A. Sethi and D. G. Boucias, PLoS One, 2011, 6, e21709 CrossRef CAS PubMed.
- A. Sethi, J. M. Slack, E. S. Kovaleva, G. W. Buchman and M. E. Scharf, Insect Biochem. Mol. Biol., 2013, 43, 91–101 CrossRef CAS PubMed.
- J. P. L. Franco Cairo, D. Cannella, L. C. Oliveira, T. A. Gonçalves, M. V. Rubio, C. R. F. Terrasan, R. Tramontina, L. S. Mofatto, M. F. Carazzolle, W. Garcia, C. Felby, A. R. L. Damasio, P. H. Walton and F. M. Squina, J. Inorg. Biochem., 2020, 111316 Search PubMed.
- B. Bissaro, A. Várnai, Å.K Røhr and V. G. H. Eijsink, Microbiol. Mol. Biol. Rev., 2018, 82, e00029–e00018 CrossRef CAS PubMed.
- Z. Yang, Y. Yu, L. Yao, G. Li, L. Wang, Y. Hu, H. Wei, L. Wang, R. Hammami, R. Razavi, Y. Zhong and X. Liang, BMC Genomics, 2011, 12, S2 CrossRef CAS PubMed.
- R. Tramontina, J. P. L. Franco Cairo, M. V. Liberato, F. Mandelli, A. Sousa, S. Santos, S. C. Rabelo, B. Campos, J. Ienczak, R. Ruller, A. R. L. Damásio and F. M. Squina, Biotechnol. Biofuels, 2017, 10, 4 CrossRef PubMed.
- G. P. Borin, C. C. Sanchez, A. P. de Souza, E. S. de Santana, A. T. de Souza, A. F. P. Leme, F. M. Squina, M. Buckeridge, G. H. Goldman and J. V. C. Oliveira, PLoS One, 2015, 10, e0129275–e0129275 CrossRef PubMed.
- WO2015069308 A1, 2015.
- I. A. Abreu and D. E. Cabelli, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804, 263–274 CrossRef CAS PubMed.
- J. C. Koningsberger, B. S. van Asbeck, E. van Faassen, L. J. Wiegman, J. van Hattum, G. P. van Berge Henegouwen and J. J. Marx, Clin. Chim. Acta, 1994, 230, 51–61 CrossRef CAS.
- M. B. Yim, P. B. Chock and E. R. Stadtman, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 5006–5010 CrossRef CAS PubMed.
- G. M. M. Rashid, X. Zhang, R. C. Wilkinson, V. Fülöp, B. Cottyn, S. Baumberger and T. D. H. Bugg, ACS Chem. Biol., 2018, 13, 2920–2929 CrossRef CAS PubMed.
- S. Boulos and L. Nyström, Analyst, 2016, 141, 6533–6548 RSC.
- V. Arantes, A. M. Milagres, T. R. Filley and B. Goodell, J. Ind. Microbiol. Biotechnol., 2011, 38, 541–555 CrossRef CAS PubMed.
- S. C. Rabelo, C. E. Vaz Rossell, G. J. de Moraes Rocha and G. Zacchi, Biotechnol. Prog., 2012, 28, 1207–1217 CrossRef CAS PubMed.
- S. M. de Vasconcelos, A. M. P. Santos, G. J. M. Rocha and A. M. Souto-Maior, Bioresour. Technol., 2013, 135, 46–52 CrossRef CAS PubMed.
- G. Siqueira, A. Várnai, A. Ferraz and A. M. F. Milagres, Appl. Energy, 2013, 102, 399–402 CrossRef CAS.
- F. Mandelli, J. P. L. Franco Cairo, A. P. S. Citadini, F. Büchli, T. M. Alvarez, R. J. Oliveira, V. B. P. Leite, A. F. Paes Leme, A. Z. Mercadante and F. M. Squina, Lett. Appl. Microbiol., 2013, 57, 40–46 CrossRef CAS PubMed.
- L. P. Zheng, L. W. Gao, J. Q. Zhou, Y. H. Sima and J. W. Wang, Afr. J. Biotechnol Search PubMed.
- I. Morgenstern-Badarau, in Bioinorganic Chemistry: An Inorganic Perspective of Life, ed. D. P. Kessissoglou, Springer Netherlands, Dordrecht, 1995, pp. 105–111 Search PubMed.
- C. G. Palivan, H. Palivan and B. A. Goodman, Proc. – R. Soc. Edinburgh, Sect. B: Biol., 1994, 102, 273–277 Search PubMed.
- J. Peisach and W. E. Blumberg, Arch. Biochem. Biophys., 1974, 165, 691–708 CrossRef CAS PubMed.
- A. Fujita, M. Hojo, T. Aoyagi, Y. Hayashi, G. Arakawa, G. Tokuda and H. Watanabe, J. Wood Sci., 2010, 56, 222–226 CrossRef CAS.
- J. Woodward, Bioresour. Technol., 1991, 36, 67–75 CrossRef CAS.
- A. Sethi, E. S. Kovaleva, J. M. Slack, S. Brown, G. W. Buchman and M. E. Scharf, Arch. Insect Biochem. Physiol., 2013, 84, 175–193 CrossRef CAS PubMed.
- B. Westereng, J. W. Agger, S. J. Horn, G. Vaaje-Kolstad, F. L. Aachmann, Y. H. Stenstrøm and V. G. H. Eijsink, J. Chromatogr. A, 2013, 1271, 144–152 CrossRef CAS PubMed.
- B. Westereng, M. Ø. Arntzen, F. L. Aachmann, A. Várnai, V. G. H. Eijsink and J. W. Agger, J. Chromatogr. A, 2016, 1445, 46–54 CrossRef CAS PubMed.
- D. Cannella, K. B. Möllers, N.-U. Frigaard, P. E. Jensen, M. J. Bjerrum, K. S. Johansen and C. Felby, Nat. Commun., 2016, 7, 11134 CrossRef CAS PubMed.
- Y. Xiong, Y. Wang, M. Li, L. Kang, J. Zhou, C. Liu, Z. Liu, Z. Zhang and S. Yuan, Food Chem., 2019, 287, 160–166 CrossRef CAS PubMed.
- S. V. Rykov, P. Kornberger, J. Herlet, N. V. Tsurin, I. N. Zorov, V. V. Zverlov, W. Liebl, W. H. Schwarz, S. V. Yarotsky and O. V. Berezina, Appl. Microbiol. Biotechnol., 2019, 103, 7553–7566 CrossRef CAS PubMed.
- J. Simões, A. S. P. Moreira, E. da Costa, D. Evtyugin, P. Domingues, F. M. Nunes, M. A. Coimbra and M. R. M. Domingues, Carbohydr. Polym., 2016, 148, 290–299 CrossRef PubMed.
- K.-i. Setsukinai, Y. Urano, K. Kakinuma, H. J. Majima and T. Nagano, J. Biol. Chem., 2003, 278, 3170–3175 CrossRef CAS PubMed.
- N. Miyake, M. Kim and T. Kurata, Biosci., Biotechnol., Biochem., 1999, 1, 54–57 CrossRef PubMed.
- K. Besser, G. P. Malyon, W. S. Eborall, G. P. da Cunha, J. G. Filgueiras, A. Dowle, L. C. Garcia, S. J. Page, R. Dupree, M. Kern, L. D. Gomez, Y. Li, L. Elias, F. Sabbadin, S. E. Mohamad, G. Pesante, C. Steele-King, E. R. de Azevedo, I. Polikarpov, P. Dupree, S. M. Cragg, N. C. Bruce and S. J. McQueen-Mason, Nat. Commun., 2018, 9, 5125 CrossRef PubMed.
- S. J. B. Mallinson, M. M. Machovina, R. L. Silveira, M. Garcia-Borràs, N. Gallup, C. W. Johnson, M. D. Allen, M. S. Skaf, M. F. Crowley, E. L. Neidle, K. N. Houk, G. T. Beckham, J. L. DuBois and J. E. McGeehan, Nat. Commun., 2018, 9, 2487 CrossRef PubMed.
- E. Masai, A. Ichimura, Y. Sato, K. Miyauchi, Y. Katayama and M. Fukuda, J. Bacteriol., 2003, 185, 1768–1775 CrossRef CAS PubMed.
- A. Geng, Y. Cheng, Y. Wang, D. Zhu, Y. Le, J. Wu, R. Xie, J. S. Yuan and J. Sun, Biotechnol. Biofuels, 2018, 11, 24 CrossRef PubMed.
- A. Ebert and A. Brune, Appl. Environ. Microbiol., 1997, 63, 4039–4046 CrossRef CAS PubMed.
- J. Ke and S. Chen, in Biological Concerstion of Biomass for Fuels and Chemicals: Exploration from Natural Utilization Systems, ed. J. Sun, S.-Y. Ding and J. D. Peterson, The Royal Society of Chemistry, Cambridge, 2013, vol. 1, pp. 177–191 Search PubMed.
- G. Sousa, A. C. P. Gandara, P. L. Oliveira, F. M. Gomes, A. C. Bahia and E. A. Machado, Sci. Rep., 2019, 9, 670 CrossRef PubMed.
- R. Barbehenn, T. Dodick, U. Poopat and B. Spencer, Arch. Insect Biochem. Physiol., 2005, 60, 32–43 CrossRef CAS PubMed.
- R. V. Barbehenn, R. E. Maben and J. J. Knoester, Environ. Entomol., 2008, 37, 1113–1118 CrossRef PubMed.
- T. J. Simmons, K. E. H. Frandsen, L. Ciano, T. Tryfona, N. Lenfant, J. C. Poulsen, L. F. L. Wilson, T. Tandrup, M. Tovborg, K. Schnorr, K. S. Johansen, B. Henrissat, P. H. Walton, L. L. Leggio and P. Dupree, Nat. Commun., 2017, 8, 1064 CrossRef CAS PubMed.
- J. W. Agger, T. Isaksen, A. Varnai, S. Vidal-Melgosa, W. G. T. Willats, R. Ludwig, S. J. Horn, V. G. H. Eijsink and B. Westereng, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6287–6292 CrossRef CAS PubMed.
- S. Marklund and G. Marklund, Eur. J. Biochem., 1974, 47, 469–474 CrossRef CAS PubMed.
- W. R. Rypniewski, S. Mangani, B. Bruni, P. L. Orioli, M. Casati and K. S. Wilson, J. Mol. Biol., 1995, 251, 282–296 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental, supporting results, supporting Fig. S1 to S6, supporting Tables S1 to S5, supporting references. See DOI: https://doi.org/10.1039/d1gc04519a |
| ‡ PhotoBioCatalysis unit, Crop Production and Biostimulation, Biomass Transformation lab, Université Libre de Bruxelles, Campus Plaine CP245, 1050-Brussels, Belgium. |
| § School of Chemistry, University of Nottingham, University Park, Nottingham, NG7 2RD, United Kingdom. |
| This journal is © The Royal Society of Chemistry 2022 |