Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular nanoinformatics approach assessing the biocompatibility of biogenic silver nanoparticles with channelized intrinsic steatosis and apoptosis†
Pritam Kumar
Panda‡
 a,
Puja
Kumari‡
bc,
Paritosh
Patel
d,
Shailesh Kumar
Samal
e,
Suman
Mishra
d,
Murtaza M.
Tambuwala
f,
Ateet
Dutt
g,
Klára
Hilscherová
b,
Yogendra Kumar
Mishra
a,
Puja
Kumari‡
bc,
Paritosh
Patel
d,
Shailesh Kumar
Samal
e,
Suman
Mishra
d,
Murtaza M.
Tambuwala
f,
Ateet
Dutt
g,
Klára
Hilscherová
b,
Yogendra Kumar
Mishra
 h,
Rajender S.
Varma
h,
Rajender S.
Varma
 i,
Mrutyunjay
Suar
i,
Mrutyunjay
Suar
 *d,
Rajeev
Ahuja
*d,
Rajeev
Ahuja
 *aj and
Suresh K.
Verma
*aj and
Suresh K.
Verma
 *ad
*ad
aCondensed Matter Theory Group, Materials Theory Division, Department of Physics and Astronomy, Uppsala University, Box 516, Uppsala, SE-751 20, Sweden. E-mail: sureshverma22@gmail.com; rajeev.ahuja@physics.uu.se
bRECETOX, Faculty of Science, Masaryk University, Kotlarska 2, Brno 61137, Czech Republic
cAdvance Science and Technology Research Centre, Vinoba Bhave University, Hazaribagh 825001, India
dSchool of Biotechnology, KIIT University, Bhubaneswar, 751024, India. E-mail: msuar@kiitbiotech.ac.in
eSection of Immunology and Chronic Disease, Institute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden
fSchool of Pharmacy and Pharmaceutical Science, Ulster University, Coleraine BT52 1SA, Northern Ireland, UK
gInstituto de Investigaciones en Materiales, UNAM, 04510, CDMX, Mexico
hMads Clausen Institute, NanoSYD, University of Southern Denmark, Alsion 2, Sønderborg, DK-6400, Denmark
iRegional Centre of Advanced Technologies and Materials, Czech Advanced Technology and Research Institute, Palacký University in Olomouc, Šlechtitelů 27, 783 71 Olomouc, Czech Republic
jApplied Materials Physics, Department of Materials Science and Engineering, Royal Institute of Technology (KTH), SE-100 44, Stockholm, Sweden
First published on 22nd December 2021
Abstract
The developmental rapidity of nanotechnology poses higher risks of exposure to humans and the environment through manufactured nanomaterials. The multitude of biological interfaces, such as DNA, proteins, membranes, and cell organelles, which come in contact with nanoparticles, is influenced by colloidal and dynamic forces. Consequently, the ensued nano-bio interface depends on dynamic forces, encompasses many cellular absorption mechanisms along with various biocatalytic activities, and biocompatibility that needs to be investigated in detail. Addressing the issue, the study offers a novel green synthesis strategy for antibacterial AgNPs with higher biocompatibility and elucidates the mechanistic in vivo biocompatibility of silver nanoparticles (AgNPs) at the cellular and molecular levels. The analysis ascertained the biosynthesis of G-AgNPs with the size of 25 ± 10 nm and zeta potential of −29.2 ± 3.0 mV exhibiting LC50 of 47.2 μg mL−1 in embryonic zebrafish. It revealed the mechanism as a consequence of abnormal physiological metabolism in oxidative stress and neutral lipid metabolism due to dose-dependent interaction with proteins such as he1a, sod1, PEX protein family, and tp53 involving amino acids such as arginine, glutamine and leucine leading to improper apoptosis. The research gave a detailed insight into the role of diverse AgNPs-protein interactions with a unique combinatorial approach from first-principles density functional theory and in silico analyses, thus paving a new pathway to comprehending their intrinsic properties and usage.
Introduction
The comprehension of nanotechnology has led its utility to a variety of applications. It includes detailed investigations and explorations about different types of nanoparticles, comprising inorganic and organic nanoparticles. Inorganic nanoparticles have been a cynosure among scientists because of their peculiar chemical and physical properties.1 Some most common metallic nanoparticles include silver (Ag), gold (Au), CuO, ZnO, TiO2 nanoparticles that have been investigated for their diverse applications. Silver nanoparticles (AgNPs) are among the most studied and applied for different biological and chemical applications, including their usage as antibacterial agents,2 catalytic agents,3 or in food packaging,4 among others.With extensive enhancement in the usage of AgNPs, concerns have been raised regarding the need for regulation of their eco-compatibility and biosafety. Increased demand and application of AgNPs have also led to their enhanced disposal into environmental matrices such as soil, water, and air;5 discarded nanomaterials as a contaminant further affect human and aquatic health.6 Hence, the detailed exploration of the toxicological impacts of natural and artificial AgNPs has become the need of the hour, which has also prompted the search for potent solutions for their green and sustainable synthesis. Numerous studies have been conducted to explain the biocompatibility of AgNPs using various in vitro and in vivo biological models;7 their toxic properties have been expounded via different in vitro8 and in vivo models.9 Assorted in vitro models have been applied for the determination of cytotoxicity on various cell lines such as epithelial, colon, and oral squamous cell lines among many others, which have been chosen based on their origin.10 However, issues have been raised regarding the fate of nanoparticles under the exact physiological conditions in humans and other living organisms. Considering the in vivo models, zebrafish has been proven as one of the promising options for toxicological assessment;11their embryonic stages are the choices for different experimental studies because of their high genetic similarities with humans and the ease of rearing at the lab scale. Thanks to their high fecundity, transparency of eggs and embryos, and small size and fast development, zebrafish embryos provide many benefits for use as a model in the laboratory for toxicity evaluation.12 Hence, the toxicological assessment of nanoparticles in zebrafish models can provide useful information for toxicity aspects for the aquatic ecosystem as well as human health.
The exploration of toxicity in zebrafish embryos and adult zebrafish has revealed that AgNPs exhibits cytotoxicity at molecular and atomic levels by inducing oxidative stress and apoptosis as a consequence of their dose-dependent internalization that influences protein machinery through their molecular interaction and accumulation at the surface of cells.13 Other metal and metal oxide nanoparticles have shown similar toxicity mechanisms.5,14 However, knowledge about their initial interaction with membrane lipids leading to their internalization is limited. It was shown that TiO2 NPs affect the metabolism of lipids in zebrafish thus playing a crucial role in determining toxicity.15 Considering these facts, it can be hypothesized that AgNPs would interfere with lipid metabolism and induce steatosis with further consequences on other physiological processes like oxidative stress and apoptosis. Addressing the issue, this study explores the detailed mechanism of biocompatibility of AgNPs concerning lipid metabolism through the combination of experimental and computational approaches.
The high demand and utility of AgNPs have urged the production of biocompatible and eco-compatible nanoparticles and attempts has been made to classify them in terms of greenness.16–18 Based on the techniques and types of substrate deployed, their synthesis can be categorized into chemical,19 physical,20 and biological routes.21 The chemical route uses harmful chemicals for synthesis. In contrast, the physical route of synthesis, such as high energy ball milling methods22,23 entails a cumbersome process, especially at the purification stage.15 The biological route has been recognized as the eco-compatible, greener and bio-compatible option24 as biological agents serve the dual role for reducing and stabilizing nanoparticles. Numerous plants and biological entities have been deployed for the synthesis of AgNPs.25–28 However, the quest for useful and abundant plants is incessant. Herein, a successful synthesis of AgNPs has been accomplished by deploying abundant and innocuous plant species commonly referred to as silver grass (Miscanthus sinensis) that is native to Eastern Asia and the American continent and widely cultivated as an ornamental plant and also deployed for bioenergy production.29 Its widespread availability and renewability make it an ideal choice for the deployment of nanoparticle synthesis and its comparison with the traditional chemical methods.
It can be hypothesized that nanoparticles synthesized from biomolecules of this plant in a newer approach will have higher biocompatibility and eco-compatibility;30 greener-synthesized AgNPs using the leaf extract of silver grass were termed as “G-AgNPs”. They were characterized for their physicochemical properties and were evaluated for their biocompatibility with zebrafish embryos in comparison to the commercially available AgNPs (labeled C-AgNP). The molecular mechanism of toxicity was determined using different experimental and computational approaches with the aim of characterizing their mechanistic biocompatibility at cellular and molecular levels. This is particularly timely as there is a tremendous ongoing activity in the field of biological preparation of nanoparticles deploying plant extracts and other biological means, especially in the biomedical arena.
Materials and methods
Green synthesis of silver nanoparticles
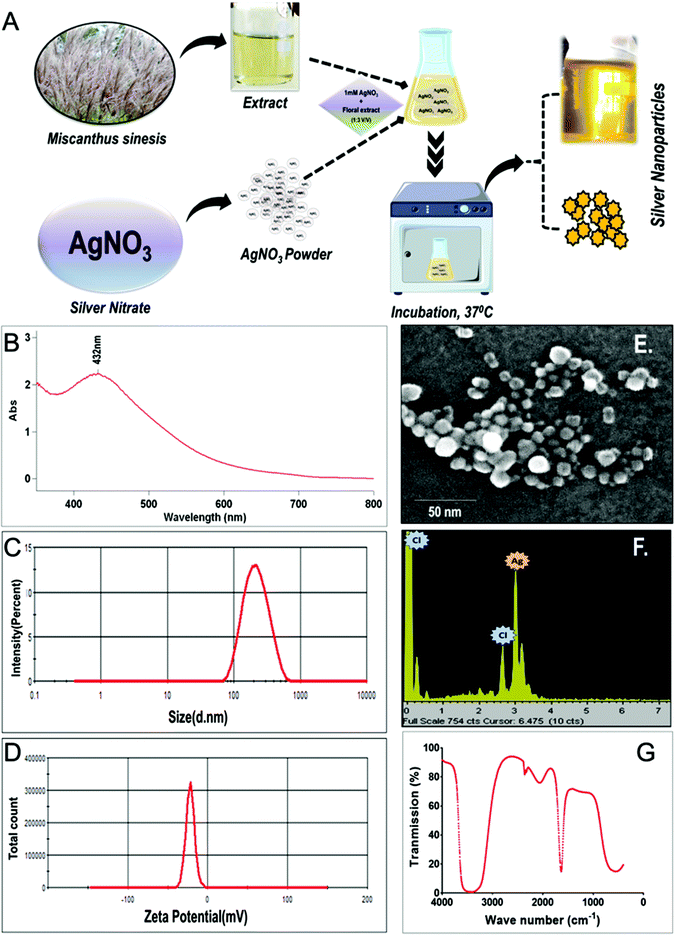
As shown in Fig. 1A, silver nanoparticles (G-AgNPs) were synthesized using silver nitrate (AgNO3) as a precursor and aqueous extract of silver grass as reducing and stabilizing agent. Silver grass leaves were collected from the premises of school of biotechnology, KIIT University, Bhubaneswar, India.31 The aqueous leaf extract was prepared by boiling 10 g of chopped leaves of silver grass in 500 mL of water for 20 minutes followed by filtration deploying a muslin cloth. The prepared extract was cooled and analyzed using GC-MS to access the role of biomolecules in AgNPs synthesis. The synthesis reaction was set by incubating the leaf extract with 1 mM AgNO3 solution in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 V/V at 37 °C under shaking conditions; the color change to yellow was recognized as the indication of AgNPs. The solution containing the synthesized G-AgNP was centrifuged at 10
4 V/V at 37 °C under shaking conditions; the color change to yellow was recognized as the indication of AgNPs. The solution containing the synthesized G-AgNP was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min to separate AgNPs from the solution, lyophilized and suspended in distilled water for further physical and biological characterizations and assays.
000 rpm for 5 min to separate AgNPs from the solution, lyophilized and suspended in distilled water for further physical and biological characterizations and assays.
GC-MS analysis
To identify bioactive compounds in the leaf extract of silver grass, GC-MS analysis was performed using an Agilent chromatograph, before and after the synthesis.32 The sample was prepared by dissolving the dry lyophilised extract in methanol. Agilent 5963 N mass selective detector was coupled with the chromatograph for the detection of molecules. The chromatograph containing HP-5MS capillary column (30 m Å ∼ 0.25 mm Å ∼ 1.0 μl) was conditioned at 40–70 °C for 10 min for the separation of sample constituents. A temperature of 280 °C was maintained in the transfer line. The optimisation of other parameters was performed as a mass scanning range: m/z 29–540; source temperature: 230 °C; the scanning rate of 2.88 scans per S and electronic impact (EI) mode at 70 eV. The career gas was helium at a flow rate of 1.0 mL per min. Obtained mass spectra were compared with the data system library (NIST 98) and the identification of the tentative volatile components in the extract was supported by the retention index data.Characterization of silver nanoparticles (G-AgNP)
Synthesized silver nanoparticles (G-AgNPs) were characterized for their physicochemical properties using standard methods. The size of nanoparticles was determined using FESEM (Zeiss, Model EVO 60) equipped with an Oxford Inca Energy dispersive X-Ray spectrophotometer (EDS). Hydrodynamic size and zeta potential of the synthesized G-AgNP in an aqueous medium were determined using dynamic light scattering (Zetasizer, Malvern, UK). Optical properties were characterized using UV-Vis spectroscopy in the range of 300–800 nm using a spectrophotometer (Cary 5000). Fourier transform infrared (FTIR) measurements on the nanoparticles were performed in the complete aqueous medium and FTIR spectrum was recorded on a PerkinElmer FT-IR spectrometer RX-I instrument with the ATR attachment. Measurements were carried out in the range of 400–4000 cm−1 at a resolution of 4 cm−1. The zeta potential and hydrodynamic diameter of commercially purchased C-AgNP were measured to have a comparison of their physicochemical properties with G-AgNP.Antibacterial activity of nanoparticles
To assess the antibacterial activities of G-AgNPs, well diffusion assay and growth curve analysis were performed using E. coli. Well-diffusion assay was performed using the modified disc diffusion method.33,34 The bacterial strain was cultured in Luria Bertani broth at 35 ± 2 °C and incubated on a shaker. Overnight culture of CFU 106 ml−1 was plated on an agar plate followed by incubation. Around 8 mm well was punched with 200 μL sterile micropipette tips. 80 μL of the synthesized nanoparticles were poured into each well. After overnight incubation zone of inhibition was observed and recorded. For the growth curve analysis, the OD of the cultures incubated with different concentrations of G-AgNPs was recorded in 2 h intervals for 12 hours.Zebrafish maintenance and embryo production
The culture and maintenance of adult zebrafish were performed in an overflow system (Aquaneering, USA) containing fish water (75 g NaHCO3, 18 g sea salt, 8.4 g CaSO4 per 1000 ml).35 Fish were kept in 12/12 h light and dark photoperiod conditions and fed three times a day with fish food containing dried bloodworm. To obtain embryos, the breeding of the fish was performed in a breeding container with a net partition. The breeding was performed with male and female zebrafish at the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The obtained embryos were then separated to select viable ones and washed with Holtfreter (HF) buffer.36,37 All toxicological experiments were performed using embryos reared in a filter sterilized HF medium. All chemicals utilized for the buffer preparation were purchased from Merck. All animal procedures were approved by the Institutional Animal Ethics Committee (IAEC) of KIIT University. All experiments were performed in accordance with the relevant animal practice guidelines and regulations of IAEC, KIIT University.
1. The obtained embryos were then separated to select viable ones and washed with Holtfreter (HF) buffer.36,37 All toxicological experiments were performed using embryos reared in a filter sterilized HF medium. All chemicals utilized for the buffer preparation were purchased from Merck. All animal procedures were approved by the Institutional Animal Ethics Committee (IAEC) of KIIT University. All experiments were performed in accordance with the relevant animal practice guidelines and regulations of IAEC, KIIT University.
Toxicological assessment
The comparative biocompatibility assessments of green-synthesized G-AgNP and commercially purchased C-AgNPs with equivalent size and charge were performed with zebrafish embryos by exposing them until 72 hpf. A suspension of G-AgNPs and C-AgNPs was prepared in a filtered HF medium for the exposure and the analysis was performed according to the protocol described by Verma et al.38 In brief, twenty 3–4 hours post fertilized (hpf) embryos were exposed to different concentrations (5, 10, 25, 50, and 100 μg mL−1) of G-AgNPs and C-AgNPs suspended in 500 μL of HF medium in 24-well plates. The plates were then incubated for 72 h with a photoperiod of 12/12 h light/dark at 28 ± 1 °C. Untreated embryos were considered as a control. The defects in morphological development such as notochord malformation, swollen yolk and pericardial edema, were analyzed under a stereomicroscope. The hatching rate was calculated by determining the ratio of the number of hatched embryos at 72 hpf in treatments compared to the untreated group. OECD 236 guidelines were followed for the evaluation of LC50 and the mortality rate. Heart rate was measured as the heartbeat count per minute. All experiments were performed in triplicate and repeated three times.Uptake analysis
The uptake of G-AgNPs and C-AgNPs by embryonic zebrafish cells was assessed by determining the mean side scatter of the treated and untreated cellular suspension with the help of flow cytometry.39 The 72 h NP exposed embryos were collected and rinsed with sterilized HF buffer. The embryos were sacrificed through tricaine treatment and their cell suspensions were prepared by mild sonication in HF buffer.35 The cells were concentrated by centrifugation at 5000 rpm for 10 min to make the suspension debris free. The cellular suspension was further analyzed in a flow cytometer for the change in mean side scatter of exposed embryo cells compared with the untreated ones. All the experiments were repeated three times for statistical accuracy.Oxidative stress and neutral lipid analysis
The mechanistic elucidation of comparative cellular and molecular biocompatibility was accomplished by the determination of oxidative stress and neutral lipid metabolism in embryos exposed to G-AgNPs and C-AgNPs for 72 h. The analysis was performed using flow cytometry and fluorescence microscopy. For microscopy analysis, the 72 h exposed untreated and treated embryos were rinsed with sterilized HF buffer and stained with 1.25 mg L−1 H2DCFDA dye for 20 min in dark for ROS assessment.40 Neutral lipid analysis was assessed by staining the embryos with 2 mg L−1 of lipidTox™ (Thermo Scientific, USA) for 20 min in dark.15 After staining, the extra stain was removed by washing the embryos thrice with sterilized HF buffer. The stained untreated and treated embryos were examined using green and red filters of the EVOS inverted fluorescence microscope (Thermo Scientific, USA) for ROS and neutral lipid analysis, respectively. For flow cytometry analysis, cellular suspensions were prepared as per the protocol mentioned in uptake analysis and were stained with 1.25 mg L−1 H2DCFDA and lipidTox™ dye. The analysis was performed using Attune acoustic focusing cytometer (Applied Biosystems, Life Technologies) equipped with a 488 nm argon laser. Data analysis was performed using FCS Express 7 (Denovo, CA). The fluorescence of H2DCFDA and lipidTox™ was measured using BL1 and BL2 filters of the cytometer. All measurements were performed in triplicate by taking embryos from three different plates and statistical analysis was performed using GraphPad Prism 7.Apoptosis analysis
The apoptosis induced by nanoparticles in exposed embryos was determined using flow cytometry and fluorescence microscopy. Acridine orange (AO) was used to stain the embryos for apoptosis analysis.41 All the exposed untreated and treated zebrafish embryos were stained with 5 μg mL−1 AO (dissolved in HF) for 20 min. After staining, the embryos were washed three times to remove the extra stain using HF buffer. The EVOS inverted fluorescent microscope (Thermo Scientific, USA) was used to capture images in the green channel of the microscope. For flow cytometry analysis, the cellular suspensions were stained with 5 μg mL−1 AO (dissolved in HF) for 20 min and analyzed using the BL1 filter of the Attune acoustic focusing cytometer (Applied Biosystems, Life Technologies). All measurements were performed in triplicate by taking embryos from three different plates and statistical analysis was performed using GraphPad Prism 7.Combinatorial DFT and in silico analyses
DFT formalism
The structure of AgNP was studied using atomic simulation environment (ASE) and its geometry was optimized with DFT formalism using the first principles density functional theory within the framework of VASP package.42–44 The Hamiltonian ion-electron interactions were dealt with by the generalization of the pseudopotential in the linear augmented-plane-wave (PAW-PBE) procedure. In contrast, the exchange and correlation parts inside the generalized gradient approach (GGA) were estimated in the parameterization of Perdew–Burke–Ernzerhof (PBE).45 We elucidated the AgNPs deformation mechanism using DFT formalism.Molecular dynamics simulation (ab initio and classical dynamics)
AgNPs in the solvent environment were simulated to observe the dynamics in a solvent (water) simulated environment using Gromacs 2019v.46 The setup was prepared using CHARMM-GUI47 sever where the parameterization was obtained from CHARMM36 forcefield.48 The Ag (100) surface was chosen to construct spherical-shaped nanoparticles with a radius of 10 Å. A total of 0.005 ns were simulated with a time step of 1 fs for energy minimization. Neighbor searching was performed every 10 steps. The PME algorithm was used for electrostatic interactions with a cut-off of 1.2 nm. A reciprocal grid of 40 × 40 × 40 cells was used with 4th order B-spline interpolation. A single cut-off of 1.26 nm was used for van der Waals interactions. For the equilibration phase, a total of 0.125 ns were simulated with a time step of 1 fs. Neighbor searching was performed every 20 steps. The PME algorithm was used for electrostatic interactions with a cut-off of 1.2 nm. A reciprocal grid of 40 × 40 × 40 cells was used with 4th order B-spline interpolation. A single cut-off of 1.2 nm was used for van der Waals interactions. Temperature coupling was performed using the Nose–Hoover algorithm. For the final production run, a total of 10 ns were simulated with a time step of 2 fs. Neighbor searching was performed every 20 steps. The PME algorithm was used for electrostatic interactions with a cut-off of 1.2 nm. A reciprocal grid of 40 × 40 × 40 cells was used with 4th order B-spline interpolation. A single cut-off of 1.22 nm was used for van der Waals interactions. Temperature coupling was performed with the Nose–Hoover algorithm. Pressure coupling was performed using the Parrinello–Rahman algorithm. Moreover, to further investigate the dynamics and stability of the AgNP cluster and AgNP in combination with lipid molecules in a vacuum, DFT formalism was employed (ab initio molecular dynamics simulation with VASP package) using the NVT ensemble at T = 300 K via the Nosé–Hoover method.Molecular docking
The excavation of cellular and molecular mechanisms was extended by in silico approach through molecular interaction studies with AgNP, which was performed with the help of HEX docking program49 with AgNP as the ligand and different proteins as the receptor. Hex docking correlations leverage SPF shape–density representations to build a list of up to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 possible solutions quickly was performed using the polynomial-order N = 20 function. As the data shows, virtually all the top 3000 scoring orientations include some near-native orientations, but using a bigger list avoids the problem of rejecting good candidates in circumstances where remarkable results are expected. These solutions are scored based on shape (i.e., no matter the number of dimensions) and shape + electrostatic correlations (e.g., polynomials ordered to 25 or 30 dimensions). Requesting polynomial (N = 25) returns representations of each protein that are soft, whereas polynomial (N = 30) results in more distinct representations.
000 possible solutions quickly was performed using the polynomial-order N = 20 function. As the data shows, virtually all the top 3000 scoring orientations include some near-native orientations, but using a bigger list avoids the problem of rejecting good candidates in circumstances where remarkable results are expected. These solutions are scored based on shape (i.e., no matter the number of dimensions) and shape + electrostatic correlations (e.g., polynomials ordered to 25 or 30 dimensions). Requesting polynomial (N = 25) returns representations of each protein that are soft, whereas polynomial (N = 30) results in more distinct representations.
HeLa–AgNP interaction study was chosen to understand the hatching rate abnormalities. PEX5 and PEX14 were chosen from PEX families to understand the uptake and internalization of AgNP,50,51 using the HEX docking algorithm (with default parameters and shape correlation type). MTTP and Apoa1 were chosen to determine the effect of AgNPs on neutral lipid metabolism,15 while sod1 and tp53 were chosen to know the probable cause of abnormal oxidative stress and apoptosis in zebrafish embryos exposed to AgNP.52 Furthermore, using HEX docking, we have elucidated the interaction mechanism of AgNPs with dipalmitoylphosphatidylcholine (DPPC) to precisely illustrate the binding affinity of AgNPs. UCSF Chimera53 was used for energy minimization of receptor proteins. The parameterization for the Ag atom was derived from DFT studies. Post-docking analyses were carried out for conformational clustering and visualized using UCSF Chimera and Discovery Studio Visualizer.54
Statistical analyses
Statistical analysis was performed using GraphPad Prism v6.01 (San Diego, California). The LC50 values and respective confidence intervals (CI) were calculated by the non-linear fit of the sigmoidal dose–response curve. One-way ANOVA followed by Tukey test with significance set at P < 0.05 was performed for the data analysis. A comparison between the results of G-AgNPs and C-AgNPs was performed at each concentration. Correlation analysis was performed between uptake, ROS, neutral lipid, and apoptosis data by computing the non-parametric Spearman correlation.Results and discussion
Green synthesis of silver nanoparticles (G-AgNPs) and their characterization
Green synthesis of silver nanoparticles (G-AgNP) was performed using the extract of silver grass as shown in Fig. 1A; visualization of the yellow color after the incubation indicated the formation of G-AgNPs, which was further characterized for their physicochemical properties using different physical methods. As shown in Fig. 1B, the UV-Vis spectrum showed SPR (surface plasmon resonance) peak at 432 nm, which has been established as an important property of AgNPs. The previously documented reports have also mentioned the SPR peak of AgNPs in a range of 400–500 nm.55 Further, the hydrodynamic size and stability of G-AgNPs and C-AgNPs were determined in the different media used for further toxicological assays. As shown in Fig. 1C and Table 1, the hydrodynamic size of the nanoparticles was found to vary in different media. The hydrodynamic diameter of G-AgNPs was found to be 124.3 ± 12.2 nm and 131.6 ± 18 nm in aqueous and Holtfreter medium (zebrafish rearing medium), respectively, while C-AgNPs was possessing the hydrodynamic diameter of 102.2 ± 15.3 and 124.4 ± 14.0 nm in aqueous and Holtfreter medium, respectively. The data indicated a non-significant difference between hydrodynamic diameters of both types of nanoparticles. The difference in size and hydrodynamic diameter of G-AgNPs estimated using FESEM and Zetasizer can be attributed to the presence of water and salt molecules around nanoparticles in the specific medium.56 The same can be reasoned for the variation in the hydrodynamic diameter and zeta potential of the nanoparticles in two (aqueous and HF) different media. The results are in line with those from previous reports4 and confirmed the successful stable synthesis of AgNPs using the leaf extract of silver grass. The stability of nanoparticles was further estimated by measuring their zeta potentials in different media. Fig. 1D and Table 1 show that the zeta potential of G-AgNPs and C-AgNPs in an aqueous medium was −38.6 ± 4.0 and −29.2 ± 3.0 mV, while it was −31.8 ± 6.2 and −30.2 ± 4.2 mV in the Holtfreter medium. The measurements inferred an equivalency in size and stability of both types of nanoparticles as an eligible criterion for their comparative study of their biological effects. To have a clear estimate of the size and identity of G-AgNPs, the nanoparticles were subjected to FESEM visualization with EDX analysis. As shown in Fig. 1E and F, FESEM analysis documented the size of nanoparticles in the range of 25 ± 10 nm, while EDX analysis confirmed the presence of AgNPs in the sample. The optical and chemical properties of the synthesized G-AgNPs were further analyzed using FTIR. As shown in Fig. 1G, FTIR analysis showed stretching of –OH bond at different transmission points of 3520–3268 cm−1, 1705–1633 cm−1, which can be attributed to –OH molecules present in different amino acids of proteins present in plant extract that has been utilized for capping reduced AgNPs.| Nanoparticles | Size (nm) by FESEM | Hydrodynamic diameter (nm) | Zeta potential (mV) | ||
|---|---|---|---|---|---|
| Aq. | HF | Aq. | HF | ||
| G-AgNPs | 22.0 ± 8.0 | 124.3 ± 12.2 | 131.6 ± 18 | −38.6 ± 4.0 | −31.8 ± 6.2 |
| C-AgNPs | 20.0 ± 4.0 | 102.2 ± 15.3 | 124.4 ± 14 | −29.2 ± 3.0 | −30.2 ± 4.2 |
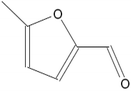
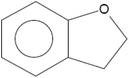
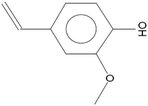
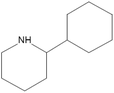
The synthesis of G-AgNPs was hypothesized to be mediated by the presence of biomolecules such as proteins, polysaccharides, and other molecules in aqueous plant extract.13 Hence, to reveal the role of different biomolecules, GC-MS analysis was performed with pre and post-synthesis aqueous plant extracts. As shown in Table 2, the aqueous plant extract of silver grass indicated the presence of different biomolecules at different retention times. Some key detected molecules were 5-methyl furfural, 2,3-dihydrobenzofuran, 2-methoxy 4-vinyl phenol and 2-cyclohexylpiperidine at retention times of 6.5, 14.1, 18.0 and 38.1 min, respectively. Fig. S1† shows the disappearance of significant peaks in “after synthesis” plant extract compared to “before synthesis” indicating the utilization of these biomolecules for the reduction and stability of G-AgNPs during the synthesis process. With reference to the data, the synthesis reaction can be represented as:
| AgNO3 + reducing agent (plant biomolecules) → Ag+ + NO3− |
| Ag+ + capping agent (plant biomolecules) → AgNP (G-AgNP) |
Antibacterial activity of G-AgNP
The synthesized G-AgNPs were speculated to possess the typical antibacterial properties38 which was affirmed with the help of different experimental assays. As shown in Fig. S2A and S2B,† formation of the zone-of-inhibition by AgNPs against E. coli culture was clearly observed in well diffusion assay compared to the untreated control. Interestingly, the inhibition was found to be higher in G-AgNPs than in C-AgNPs (Fig. S2B and C†). Furthermore, growth curve analysis showed declining growth of bacteria with an increase in the concentration of AgNPs (Fig. S2D and E†). The results confirmed the antibacterial activity of G-AgNPs and indicated their enhanced effect with an increase in concentration compared to C-AgNPs. Previous literature has reported similar effects and attributed the probable cause of the phenomenon to the membrane damage of bacteria due to the interaction of AgNPs leading to their death.7,57,58In vivo biocompatibility of G-AgNPs
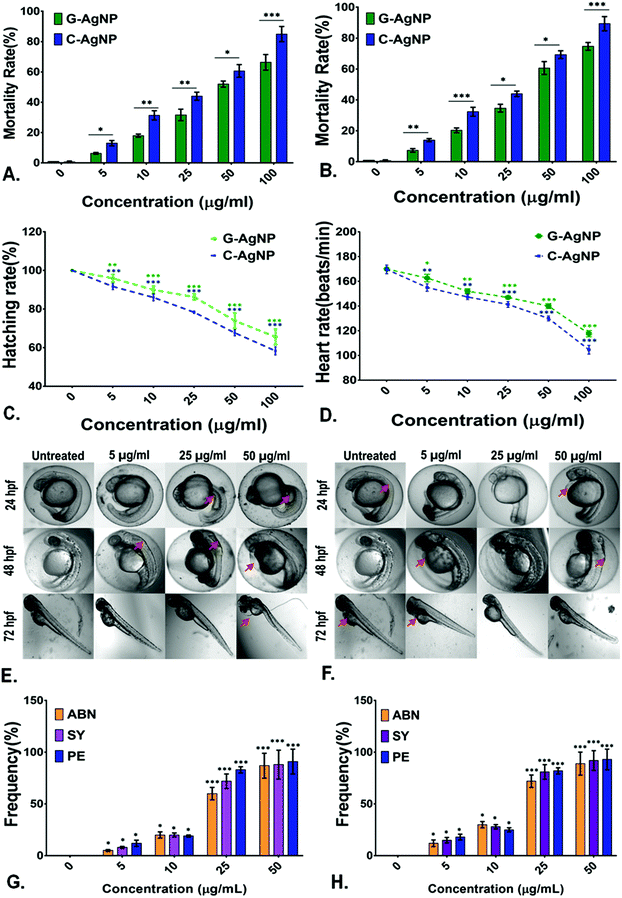
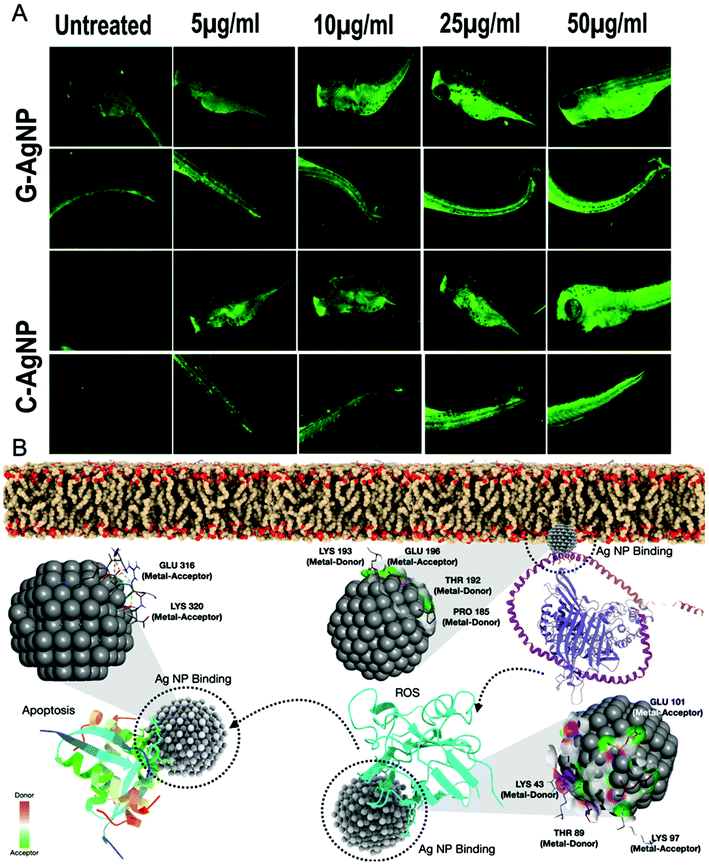
Evaluation of biocompatibility of nanoparticles is an important criterion to determine their utility for biomedical and environmental purposes.59 After being used, the nanoparticles can be released into the environment and come in contact with organisms, especially in the water. Hence, an aquatic model was chosen to determine the biocompatibility of G-AgNPs. The comparative in vivo biocompatibility of the synthesized G-AgNPs and C-AgNPs were estimated at the cellular and molecular levels using embryonic zebrafish. As shown in Fig. 2A and B, the mortality rate of the embryos was found to be concentration and time-dependent on the nanoparticle exposure. Interestingly, the mortality rate was higher in the case of C-AgNPs exposure compared to that with G-AgNPs. As calculated, the LC50 was found to be 35.6 ± 1.2 μg ml−1 in the case of C-AgNPs and 47.2 ± 1.6 μg mL−1 for G-AgNPs at 24 h of exposure, indicating higher biocompatibility of G-AgNPs compared to that of C-AgNPs. The hatching rate and heartbeat rate of the exposed embryos were also found to be declining with an increase in the exposure concentration of both G-AgNPs and C-AgNPs (Fig. 2C and D). However, C-AgNPs were found to be more potent in decreasing the hatching and heartbeat rate in exposed embryos compared to G-AgNPs. Abnormalities in the hatching rate of zebrafish embryos exposed to different types of metallic nanoparticles have been reported in previous studies.11 The irregularities can be attributed to the hardening of chorion due to the structural and functional dysfunctionality of proteins such as He1a because of interaction with exposed nanoparticles.60 Similarly, abnormal heart rate of embryos can be endorsed by hypoxic conditions created due to blockage of chorion due to the accumulation of nanoparticles at the surface.61 Further, it was postulated that the interactions of internalized AgNPs would trigger abnormalities in the morphological and anatomical development of embryos. The hypothesis was confirmed by analyzing the morphological abnormalities in embryos exposed to different concentrations of G-AgNPs and C-AgNPs. As shown in Fig. 2E and F, significant time and concentration-dependent morphological abnormalities were observed in zebrafish embryos exposed to both C-AgNPs and G-AgNPs. Significant defects such as pericardial edema, swollen yolk and abnormal notochord formation were found in embryos at 48 hpf and 72 hpf exposed to 25 μg ml−1 and 50 μg ml−1 exposure. Interestingly, a higher frequency has been observed in the case of C-AgNPs exposure compared to G-AgNPs (Fig. 2G and H); similar results have been reported by our group and other researchers in zebrafish embryos exposed to AgNPs synthesized through other routes. The observed results can be ascribed to the influential effect of nanoparticles on molecular machinery responsible for the phenotypic expression at the cellular level.62 Moreover, the impact of physiological parameters such as size and charge has been described for the reported toxicity of AgNPs.63 It can be argued that the comparatively bigger size and higher stability of G-AgNPs due to their biomolecular coating have a significant influence on their accumulation and internalization for the comparative physiological abnormalities in zebrafish embryos.Hence, a computational approach was undertaken to understand the influence of AgNP interaction with proteins responsible for hatching and other processes through molecular docking.
Structural conformation of AgNPs
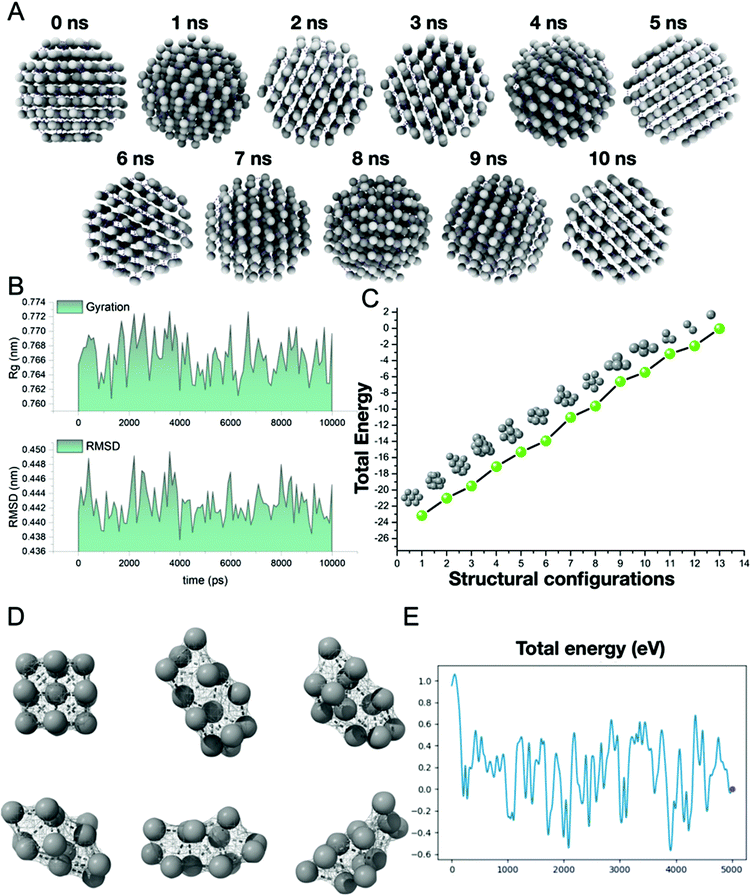
Before evaluating the interaction mechanisms, we delved into constructing AgNPs using molecular modeling software (described in the methods). AgNPs were simulated in a solvated medium and vacuum using the combinatorial molecular dynamics simulation methods, i.e., classical mechanics and DFT formalism, respectively. The 10 ns simulated AgNPs (Fig. 3A) in the solvent medium, which was evaluated using classical mechanics, showed no substantial changes in the geometry of the structure. As depicted in Fig. 3B, the radius of gyration (Rg) showed a minor deviation (0.76–0.77 nm) in the compactness of the structure due to the influence of the solvent medium. The root means square deviation (RMSD) also showed a similar pattern with no substantial changes in the structure i.e., 0.44–0.45 nm. After evaluating the solvent effect on AgNPs, we then shifted our focus to illustrate the effect of vacuum on AgNPs with DFT formalism. The unique strategy was adopted as we tested the total energy variation of each Ag atom when isolated from the AgNPs cluster sequentially. The plot (Fig. 3C) depicts the structural configurations of the aforementioned strategy and the total energy variation. As shown in the plot, the full AgNPs cluster showed ∼−24 eV and varied at a sequential rate of ∼2 eV until all Ag atoms were removed from the cluster, which means that each Ag atom possesses 2 eV as the valence of the Ag atom is +2. Further, the dynamics of the cluster have also been evaluated using the DFT formalism in a vacuum (Fig. 3D). As shown in Fig. 3E, the total energy variation has substantially affected the cluster in a vacuum environment. The cluster configuration has been substantially affected and it does not possess the native conformation, as revealed in the ESI (SV1).†AgNPs uptake through channelized interaction with lipid biomolecules
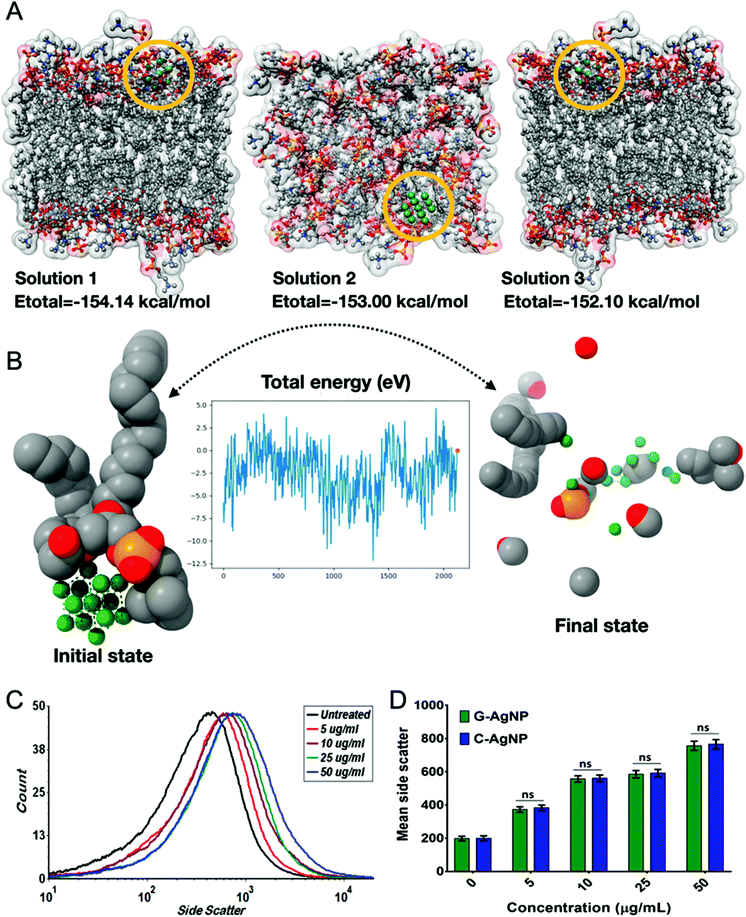
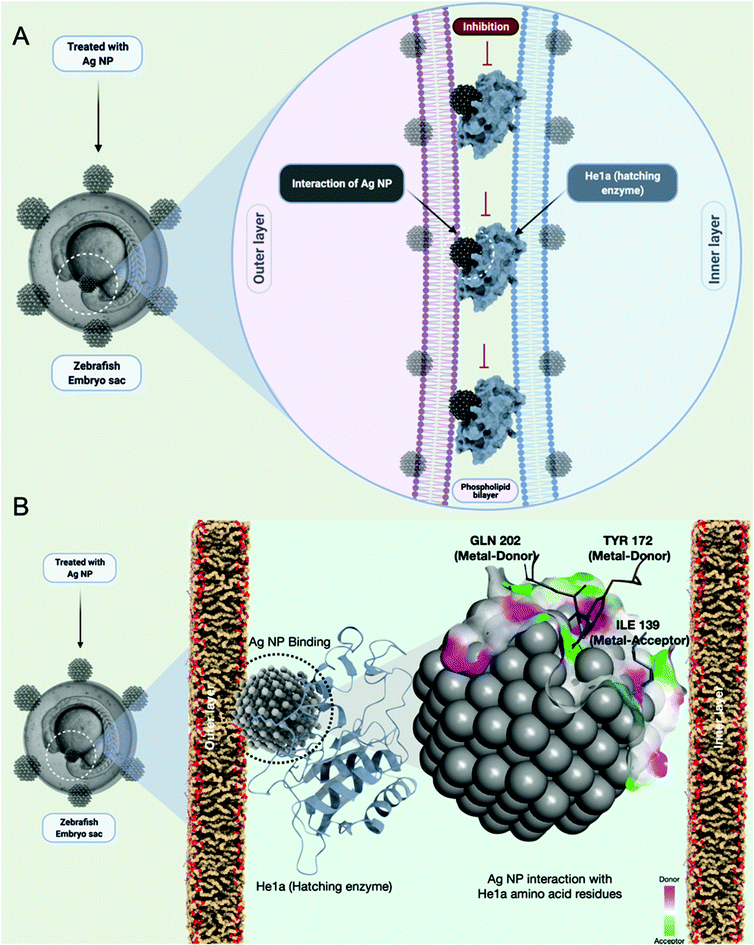
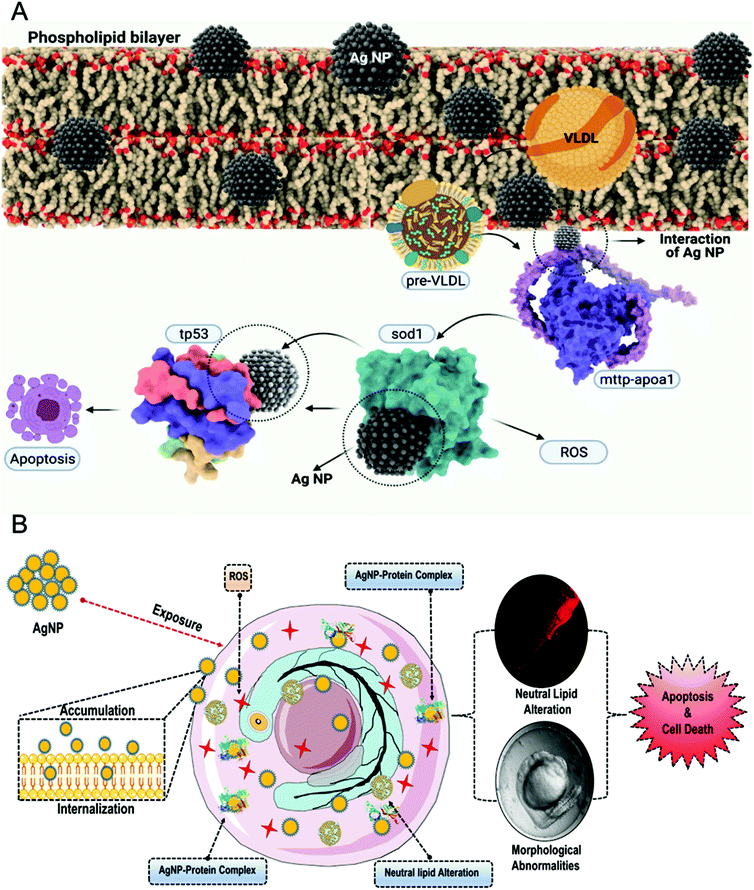
Before delving into the actual mechanism of how AgNPs interact with specific proteins and invade the phospholipid bilayer membrane, first, the interaction was predicted between the specific lipid membrane, i.e, DPPC using the HEX docking program. 3000 solutions were modelled of which the highest binding energies were selected, i.e., −154.14 kcal mol−1, −153 kcal mol−1 and −152.10 kcal mol−1 as illustrated in Fig. 4A. Since rigid docking was performed in a vacuum environment, the interacting molecules were taken into consideration using DFT formalism to study the effect of the interaction. The total energy variation was substantially high when dynamics were performed for 2000 steps. As shown in Fig. 4B, the initial state of the interaction was substantially altered during the course of the simulation. The AgNPs cluster was dissociated in the final state, while the interaction with the lipid molecules remains intact (ESI SV1†). The same has been evaluated using in vivo experiments through uptake analysis. The exposed AgNPs (G-AgNPs and C-AgNPs) were hypothesized to exhibit biotoxicity through a channel of cellular processes initiated by the accumulation at the surface and subsequent uptake of nanoparticles.13 The uptake of nanoparticles was estimated through flow cytometry analysis. The change in the granularity of cells has been described to be an important factor to assess the uptake of nanoparticles.39 The granularity change was depicted by the measurement of side scatter in flow cytometry. As shown in Fig. 4C, mean side scatter of embryonic cells exposed to G-AgNPs and C-AgNPs were found to increase with an increase in exposure concentrations of 0, 5, 10, 25 and 50 μg ml−1. Comparatively, there was a non-significant difference of mean side scatter in cells exposed to C-AgNPs and G-AgNPs at each concentration of exposure (Fig. 4D). The data indicated an equivalent uptake of both types of nanoparticles in the embryonic cells at each concentration. However, it was important to unravel the mechanism of uptake of nanoparticles in zebrafish cells. As shown in Fig. 5A and B, AgNPs were found to interact with He1a through amino acid residues such as glutamine (GLN), tyrosine (TYR) and isoleucine (ILE) due to their metal donor and metal acceptor properties. This interaction can be associated with the structural and functional dysfunctionality of He1a leading to irregular and late hardening of chorion for hatching rate abnormalities.AgNPs interaction with peroxisomal protein family: Import mechanism
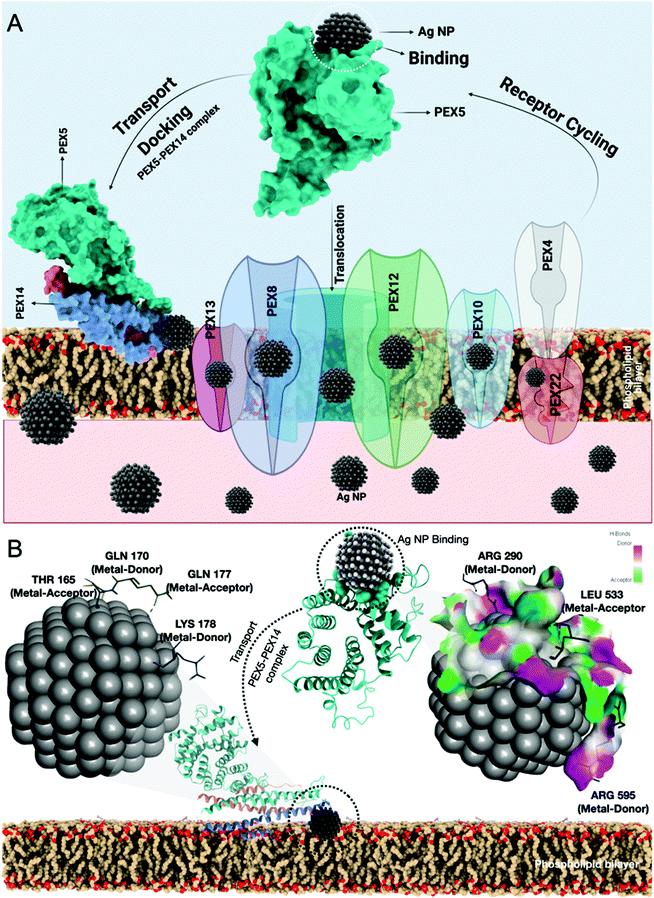
Previous reports have suggested the role of many protein receptor molecules at the surface of the cell membrane for translocation.51 To gain insight into the molecular mechanisms of matrix-protein import, we focused our attention on those aspects of peroxisomal-matrix-protein import for which the biochemical data are most reliable, starting with the recognition of PTS1-containing enzymes by PEX5, the step-in import that has been subjected to the most rigorous biochemical analysis. The peroxisomal biogenesis receptor protein family (PEX) has been reported to play an important role in the transfer of proteins and other molecules for metabolic processes like β-oxidation.50 Hence, it was hypothesized that the PEX family proteins were also playing an important role in the uptake of nanoparticles across the membrane. To understand the mechanism, in silico assessment was conducted through a molecular docking approach to analyze the interaction of AgNPs with the PEX protein family. It was estimated that PEX5 protein was interacting with AgNPs and helped in the translocation of nanoparticles directly through the membrane and indirectly through a channelized association with PEX14 along with other PEX family members like PEX13,8,12,10 and PEX 4 (Fig. 6A). As shown in Fig. 6B, docking analysis showed AgNPs interaction with PEX5 via amino acids arginine (ARG290, ARG595) and leucine (LEU533) inducing metal acceptor and donor effects. The PEX5–PEX14 complex as deciphered in the mechanism has been involved in the transport mechanism. AgNPs were found to have a firm interaction with only PEX14 from the PEX5–PEX14 complex via amino acids glutamine (GLN170, GLN177), threonine (THR165) and lysine (LYS178). The computational data enlightened the mechanism of uptake of nanoparticles through the PEX protein family.In vivo oxidative stress mechanism induced by AgNPs
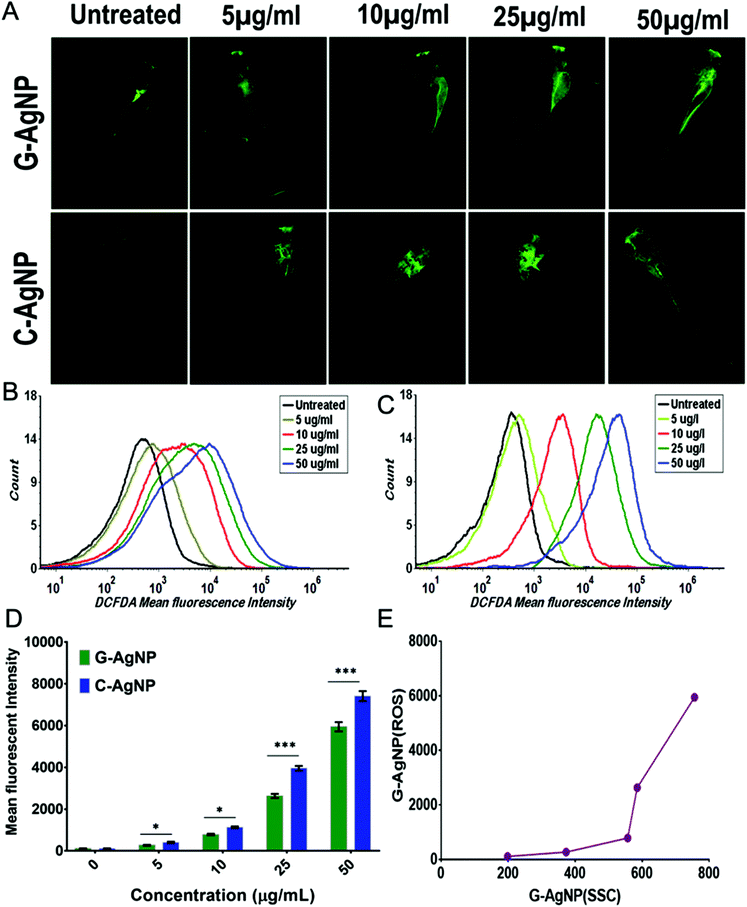
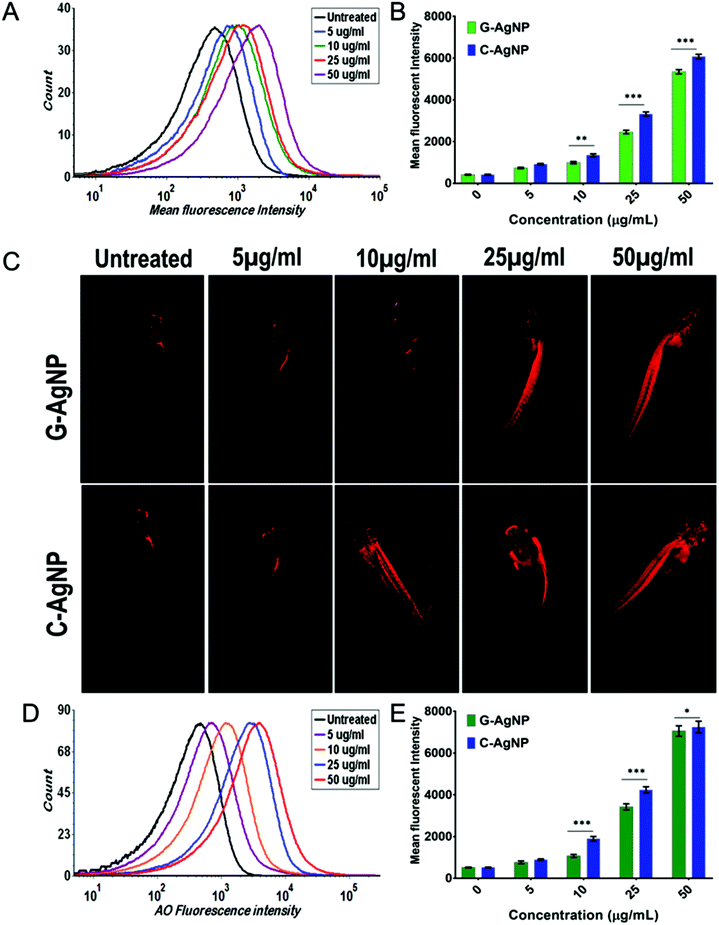
AgNPs have been reported to cause cytotoxicity by dysregulating the metabolic function through induction of oxidative stress.13,64 Hence, it was postulated that internalized AgNPs were inducing oxidative stress by enhancing the production of ROS, which further causes imbalanced activities like lipid alteration and apoptosis. G-AgNPs and C-AgNPs were assessed for their potential to induce oxidative stress and anomaly in metabolic processes. As shown in Fig. 7A, DCFDA staining in untreated and exposed embryos revealed the induction of reactive oxygen species after G-AgNPs and C-AgNPs exposure.40 The green fluorescence was found to be increasing with an increase in the exposure concentration of both types of nanoparticles. However, it was at a higher intensity in the case of C-AgNPs exposure at each concentration (Fig. 7D). The increase in green fluorescence indicated the induction of a higher number of oxidative radicals (ROS) with an increase in exposure concentration. The fluorescence indicating the production of ROS was further quantified using flow cytometry. As shown in Fig. 7B and C, the mean fluorescence intensity of DCFDA was enhanced with an increase in the concentration of G-AgNPs and C-AgNPs. Comparatively, the intensity was found to be higher in the case of C-AgNP at each exposure concentration (Fig. 7D). A relationship between the mean side scatter and the mean fluorescence intensity of DCFDA confirm the ROS induction due to the uptake of nanoparticles (Fig. 7E). The analysis confirmed the concentration-based induction of ROS after exposure to G-AgNPs and C-AgNPs.The enhancement of ROS production after exposure to nanoparticles has been discussed in relation to the imbalance in other metabolic functions such as lipid metabolism and apoptosis.64,65 Hence, it was hypothesized that exposure to nanoparticles inducing ROS altered these metabolic activities. Alteration in neutral lipid metabolism commonly called “Steatosis” was checked using flow cytometry by measuring the red fluorescence intensity of LipidTox™ in embryonic cells exposed to different concentrations of G-AgNPs and C-AgNPs.15 LipidTox™ has been recognized to stain the neutral lipid molecules like LDL and VLDL in cells.15 As shown in Fig. 8A and Fig. S3,† mean fluorescence intensity was found to increase with increasing exposure concentration of both C-AgNPs and G-AgNPs from 5 μg mL−1 to 50 μg mL−1. Comparatively, it was found to be higher in the case of C-AgNPs exposure (Fig. 8B) and the result was further verified using fluorescence microscopy. Fig. 8C shows the red fluorescence intensity of LipidTox™ staining VLDL and LDL molecules in embryos exposed to C-AgNPs and G-AgNPs. Enhanced fluorescence was found with the increase in exposure concentrations of nanoparticles. The data indicated enhanced accumulation of neutral lipids in the head, tail, and trunk region of the exposed embryos and was in line with the flow cytometry analysis. The differential alteration in neutral lipids due to exposure to G-AgNPs and C-AgNPs can be attributed to the differential physiochemical properties due to their origin of synthesis.37 Further, induced neutral lipid alteration and oxidative stress (ROS) were hypothesized to induce cytotoxic effects in cells leading to apoptosis. This hypothesis was examined by experimental analysis of apoptosis in C-AgNPs and G-AgNPs exposed embryos through acridine orange (AO) staining using fluorescence microscopy and flow cytometry. AO has been recognized to stain apoptotic cells.66 As shown in Fig. 8D and Fig. S4,† flow cytometry analysis of AO fluorescence showed a concentration-dependent increase of apoptotic cells in zebrafish embryos exposed to C-AgNPs and G-AgNPs. In comparison to G-AgNPs, C-AgNPs were found to induce higher apoptosis at each exposure concentration (Fig. 8E). The analysis was further visualized using fluorescence microscopy. Fig. 9A and Fig. S5† show the increase in green fluorescence intensity of AO representing a higher number of apoptotic cells with an increase in exposure concentration of both C-AgNPs and G-AgNPs in the head and trunk regions of the exposed embryos. Interestingly, the fluorescence intensity was higher in the case of C-AgNPs at each concentration, which correlates with the data from flow cytometry analysis. The results advocated the lower toxicity of biogenic G-AgNPs compared to commercial C-AgNPs and can be reasoned to physicochemical properties of G-AgNPs like size, stability, and biomolecule coating.
Further, a correlation analysis was performed with the experimental data obtained through uptake, neutral lipid alteration and apoptosis analysis to understand the impact of internalized AgNPs on different physiological processes. As shown in Fig. S6A and 6B,† mean side scatter was found to be correlated with the fluorescence of neutral lipid intensity and apoptotic cells, thereby confirming the effect of AgNPs internalized during exposure. Moreover, correlation analysis of ROS induction with lipid and apoptosis also showed a clear relation depicting the effect of induced ROS in dysregulation of these physiological activities (Fig. S6C and 6D†). Interestingly, neutral lipid accumulation and apoptosis analysis also showed a well-founded relationship between them inferring to have dependent activity on each other (Fig. S6E†).
The experimental analysis confirmed the impact of internalized AgNPs on neutral lipid alteration and apoptosis of cells due to ROS induction.67 However, the molecular mechanism needs to be unraveled. To excavate the molecular mechanism, computational docking analysis was performed to study the interaction with different proteins responsible for ROS induction, steatosis, and apoptosis. Superoxide dismutase (sod1) is known for its key role in oxidative stress.68 As shown in Fig. 9B and Fig. S7,† molecular docking analysis of sod1 with AgNPs showed interaction via amino acids lysine (LYS43, LYS97), glutamic acid (GLU101) and threonine (THR89) through the hydrogen bond with an average energy of −309.55 kcal mol−1. It has been reported that apolipoproteins (apoa1a) are synthesized by the yolk syncytial layer during embryonic development. These apolipoproteins further form very-low-density lipid droplets (VLDL) and cytoplasmic LD from yolk lipids, which enter into the circulatory system for the deliverance of lipids to different tissues.69 The transfer of the VLDLs and LDs is regulated by microsomal triglyceride transfer protein (MTTP) via the formation of MTTP-apoa1a complex.70 Docking analysis of AgNPs with MTTP-apoa1a complex showed interaction via amino acids lysine (LYS193), glutamic acid (GLU316), threonine (THR192) and proline (PRO195) through H-bonding with energy of −12.21 kcal mol−1 (Fig. 9B and Fig. S7†). The apoptotic activity in cells has been reported to be regulated by different proteins such as tp53 and caspase. For our study, tp53 was docked with AgNPs to understand their interaction. Fig. 9B shows the interaction of tp53 with AgNPs via glutamic acid (GLU 316) and lysine (LYS320) with an average energy of −393.82 kcal mol−1. With reference to the analysis, it can be interpreted that the AgNPs were influencing the structural and functional activity of sod1, mttp-apoa1a complex and tp53 proteins, thus affecting the process of ROS production, neutral lipid accumulation and apoptosis. Previous studies have also shown the effect of AgNPs on sod1 and Tp53.7 Moreover, nanoparticles such as TiO2 NP and carbon NP have been shown to have a significant effect on steatosis, lipid peroxidation and lipid alteration in the brain and gastrointestinal tissues.71 Hence, based on the experimental results and the literature, it can be estimated that AgNPs were inducing their cytotoxic effect by altering the cellular metabolic process at the molecular level.
Mechanism
With reference to the experimental and computational analysis, the mechanism of the biocompatibility of G-AgNPs can be outlined as the following: exposure of AgNPs to the zebrafish embryos leads to their accumulation at the surface of the chorion leading to blockage of the pores as well as delay in hardening of the chorion due to inhibition in he1a enzyme. The dysfunctionality can be associated with the interaction with AgNPs. The molecular imbalance causes abnormalities in the hatching rate of the embryos. Moreover, the blockage of chorion pores leads to a hypoxic condition in embryos leading to the abnormal production of ROS to neutralize oxygen deficiency.61 Some of the accumulated nanoparticles get internalized inside the embryos and cause effects on early and late developmental processes. The nanoparticles interact with receptor proteins such as PEX5 and PEX14 and get internalized inside the cellular cytosol. The internalized nanoparticles further interact with antioxidant and metabolic proteins such as sod1, apoa1-mttp complex, and tp53 influencing their structural and functional integrity. Impact on sod1 leads to oxidative stress. The ROS formed due to the hyper-regulation of different cytochrome systems further leads to abnormal consequences in the functionality of other metabolic functions.72 Apart from that, the internalized nanoparticles interact with the apoa1-mttp protein complex to affect the formation of VLDL and LDL leading to irregular transfer to the system and steatosis metabolism (Fig. 10). Moreover, AgNPs also interact with tp53 proteins to disturb the regulated program cell death (apoptosis) along with factors such as ROS induction and improper apoa1-mttp function. Thus, as a combinatorial effect of these metabolic irregularities, cellular apoptosis fastens up with concentration-dependent exposure of AgNPs leading to their cytotoxicity. Thus, the study outlined the mechanism of molecular interaction biocompatibility of AgNPs along with a determination of the differential nature of biocompatibility in G-AgNPs and C-AgNPs. Moreover, it advocates and recommends a green technique to produce more eco-compatible and biocompatible silver nanoparticles for biomedical and ecological applications.Conclusions
Since tremendous activity is ongoing in the field of biological preparation of nanoparticles, especially in the biomedical arena, it is important to delineate the reasons to validate the claims unearthed in these investigations. In brief, the study disclosed the antibacterial potential of green synthesized G-AgNPs with a detailed assessment of their comparative biocompatibility with commercially available C-AgNPs in the embryonic zebrafish model. Successful poly-dispersed, stable silver nanoparticles with a size of 25–35 nm were synthesized with the help of silver grass (Miscanthus sinensis) leaves extract with an SPR at 432 nm. G-AgNPs showed significantly lower toxicity to developing embryos than C-AgNPs. The mechanistic evaluation of the biocompatibility showed firm uptake of AgNPs with a significant effect on ROS induction, neutral lipid alteration and apoptosis. The computational and experimental analysis elucidated the mechanism as an impact of concentration-dependent accumulation and internalization of AgNPs leading to the enhancement of oxidative stress and abnormalities in metabolic processes due to intrinsic atomic interaction of internalized AgNPs with metabolic proteins like sod1, apoa1-mttp and tp53. Moreover, the study presented a novel way for green synthesis of AgNPs with the help of silver grass for biomedical and ecological applications with higher biocompatibility and eco-compatibility.Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the ESI.† Additional data related to this paper may be requested from the authors.Author contributions
All authors have given approval to the final version of the manuscript. P. K. P and P. K contributed equally as the first author. Conceptualization: P. K. P., P. K., and S. K. V.; computational analyses: P. K. P., S. M.; illustrations: P. K. P, S. K. V; experimental analyses: P. K., S. K. V, P. P, K. H. writing (original draft preparation): S. K. V, P. K. P, R. V, K. H.; writing (review and editing): P. K. P., P. K., R. A., Y. K. M., P. P, S. M., M. M. T, A. D., S. K. S., S. K. V., Y. K. M., and M. S.; supervision: R. A. P. K. P., M. S, K. H and S. K. V.; funding acquisition: R. A., M. S, K. H, S. K. V.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
SNIC and HPC2N have been acknowledged for providing computational resources. The schematic figures were created with BioRender.com.The authors acknowledge the financial support from the Swedish Research Council (VR grant no. 2016-06014) and Carl Tryggers Stiftelse for Vetenskaplig Forskning. Authors thank Research Infrastructure RECETOX RI (No. LM2018121) financed by the Ministry of Education, Youth and Sports, and Operational Programme Research, Development and Innovation – project CETOCOEN EXCELLENCE (No. CZ.02.1.01/0.0/0.0/17_043/0009632) for supportive background. YKM acknowledges funding by Interreg Deutschland–Denmark with money from the European Regional Development Fund, project number 096-1.1-18 (Access and Acceleration). MS and SKV acknowledge DBT-BUILDER program (BT/INF/22/SP42155/2021) for infrastructure support at KIIT UNIVERSITY.
References
- V. Pareek, A. Bhargava, R. Gupta, N. Jain and J. Panwar, Adv. Sci., Eng. Med., 2017, 9, 527–544 CrossRef CAS.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS PubMed.
- L. Wei, J. Lu, H. Xu, A. Patel, Z.-S. Chen and G. Chen, Drug Discovery Today, 2015, 20, 595–601 CrossRef CAS PubMed.
- K. M. M. Abou El-Nour, A. Eftaiha, A. Al-Warthan and R. A. A. Ammar, Arabian J. Chem., 2010, 3, 135–140 CrossRef CAS.
- M. Wypij, T. Jędrzejewski, M. Ostrowski, J. Trzcińska, M. Rai and P. Golińska, Molecules, 2020, 25(13), 3022 CrossRef CAS PubMed.
- B. Fadeel, L. Farcal, B. Hardy, S. Vázquez-Campos, D. Hristozov, A. Marcomini, I. Lynch, E. Valsami-Jones, H. Alenius and K. Savolainen, Nat. Nanotechnol., 2018, 13, 537–543 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, P. K. Panda, A. Thirumurugan, S. Patro, S. K. S. Parashar and M. Suar, Mater. Sci. Eng., C, 2018, 92, 807–818 CrossRef CAS PubMed.
- A. Gliga, S. Skoglund, I. Odnevall Wallinder, B. Fadeel and H. Karlsson, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- P. V. Asharani, Y. Lian Wu, Z. Gong and S. Valiyaveettil, Nanotechnology, 2008, 19, 255102 CrossRef CAS PubMed.
- S. R. Satapathy, P. Mohapatra, R. Preet, D. Das, B. Sarkar, T. Choudhuri, M. D. Wyatt and C. N. Kundu, Nanomedicine, 2013, 8, 1307–1322 CrossRef CAS PubMed.
- S. K. Verma, A. Nandi, A. Sinha, P. Patel, E. Jha, S. Mohanty, P. K. Panda, R. Ahuja, Y. K. Mishra and M. Suar, Precis. Nanomed., 2021, 4, 21978 Search PubMed.
- A. J. Hill, H. Teraoka, W. Heideman and R. E. Peterson, Toxicol. Sci., 2005, 86, 6–19 CrossRef CAS PubMed.
- S. Kumari, P. Kumari, P. K. Panda, P. Patel, E. Jha, M. A. Mallick, M. Suar and S. K. Verma, Nanomedicine, 2020, 15, 2119–2132 CrossRef CAS PubMed.
- L. Qiang, Z. H. Arabeyyat, Q. Xin, V. N. Paunov, I. J. F. Dale, R. I. L. Mills, J. M. Rotchell and J. Cheng, Int. J. Mol. Sci., 2020, 21, 1876 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, P. K. Panda, M. Mukherjee, A. Thirumurugan, H. Makkar, B. Das, S. K. S. Parashar and M. Suar, Toxicol. Res., 2018, 7, 244–257 CrossRef CAS PubMed.
- M. Cinelli, S. R. Coles, M. N. Nadagouda, J. Błaszczyński, R. Słowiński, R. S. Varma and K. Kirwan, J. Cleaner Prod., 2017, 162, 938–948 CrossRef CAS PubMed.
- M. Cinelli, S. R. Coles, M. N. Nadagouda, J. Błaszczyński, R. Słowiński, R. S. Varma and K. Kirwan, Green Chem., 2015, 17, 2825–2839 RSC.
- M. Kadziński, M. Cinelli, K. Ciomek, S. R. Coles, M. N. Nadagouda, R. S. Varma and K. Kirwan, Eur. J. Oper. Res., 2018, 264, 472–490 CrossRef PubMed.
- N. R. Jana, L. Gearheart and C. J. Murphy, Chem. Commun., 2001, 7, 617–618 RSC.
- Q. H. Tran, V. Q. Nguyen and A.-T. Le, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2013, 4, 033001 CAS.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS PubMed.
- N. Salah, S. S. Habib, Z. H. Khan, A. Memic, A. Azam, E. Alarfaj, N. Zahed and S. Al-Hamedi, Int. J. Nanomed., 2011, 6, 863–869 CrossRef CAS PubMed.
- B. K. Das, S. K. Verma, T. Das, P. K. Panda, K. Parashar, M. Suar and S. K. S. Parashar, Chem.-Biol. Interact., 2019, 297, 141–154 CrossRef CAS PubMed.
- Q. H. Tran, V. Q. Nguyen and A.-T. Le, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2013, 4, 20 Search PubMed.
- D. Hebbalalu, J. Lalley, M. N. Nadagouda and R. S. Varma, ACS Sustainable Chem. Eng., 2013, 1, 703–712 CrossRef CAS.
- M. C. Moulton, L. K. Braydich-Stolle, M. N. Nadagouda, S. Kunzelman, S. M. Hussain and R. S. Varma, Nanoscale, 2010, 2, 763–770 RSC.
- B. Baruwati, V. Polshettiwar and R. S. Varma, Green Chem., 2009, 11, 926–993 RSC.
- M. N. Nadagouda and R. S. Varma, Green Chem., 2008, 10, 859–886 RSC.
- A. García, A. Toledano, M. Á. Andrés and J. Labidi, Process Biochem., 2010, 45, 935–940 CrossRef.
- R. Sheel, P. Kumari, P. K. Panda, M. D. Jawed Ansari, P. Patel, S. Singh, B. Kumari, B. Sarkar, M. A. Mallick and S. K. Verma, Environ. Pollut., 2020, 267, 115482 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, K. J. Kiran, S. Bhat, M. Suar and P. S. Mohanty, Mater. Today: Proc., 2016, 3, 1949–1957 Search PubMed.
- E. Jha, P. K. Panda, P. Patel, P. Kumari, S. Mohanty, S. K. S. Parashar, R. Ahuja, S. K. Verma and M. Suar, Nanomedicine, 2021, 16, 307–322 CrossRef CAS PubMed.
- S. Lee, H. Kim, L. R. Beuchat, Y. Kim and J. H. Ryu, Food Microbiol., 2020, 90, 103489 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, P. K. Panda, J. K. Das, A. Thirumurugan, M. Suar and S. Parashar, Nanomedicine, 2018, 13, 43–68 CrossRef CAS PubMed.
- P. Patel, P. K. Panda, P. Kumari, P. K. Singh, A. Nandi, M. A. Mallick, B. Das, M. Suar and S. K. Verma, Ecotoxicol. Environ. Saf., 2020, 192, 110321 CrossRef CAS PubMed.
- Modified Holtfreter’s solution, Cold Spring Harb. Protoc., 2009, 2009, DOI:10.1101/pdb.rec11902.
- S. K. Verma, P. K. Panda, P. Kumari, P. Patel, A. Arunima, E. Jha, S. Husain, R. Prakash, R. Hergenröder, Y. K. Mishra, R. Ahuja, R. S. Varma and M. Suar, Green Chem., 2021, 23, 3439–3458 RSC.
- S. K. Verma, E. Jha, P. K. Panda, A. Mishra, A. Thirumurugan, B. Das, S. K. S. Parashar and M. Suar, Toxicol. Sci., 2018, 161, 125–138 CrossRef CAS PubMed.
- R. M. Zucker, E. J. Massaro, K. M. Sanders, L. L. Degn and W. K. Boyes, Cytometry, Part A, 2010, 77, 677–685 CrossRef CAS PubMed.
- E. Eruslanov and S. Kusmartsev, in Methods in Molecular Biology, 2010, vol. 594, pp. 57–72 Search PubMed.
- S. Kumari, P. kumari, P. K. Panda, N. Pramanik, S. K. Verma and M. A. Mallick, Environ. Nanotechnol. Monit. Manag., 2019, 11, 100201 Search PubMed.
- M. Marqués Arias, Vienna Ab-initio Simulation Package, Univ. Edinburgh., 2009 Search PubMed.
- J. Hafner, J. Comput. Chem., 2008, 29, 2044–2078 CrossRef CAS PubMed.
- G. Kresse and M. Marsman, VASP the GUIDE, 2012 Search PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS PubMed.
- S. Jo, X. Cheng, J. Lee, S. Kim, S.-J. Park, D. S. Patel, A. H. Beaven, K. Il Lee, H. Rui, S. Park, H. S. Lee, B. Roux, A. D. J. MacKerell, J. B. Klauda, Y. Qi and W. Im, J. Comput. Chem., 2017, 38, 1114–1124 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- D. W. Ritchie, Proteins, 2003, 52, 98–106 CrossRef CAS PubMed.
- W. Girzalsky, D. Saffian and R. Erdmann, Biochim. Biophys. Acta, Mol. Cell Res., 2010, 1803, 724–731 CrossRef CAS PubMed.
- P. K. Kim and E. H. Hettema, J. Mol. Biol., 2015, 427, 1176–1190 CrossRef CAS PubMed.
- S. K. Verma, K. Nisha, P. K. Panda, P. Patel, P. Kumari, M. A. Mallick, B. Sarkar and B. Das, Sci. Total Environ., 2020, 713, 136521 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- D. S. Biovia, H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, P. E. Bourne, T. Darden, D. York, L. G. Pedersen, G. Bussi, D. Donadio, M. Parrinello, U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee, L. G. Pedersen, M. Parrinello, A. Rahman, V. Hornak, R. Abel, A. Okur, B. Strockbine, A. Roitberg, C. Simmerling, M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess, E. Lindah, H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren, A. Dinola, J. R. Haak, R. W. Hockney, S. P. Goel, J. W. Eastwood, C. A. Davey, D. F. Sargent, K. Luger, A. W. Maeder and T. J. Richmond, J. Chem. Phys., 2000, 10, 21–9991, DOI:10.1016/0021-9991(74)90010-2.
- P. Patel, P. Kumari, S. K. Verma and M. A. Mallick, Eng. Nanomater. – Heal. Saf., 2019, 70677 Search PubMed.
- R. Xu, Particuology, 2008, 6, 112–115 CrossRef CAS.
- S. Husain, S. K. Verma, Hemlata, M. Azam, M. Sardar, Q. M. R. Haq and T. Fatma, Mater. Sci. Eng., C, 2021, 122, 111888 CrossRef CAS PubMed.
- S. K. Verma, E. Jha, B. Sahoo, P. K. Panda, A. Thirumurugan, S. K. S. Parashar and M. Suar, RSC Adv., 2017, 7, 40034–40045 RSC.
- S. Kumari, P. Kumari, P. K. Panda, P. Patel, E. Jha, M. A. Mallick, M. Suar and S. K. Verma, Nanomedicine, 2020, 15, 22 CrossRef PubMed.
- K. J. Ong, X. Zhao, M. E. Thistle, T. J. Maccormack, R. J. Clark, G. Ma, Y. Martinez-Rubi, B. Simard, J. S. C. Loo, J. G. C. Veinot and G. G. Goss, Nanotoxicology, 2014, 8, 295–304 CrossRef CAS PubMed.
- S. K. Verma, P. K. Panda, E. Jha, M. Suar and S. K. S. Parashar, Sci. Rep., 2017, 7, 13909 CrossRef PubMed.
- S. K. Verma, E. Jha, P. K. Panda, P. Kumari, N. Pramanik, S. Kumari and A. Thirumurugan, Artif. Cells, Nanomed., Biotechnol., 2018, 46, S671–S684 CrossRef CAS PubMed.
- S. Husain, S. K. Verma, D. Yasin, Hemlata, M. M. A. Rizvi and T. Fatma, Bioorg. Chem., 2021, 107, 104535 CrossRef CAS PubMed.
- P. Kumari, P. K. Panda, E. Jha, K. Kumari, K. Nisha, M. A. Mallick and S. K. Verma, Sci. Rep., 2017, 7, 16284 CrossRef PubMed.
- A. Manke, L. Wang and Y. Rojanasakul, BioMed Res. Int., 2013, 2013, 942916 Search PubMed.
- H. A. Elfawy, S. Anupriya, S. Mohanty, P. Patel, S. Ghosal, P. K. Panda, B. Das, S. K. Verma and S. Patnaik, Sci. Total Environ., 2021, 789, 147989 CrossRef CAS PubMed.
- P. K. Panda, S. K. Verma and M. Suar, Nanomedicine, 2021, 16, 2249–2254 CrossRef CAS PubMed.
- C. F. Ken, C. T. Lin, J. F. Shaw and J. L. Wu, Mar. Biotechnol., 2003, 5, 167–173 CrossRef CAS PubMed.
- A. Schlegel and D. Y. R. Stainier, Biochemistry, 2006, 45, 15179–15187 CrossRef CAS PubMed.
- K. Minehira, S. G. Young, C. J. Villanueva, L. Yetukuri, M. Oresic, M. K. Hellerstein, R. V. Farese, J. D. Horton, F. Preitner, B. Thorens and L. Tappy, J. Lipid Res., 2008, 49, 2038–2044 CrossRef CAS PubMed.
- E. Gorrochategui, J. Li, N. J. Fullwood, G.-G. Ying, M. Tian, L. Cui, H. Shen, S. Lacorte, R. Tauler and F. L. Martin, Mutagenesis, 2017, 32, 91–103 CrossRef CAS PubMed.
- R. Eram, P. Kumari, P. K. Panda, S. Singh, B. Sarkar, M. A. Mallick and S. K. Verma, Cellular Investigations on Mechanistic Biocompatibility of Green Synthesized Calcium Oxide Nanoparticles with Danio rerio, Journal of Nanotheranostics, 2021, 2, 51–62, DOI:10.3390/jnt2010004.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1gc04103g |
| ‡ Authors with equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |