Open Access Article
Open Access ArticleTargeting proteases involved in the viral replication of SARS-CoV-2 by sesquiterpene lactones from chicory (Cichorium intybus L.)
María Ángeles
Ávila-Gálvez
 ab,
Carlos
Rafael-Pita
ab,
Naiara
Fernández
a,
João
Baixinho
ac,
José D.
Anastácio
ab,
Katarina
Cankar
ab,
Carlos
Rafael-Pita
ab,
Naiara
Fernández
a,
João
Baixinho
ac,
José D.
Anastácio
ab,
Katarina
Cankar
 d,
Dirk
Bosch
d and
Cláudia
Nunes dos Santos
*ab
d,
Dirk
Bosch
d and
Cláudia
Nunes dos Santos
*ab
aInstituto de Biologia Experimental e Tecnológica (iBET), Av. República, Qta. Marquês, 2780-157 Oeiras, Portugal
bNOVA Medical School|Faculdade de Ciências Médicas, NMS|FCM, Universidade Nova de Lisboa, Lisboa, Portugal. E-mail: claudia.nunes.santos@nms.unl.pt
cInstituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, 2780-157 Oeiras, Portugal
dWageningen University and Research, Wageningen Plant Research, BU Bioscience, Droevendaalsesteeg 1, 6708PB Wageningen, The Netherlands
First published on 8th August 2022
Abstract
SARS-CoV-2 is a highly transmissible and pathogenic coronavirus causing a respiratory disease that emerged in 2019, leading to a public health emergency situation which continues to date. The treatment options are still very limited and vaccines available are less effective against new variants. SARS-CoV-2 enzymes, namely main protease (Mpro) and papain-like protease (PLpro), play a pivotal role in the viral life cycle, making them a putative drug target. Here, we described for the first time the potential inhibitory activity of chicory extract against both proteases. Besides, we have identified that the four most abundant sesquiterpene lactones in chicory inhibited these proteases, showing an effective binding in the active sites of Mpro and PLpro. This paper provides new insight for further drug development or food-based strategies for the prevention of SARS-CoV-2 by targeting viral proteases.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was named coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO).1 This respiratory virus has caused a severe situation worldwide and is causing lasting negative effects in both the adult and children populations.2,3 SARS-CoV-2 belongs to a family of enveloped positive-strand RNA viruses infecting vertebrates.4 Its genome consists of 14 functional open reading frames (ORFs), which include the regions encoding for non-structural proteins (NSPs), and also accessory and structural proteins.5 When this virus infects the cells of the host, ORF1a and ORF1b are translated through a ribosomal frameshift to the polyproteins pp1a and pp1ab, respectively.6 Then, NSPs are formed by the action of two viral proteases, namely main protease (Mpro), which is also known as chymotrypsin-like protease 3CLpro, and a papain-like protease (PLpro). Lastly, NSPs with a few host factors are the main players for viral genome replication and transcription of the virus into the cells. Currently, vaccines are available to prevent the expansion of this disease, although they do not protect completely, and their efficacy seems to be strain dependent. Besides, this severe situation worldwide requires constant monitoring to prevent and control its highly transmissible spread.7 Overall, proteases involved in the viral replication have become an attractive target to search for bioactive compounds or drugs that limit the SARS-CoV-2 replication.8–10In the early 2000s, SARS-CoV caused a huge outbreak leading to the screening of a large variety of molecules and natural products for the inhibition of the SARS-CoV Mpro activity.11 One example was the identification of PF-00835231 as a potent inhibitor of recombinant Mpro of SARS-CoV-1, which has been also described as specific inhibitor of the SARS-CoV-2 Mpro activity.12 This finding has served as a basis to find out new effective molecules (with similar structures) to inhibit SARS-CoV Mpro.13 Moreover, the additive/synergistic activity of this Mpro inhibitor in combination with remdesivir has been proven.13 While some protease inhibitors for COVID-19 have been authorized by the Food and Drug Administration (FDA) in the condition of Emergency Use Authorization (EUA), remdesivir is the unique drug against SARS-CoV-2 that can act as a Mpro inhibitor and has been approved for the treatment of COVID-19 so far.14 The urgent need to tackle the replication of SARS-CoV-2 has given rise to numerous investigations using FDA-approved protease inhibitors against other viruses. This fact led to the discovery of the inhibitory activity of GC376 towards SARS-CoV-2 Mpro.15,16 In addition to the identification of previous protease inhibitors, different plant extracts were identified as the inhibitors of the SARS-CoV Mpro.17,18 A similar research strategy was followed related to PLpro, which has been established as the other potentially targetable protease for antiviral drug design and the inhibitor of SARS-CoV-2.19 The PLpro enzyme is essential for viral replication and several inhibitors have been effective as therapeutic options against SARS-CoV infections.20 Although PLpro is a less amenable drug target, GRL0617 and different analogues, which are in the FDA-approved drug library,21 have shown to be SARS-CoV-2 PLpro inhibitors binding differently to the enzyme.22
During the last few decades, there has been growing interest in biologically active compounds from natural sources to prevent the onset of various diseases. This situation has led to an increased demand from the population, especially for natural dietary supplements. In this context, root chicory (Cichorium intybus L.) is potentially a rich source of bioactive substances for human health.23 In addition to the already commercially used dietary fibers (i.e. inulin), chicory roots also accumulate large quantities of sesquiterpene lactones (STLs). This class of natural compounds is being increasingly studied due to the large number of biological activities they demonstrate.24–27 Amongst their biological activities, STLs have exhibited activity against viruses such as the human immunodeficiency virus (HIV), hepatitis virus (HV), and influenza A virus (IAV).28–30
An essential feature that has been documented for STLs is how the specific structural elements (i.e. α-methylene-γ-lactone rings) can influence their bioactivity. However, the structure of STLs contains more than one reactive centre, which may have the capability of interacting with a broader variety of biological targets.23
Thus, in the present study, we aim to explore the in vitro potential of chicory and its STLs against the SARS-CoV-2 Mpro and PLpro, as the first step towards the development of novel natural therapeutic options for the future control of coronavirus infections. Moreover, a molecular docking analysis was performed to address the binding affinity of different STLs from chicory against both proteases.
Materials and methods
Materials
Lactucin, lactucopicrin, 11β-13-dihydrolactucin and 11β-13-dihydrolactucopicrin were acquired from Extrasynthese (Genay Cedex, France). The reagents used for extractions and purification steps and chromatographic analysis were dichloromethane (Honeywell, Riedel-de Haën, Germany), dimethyl sulfoxide (DMSO) (≥99.9%, Sigma-Aldrich, St Louis, MO, USA), ethanol LC-MS grade (Merck, Saint Louis, MO, USA), ethyl acetate (99.98%, Fisher Scientific U.K. Limited, Loughborough, UK), methanol (99.8%, Fisher Scientific U.K. Limited, Loughborough, UK), and ultrapure water purified with a Milli-Q water purification system (Merck Millipore, Billerica, MA, USA).Supercritical CO2 extractions and purification of fractions
Supercritical fluid extract (SFE) and the different fractions of SFE were extracted from Cichorium intybus L. Roots as we previously reported.31 Briefly, supercritical CO2 extraction (350 bar, 40 °C, 10% ethanol) was carried out to extract SL rich extracts from freeze-dried and milled chicory roots. The extract was further fractionated by flash column chromatography (stationary phase: silica gel 60; mobile phase: dichloromethane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol 9.5
methanol 9.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, v
0.5, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v). Five fractions (F1, F2, F3, F4 and F5) were collected, and the solvent was evaporated under reduced pressure, for further analysis.
v). Five fractions (F1, F2, F3, F4 and F5) were collected, and the solvent was evaporated under reduced pressure, for further analysis.
Characterization of SFE and SFE fractions by LC-MS
SFE and different fractions of SFE were diluted in DMSO to a concentration of 10 mg mL−1. Next a 100× dilution was prepared in the assay buffer. The diluted samples were mixed with methanol containing formic acid (FA, 0.1%) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 ratio. An UltiMate 3000 U-HPLC system (Dionex, Sunnyvale, CA, USA) was used to create a 45 min linear gradient of 5–75% acetonitrile (v/v) in 0.1% FA in water at a flow rate of 0.19 ml min−1, which was followed by 15 min of washing and equilibration. Of each extract, 5 μL was injected and compounds were separated on a Luna C18 column (2.0 × 150 mm, 3 μm; Phenomenex) at 40 °C. A Q Exactive plus-Orbitrap FTMS mass spectrometer (Thermo Fisher Scientific), operating at a resolution of 35
7 ratio. An UltiMate 3000 U-HPLC system (Dionex, Sunnyvale, CA, USA) was used to create a 45 min linear gradient of 5–75% acetonitrile (v/v) in 0.1% FA in water at a flow rate of 0.19 ml min−1, which was followed by 15 min of washing and equilibration. Of each extract, 5 μL was injected and compounds were separated on a Luna C18 column (2.0 × 150 mm, 3 μm; Phenomenex) at 40 °C. A Q Exactive plus-Orbitrap FTMS mass spectrometer (Thermo Fisher Scientific), operating at a resolution of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with positive electrospray ionization (ESI) mode over the m/z range 90–1350, was used to detect eluting compounds. The compounds were identified based on the accurate mass and where possible by comparison with commercial standards (lactucin, lactucopicrin, 11β-13-dihydrolactucin and 11β-13-dihydrolactucopicrin). 8-Deoxylactucin and 11β-13-dihydro-8-deoxylactucin were identified tentatively based on the accurate mass, retention time and MSn analysis.
000 with positive electrospray ionization (ESI) mode over the m/z range 90–1350, was used to detect eluting compounds. The compounds were identified based on the accurate mass and where possible by comparison with commercial standards (lactucin, lactucopicrin, 11β-13-dihydrolactucin and 11β-13-dihydrolactucopicrin). 8-Deoxylactucin and 11β-13-dihydro-8-deoxylactucin were identified tentatively based on the accurate mass, retention time and MSn analysis.
Docking analysis
To understand the ligand–target interactions with the protease Mpro and PLpro we used mcule.com, which is an online drug discovery platform server (mcule, Inc., Palo Alto, CA 94301, USA).32 This platform provides a collection of online drugs and discovery tools with well-curated databases of chemicals. The ‘InChIKey’ of each one of the explored compounds was retrieved from the PubChem database, and then brought to the ‘property calculator’ of the mcule platform. Upon defining the binding site of the proteases using the mcule platform, this information was exported in a file (.pdb). The molecular interactions and images of the docking were generated in Biovia Discovery Studio 2021.The crystal 3D structures of SARS-CoV-2 Mpro (PDB ID 6WQF) and SARS-CoV-2 PLpro (PDB ID 6W9C) were downloaded from the RCSB Protein DataBank.33 During the screening, the predicted 3D structures of each one of the molecules are fitted into the binding site of the modelled 3D target structure. The critical interactions of the molecules with the target are predicted based on better (more negative) docking scores and thus ranked higher. Additionally, properties that would make the studied compounds potential drugs for humans (Lipinski's rule of five) were evaluated.34
In vitro inhibition assays
SFE from chicory, the different fractions from SFE and the commercially pure STLs (lactucin, lactucopicrin, 11β-13-dihydrolactucin and 11β-13-dihydrolactucopicrin) were dissolved in DMSO at 5 times higher concentration than the final concentration tested, respectively. 3CL protease (Mpro) (SARS-CoV-2) and papain-like protease (PLpro) assay kits (BPS Bioscience, San Diego, CA, USA) were used to test the inhibitory activity against both proteases, respectively. The steps of the in vitro assays were followed according to the manufacturer's protocol. In brief, each reaction was completed with 50 μL into black 96-well plates. The final concentration in each reaction solution contained 3 ng μL1 of recombinant Mpro or 0.3 ng μL−1 of recombinant PLpro, 25 μM fluorogenic substrate, and SFE, F1, F2, F3, F4 or F5 (individually at 100 μg mL−1), or each individual STLs at the same concentrations they were found in 100 μg mL−1 of its corresponding fraction (11β,13-dihydrolactucopicrin at 7 μM; lactucopicrin at 85 μM; 11β,13-dihydrolactucin 126 μM and lactucin 174 μM).The selective inhibitors GC376 (100 μM) and GRL0617 (100 μM) were used as positive controls inhibiting Mpro and PLpro, respectively. The reaction mixtures were incubated according to the manufacturers’ recommended procedures. The fluorescence intensity of each reaction was recorded in a Synergy HT microplate reader (BioTek Instruments, Winooski, VT, USA). The excitation wavelength was 360 nm, and the detection emission wavelength was 460 nm. Each concentration of the compounds and extracts was tested in duplicate and in three different experiments. The fluorescence intensities of each reaction were converted to percentages and normalized to the control (CT) (fluorogenic substrate + recombinant protease). Differences between the controls and the treatments were tested using one-way ANOVA with Dunnett's multiple comparison. Statistically significant differences were considered at p < 0.05.
Results and discussion
Inhibitory effects of SFE against Mpro
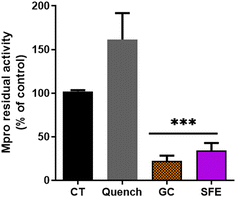
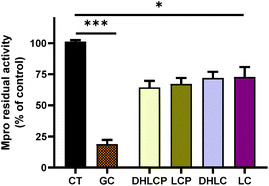
The supercritical fluid extract (SFE) containing chicory sesquiterpene lactones (STLs) was prepared as described in Materials and methods.31 Firstly, the impact of 100 μg mL−1 of the SFE from chicory against the SARS-CoV-2 Mpro was determined (Fig. 1). A significant decrease in Mpro activity was found with SFE compared to the control (p < 0.01). The compound used as the inhibitor (GC376) reduced Mpro residual activity by 20% (p < 0.01).A similar recent study has shown the potential in vitro inhibition of different plant extracts against SARS-CoV-2 Mpro.35 Guijarro et al. reported that among the 17 plant extracts tested, 7 showed a low inhibitory effect (residual Mpro activity between 72.4 and 82.8%), 5 displayed intermediate inhibitory capacity (35–55% of residual activity) and 3 extracts were found showing the highest inhibitory capacity (14.9–0%), at the concentration of 500 μg mL−1. Here, we reported that the residual Mpro activity was about 34% using 100 μg mL−1 of chicory SFE.
Several studies have focused on the research of natural extracts exhibiting antiviral activities both in silico and in vitro against Mpro, where polyphenols are particularly noteworthy.36–38 However, there is only one study that has evaluated the inhibitory activity of SARS-CoV-2 Mpro using a natural extract containing STLs.39
Dogan et al. evaluated 21 Artemisia annua L. extracts with the artemisinin content between 0.062 and 0.066%. In this study, 2 extracts out of 21 analysed reduced the activity of Mpro over 50% at 100 μM. Nonetheless, this effect could be even higher due to the autofluorescence observed with the extracts.39
Since the fluorescence quenching can be a common phenomenon with natural extracts,40,41 we employed a quenching control (Quench, Fig. 1). We ascertained that the SFE did not decrease the fluorescence quantum yield of the fluorophore by decreasing the fluorescence intensity per se. Interestingly, the quenching control even showed an increase of the fluorescence (Fig. 1) indicating that the inhibitory capacity of SFE against Mpro is underestimated. In this way, our results are in agreement with the autofluorescence observations described for Artemisia annua L. extracts.39
Characterization of SFE fractions by LC-MS
Next, the SFE extract was fractionated by flash column chromatography, leading to five fractions (F1–F5) that were analysed by LC-MS (Fig. 2). Some of the major chicory STLs could be identified in fractions F2, F3 and F4, while in fractions F1 and F5 only minor compounds are present and remained unknown (Table 1).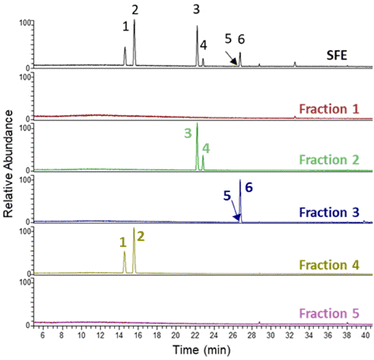 | ||
| Fig. 2 LC-MS total MS chromatogram of the detected STLs in the SFE and the five different fractions isolated from SFE. Peaks 1–6 correspond to Table 1. | ||
| Peak no. | RT (min) | Compound | [M + H]+ | Fraction |
|---|---|---|---|---|
| Peak no. corresponds to the compounds shown in Fig. 2. | ||||
| 1 | 14.60 | 11β-13-Dihydrolactucin | 279.1227 | F4 |
| 2 | 15.58 | Lactucin | 277.1071 | F4 |
| 3 | 22.20 | 8-Deoxylactucin | 261.1121 | F2 |
| 4 | 22.80 | 11β-13-Dihydro-8-deoxylactucin | 263.1279 | F2 |
| 5 | 26.39 | 11β,13-Dihydrolactucopicrin | 413.1595 | F3 |
| 6 | 26.78 | Lactucopicrin | 411.1438 | F3 |
The fractions were initially diluted in DMSO at a concentration of 10 mg mL−1. The fractions were diluted in the assay buffer and the STLs were identified by LC-MS and quantified by comparing with the standard curves made with authentic standards. The amount of lactucin and 11β,13-dihydrolactucin in F4 was found to be 4.8 mg mL−1 and 3.5 mg mL−1 in the DMSO solution, respectively. F3 contained 3.5 mg mL−1 of lactucopicrin and 0.3 mg mL−1 11β,13-dihydrolactucopicrin. The amount of 8-deoxylactucin and 11β-13-dihydro-8-deoxylactucin in F2 could not be quantified since no standards are available for these compounds.
Inhibitory effects of fractions isolated from SFE against Mpro and PLpro
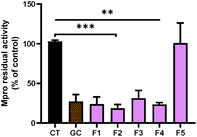
After the fractionation of SFE, we decided to explore the potential of the purified fractions from the SFE of chicory to inhibit Mpro and PLpro. In general, the individual fractions used at the final concentration of 100 μg mL−1 showed a significant inhibitory effect of Mpro (Fig. 3) except for F5, which did not exert any effect (Fig. 3).The most effective fraction was F2, exhibiting about 82% decrease of the Mpro activity compared with the control (CT) (p < 0.001). The rest of the fractions, F1, F3 and F4, showed a significant reduction (p < 0.01) of 76%, 69% and 77% compared with the CT, respectively. Remarkably, all of the fractions with inhibitory effects were more effective against Mpro than the SFE.
Moreover, our results showed that the inhibitory activity in fractions F1 to F4 was even higher compared with the most potent Mpro inhibitor (GC) known (Fig. 3). As we previously reported, using high pressure technologies a more selective and effective recovery of high-value compounds from chicory can be achieved, enabling an increase of human health-promoting applications of natural extracts.31 While in F2, F3 and F4, different STLs could be identified (Fig. 2), in F1, no compounds were identified. Remarkably, F1 showed similar inhibitory activity to F2. The effects observed with the F1 might be related with other minor STLs not detected in the LC-MS analysis due to low concentrations, or even because of the presence of other classes of compounds present in chicory roots (e.g., chlorogenic acids).
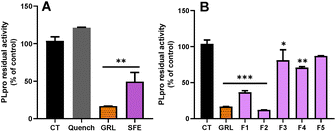
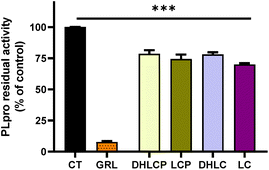
In order to explore the inhibitory activity towards the papain-like protease (PLpro), we tested both SFE and its different fractions, against PLpro. This enzyme also plays a key role in the replication of SARS-CoV-2. Targeting this protease has proven successful for designing inhibitors for older coronaviruses.42 As shown in Fig. 4A, the SFE exhibited a significant reduction of the PLpro activity compared with the control (p > 0.01). While the specific inhibitor of PLpro (GRL) reduced the activity of this enzyme to 17%, the reduction exerted for the SFE was over 50% (p > 0.01). Here, we also used the quenching control, and as shown in Fig. 4A, the inhibitory capacity of SFE against PLpro was not due to the quenching phenomenon.
In the case of the individual fractions (Fig. 4B), out of the 5 fractions tested, only F1 and F2 were able to inhibit more than 60% of the PLpro activity (p > 0.001). In contrast, F3 and F4 only inhibited PLpro activity by about 19% and 29%, respectively (Fig. 4B). These results suggest that F3 and F4 exhibit more specific inhibitory activity towards SARS-CoV-2 Mpro.
A recent in vitro study using a green tea (C. sinensis) extract reported a high variability in the inhibition of the SARS-CoV-2 PLpro activity between the crude extract and its 6 different fractions.43
In our case, F2 was the fraction with the most potent inhibitory capacity, even higher than the specific PLpro inhibitor (Fig. 4B), similarly to that observed for the inhibition of Mpro (Fig. 3). Mpro from older coronaviruses is conserved by 96% and shares a very similar 3D structure homology to SARS-CoV-2.44 Besides, Mpro has at least eleven inter-domain sites on the pp1a and pp1ab polyproteins whereas the PLpro of coronavirus cleaves at no less than two sites on the pp1a polyprotein.45 Due to these reasons, the main searches for effective molecules against COVID 19 have focused on the inhibitors of the protease Mpro. However, both Mpro and PLpro are highly conserved in SARS-CoV-2 because mutations in these key proteins are fatal to the virus.46
Our study shows that the inhibitory effects of chicory extracts were higher against Mpro compared with PLpro; however, the SFE of chicory and its fractions may act as dual-target inhibitors of SARS-CoV-2 Mpro/PLpro.
Studies looking for molecules with inhibitory effects against these proteases may represent a basis for the development of prophylactic or therapeutic plant extracts against COVID-19.
In silico docking of STLs into the active sites of Mpro and PLpro
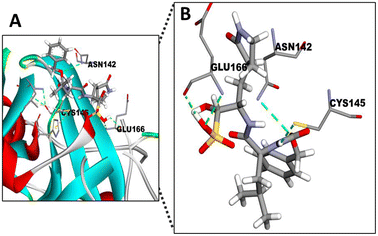
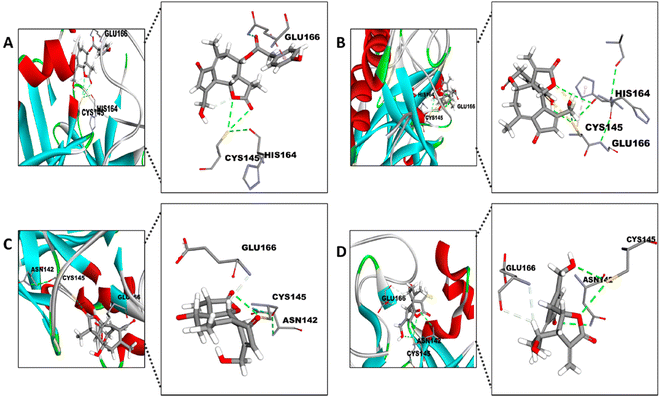
Mpro is a thiol-based protease containing a cysteine145 (CYS145), that is responsible for the activity displayed by this protease cleaving the polypeptide in functional peptides33 and also acts as a nucleophile residue.47 The known inhibitor (GC376) of this protease, which we used in our in vitro assays, binds with a covalent bond to this cysteine.48 It has also been described that several antiviral drugs bind to this specific CYS145 residue of Mpro.49 Hence, we decided to use GC376 as a molecule to verify if the in silico system mcule tool is suitable to test different STLs found in the fractions of SFE from chicory.The docking simulation of Mpro (PDB ID 6WQF) with GC376 using the mcule tool (Fig. 5) showed a good binding affinity for the protease (−6.5 kcal mol−1, Table 2) specifically by binding with the CYS145 residue. Besides, two additional important binding residues (GLU 166 and ASN142) in the active site were identified in the docking study. In agreement with these results, GLU166 has been identified as one of the most important residues to form hydrogen bonds stabilizing the binding of different Mpro inhibitors.50 The other interacting residue identified was ASN142, a polar residue contributing in the active site of Mpro, forming a series of hydrogen bonds involved in the binding of Mpro and small molecule inhibitors.51 In view of these results, we assumed the mcule tool as a good in silico tool to test the affinity of chicory compounds with the SARS-CoV-2 Mpro.
Firstly, the docking analysis was performed in the active site of Mpro with the STLs identified in different fractions from chicory SFE and that are commercially available (Fig. 6; Table 2). Among these STLs, the best binding score was found for 11β-13-dihydrolactucopicrin (PubChem CID: 315492320 and lactucopicrin (PubChem CID: 174880) with a binding affinity value of −7.6 kcal mol−1.
The potential interaction between the CYS145 residue and the different STLs is with their α-methylene-γ-lactone (aMyL) ring, as shown in Fig. 6. In this sense, the aMyL acts as a Michael acceptor, reacting with nucleophile groups such as sulfhydryl groups (R-SH) in enzymes and other proteins.52 The interaction between the R-SH from the CYS145 and aMyL may be, at least in part, responsible for the activity of STLs against SARS-CoV-2 Mpro, since this aMyL group has been described to exhibit the major centre reactivity that influences the activity of STLs.23,52 So far, one STL, lactucopicrin 15-oxalate, has been described from in silico data to exhibit an interaction with CYS145 of Mpro through van der Waals forces.53 Besides the interaction with CYS145, the ligand binding site showed other opportunities to improve the potential linking of the STLs between their aMyL group and the other residues, like HIS164 in the case of 11β-13-dihydrolactucopicrin and lactucopicrin (Fig. 6A and B), and ASN142 with lactucin and 11β-13-dihydrolactucin (Fig. 6C and D). This is in accordance with previous structure–activity studies which also described that the aMyL moiety of STLs can interact with the sulfhydryl, hydroxyl or amine groups through the epoxide ring.54 In fact, the strategy of adding an amine group to the aMyL moiety increased the solubility and selectivity of different STLs.55 Moreover, all STLs displayed a potential interaction with GLU166 by the virtue of hydrogen bonds to this residue of the active site. As discussed above, GLU166 has the ability to form hydrogen bonds stabilizing the binding of different inhibitors of Mpro.50
Moreover, we noticed that the STLs with the best binding affinity have molecular weights in the range 410–482 (Table 2), similar to the GC376 inhibitor, which has a molecular weight equal to 507.5 g mol−1. The potential reason to explain this fact may be because the size of the peptide substrate and the deeply buried protease active site demand a large molecule with many rotatable dynamics to fit into it.49
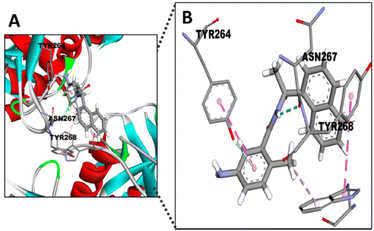
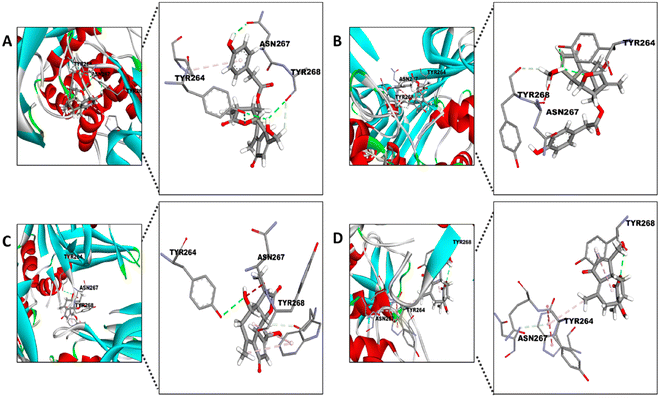
We have also conducted molecular docking with the four STLs towards SARS-CoV-2 PLpro (PDB ID 6W9C). PLpro is structurally different from Mpro. PLpro has a ubiquitin-specific protease (USP) fold, typical of the family in humans, which is topologically organized into four domains – UBL, thumb, palm, and fingers.19 In order to verify this docking, we used GRL0617, which is the most potent known inhibitor for PLpro.56 This naphthalene-based molecule interacts with the TYR268 residue of SARS-CoV-2 PLpro.57 Therefore, the docking simulation was conducted targeting TYR268 of SARS-CoV-2 PLpro. Fig. 7 shows this interaction with GRL0617 and the binding affinity was −7.5 kcal mol−1 (Table 2). Notably, 11β-13-dihydrolactrolactucopicrin and lactucopicrin showed better binding affinity values than the specific PLpro and Mpro inhibitors (Table 2).
Surprisingly, the TYR264 residue, which shows hydrophobic interactions between its ring and the 1-naphthyl group of the inhibitor (Fig. 7),56 seems to display similar binding with the STLs by hydrophobic interactions (Fig. 8). On the other hand, TYR268 seems to be the residue involved in the interaction between the aMyL group of the STLs and the amine group of the residue (Fig. 8A–D). Lastly, the ASN267 residue may construct carbon hydrogen bonds with the STLs, as shown in the docking between darunavir, an antiretroviral drug for HIV, and PLpro.58
According to our docking analysis, these STLs are very promising targets for drug development. With the aim to evaluate if these STLs are safe for human oral consumption and further drug development, the properties of Lipinski's rule of five34 were evaluated for the STLs. Based on the results, all STLs showed Lipinski's violation 0. All molecules complied with a molecular mass less than 500 g mol−1, no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, and an octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) not greater than 5.34
P) not greater than 5.34
However, other measures of drug likeness as the Polar Surface Area (PSA) and rotatable bonds (Rot B) should also be considered, since meeting the rule of five does not guarantee that a molecule is a drug-like.59 As Table 2 shows, PSA was <140 Å2 and less than 10 Rot B were found for all the STLs, thus fulfilling Veber's oral bioavailability rule.59
The chemical inhibitor of SARS-CoV-2 Mpro (GC376) showed Lipinski's violation 2 (Table 2), besides the values PSA = 179.51 Å2 and 14 Rot B. In the case of chemical SARS-CoV-2 PLpro, the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P is violated (Table 2).
P is violated (Table 2).
Bearing in mind that 11β-13-dihydrolactucopicrin, lactucopicrin, 11β-13-dihydrolactucin and lactucin satisfied the drug development principles and their values of binding affinity to each protease were similar to those of the specific inhibitors, respectively, further studies with these STLs should be addressed for drug development inhibiting the activity of SARS-CoV-2 proteases.
Inhibitory effects of the individual STLs against Mpro and PLpro
To ascertain whether the inhibitory effects of chicory SFE and its fractions against the SARS-Cov-2 Mpro and PLpro could be mediated by individual compounds, we performed enzymatic inhibition assays against both proteases using each individual SL at the same concentrations which were found in the tested fractions.In agreement with the binding affinity values obtained from the docking analysis against Mpro, the compounds with the highest effect were 11β,13-dihydrolactucopicrin and lactucopicrin. The residual activity of Mpro was 67 and 64% compared with the control, respectively. Regarding 11β,13-dihydrolactucin and lactucin, the residual activities of the protease were 72 and 73%, respectively. In all of the cases, the inhibition of SARS-CoV-2 Mpro activity with each STL was statistically significant compared with the control (p > 0.05) (Fig. 9). In contrast to the effects observed with F3 and F4, which contain these STLs, the inhibitory effects by individual STLs were 2-fold lower in the case of 11β,13-dihydrolactucopicrin and lactucopicrin than with its respective fraction (F3). Regarding 11β,13-dihydrolactucin and lactucin, the observed inhibitory effects were 3-fold lower compared to the corresponding fraction (F4). The lower activity against the Mpro of individual natural compounds versus the inhibitory capacity of the fractions from extracts where they are present has also been reported with turmeric rhizomes, mustard seeds and wall rocket extracts.35 Whereas the turmeric rhizomes extract reduced the activity of Mpro to 0%, curcumin (the major compound in turmeric) at the same concentration found in the assayed turmeric, only displayed 20% of inhibition.35 In the case of sinigrin, the major compound in mustard seeds and wall rocket extracts didn't exert any inhibitory effect against Mpro whilst mustard seeds and wall rocket extracts reduced the Mpro activity over 80%. Tito et al., 2021 reported similar findings, while the pomegranate peel extract significantly reduced the SARS-CoV-2 Mpro activity, the main compounds present in the extract (gallic and ellagic acids) exerted much less or no effect at all.38 According to these previous studies, our results suggest that other compounds present in chicory must also play a key role in inhibiting Mpro activity. Besides, the mixture of 2 STLs in each fraction (F3 and F4) may have synergistic effects potentiating the protease inhibition.
Studies assessing the in vitro inhibitory activity of STLs specifically against SARS-CoV-2 Mpro are lacking. However, recent works based on molecular docking pointed to artemisinin60 and lactucopicrin 15-oxalate53 as natural STLs displaying high capacity for binding to the active site of SARS-CoV-2 Mpro. The potential activity of artemisinins has been extensively explored.61 In human trials it was shown to reduce the time of COVID-19 infection62 and in in vitro studies this SL and its derivatives showed different mechanisms of action towards SARS-CoV-2.63
In contrast, many natural compounds have displayed great potential as SARS-CoV Mpro inhibitors such as quercetin, apigenin and luteolin (IC50 of 20–200 μM).64 In this context, our work suggests that the evaluated STLs are active in this range of concentrations. However, the current study used a targeted approach towards the replication of SARS-CoV-2.
Finally, it should be pointed out that we detect an inhibitory activity for 11β-13-dihydrolactucopicrin at a very low concentration (7 μM).
Next, we investigated the effect of the individual STLs against PLpro. The results of the inhibitory capacity against this protease (Fig. 10) revealed that all of the STLs reduced the PLpro activity, ranging between 30% and 23% of inhibitory capacity. In contrast to what was observed in the activity of individual compounds against Mpro, all STLs tested showed similar inhibitory activities towards PLpro as those in the fractions where they were present (Table 1; Fig. 4 and 10). Previous studies with natural compounds, mainly flavonoids, have been conducted for in vitro inhibitory activity against SARS-CoV PLpro, indicating IC50 values in a micromolar range.65 Moreover tanshinones, terpenoid compounds, isolated from Salvia miltiorrhiza, have demonstrated to be very potent PLpro inhibitors (IC50 of 0.8–30 μM depending on the molecule).66 Regarding the studies that are specifically focused on the inhibitory activity of natural compounds against the SARS-CoV-2 PLpro, Montone et al. reported a IC50 value equal to 11.62 ± 0.47 μg mL−1 (about 25 μM) of (−)-epicatechin-3-gallate inhibiting this protease.43 Other compounds with inhibitory activity towards this protease were ginkgolic acid and anacardic acid with IC50 values of 16.30 ± 0.64 μM and 17.08 ± 1.30, respectively.67 In our study, lactucin was the SL with the highest inhibitory activity against SARS-CoV-2 PLpro at the concentration of 174 μM, which is clearly a higher concentration than that reported for other active compounds.43,67 Nonetheless, this is the first screening study using these STLs (11β,13-dihydrolactucopicrin, lactucopicrin, 11β,13-dihydrolactucin and lactucin), so even though these compounds seem to be suitable natural inhibitors to PLpro, further studies are required to confirm their potential.
In general, previous studies that report the effect of compounds against SARS-CoV-2 PLpro are clearly minor as opposed to those targeting the Mpro. This obvious difference can be noticed in the Key Protein Targets and Related Patents in the CAS Content Collection which showed 2178 drug candidates and 49 patents related to Mpro while only 189 candidates and 4 patents to PLpro towards COVID-19.68 This fact could be explained due to several inhibitors described for SARS-CoV Mpro in previous works,68 since SARS-CoV-2 Mpro is conserved in 96% of its sequence.44 In contrast, the PLpro sequence of SARS-CoV-2 and SARS-CoV is only 83% similar, although they share similar active sites.69 Mpro has been described as the most attractive target for drug design against SARS-CoV-2,70,71 but dual-targeted drugs have the advantage of fewer adverse effects and better therapeutic outcomes than single-targeted drugs for this coronavirus.46
Conclusions
To the best of our knowledge, this is the first report showing in vitro the inhibitory potential of chicory extract and its different sesquiterpene lactones against the two main proteases involved in the SARS-CoV-2 replication. Based on the binding energies obtained, this study also contains valuable information to increase the knowledge on certain structural families of compounds with great affinities towards SARS-CoV-2 Mpro and PLpro proteases. Furthermore, these results may provide molecular details that set the basis to further propose structural modifications of some compounds to make the interaction between them and proteins even more effective.Despite all the efforts dedicated to stop the pandemic dissemination of COVID-19, there is no conclusive treatment to date. Our findings provide information that can help in the search for COVID-19 prophylactic or curative treatments, acting as a basis for future in vivo studies.
Author contributions
Conceptualization, C. N. S., D. B., and K. C.; methodology, M. A. A.-G., C. R.-P., N. F., J. B., J. D. A., and K. C.; investigation M. A. A.-G., C. R.-P. and J. D. A.; formal analysis and data curation, M. A. A-G., C. R.-P., N. F., and K. C.; funding acquisition C. N. S., D. B., and K. C.; writing – original draft preparation M. A. A.-G.; writing – review and editing C. N. S., N. F., D. B., and K. C.; visualization, M. A. A.-G. and K. C.; supervision, C. N. S. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research has been supported by the EU Horizon 2020 research & innovation programme (H2020-NMBP-BIO-2017, project CHIC grant agreement no. 760891).References
- C.-C. Lai, T.-P. Shih, W.-C. Ko, H.-J. Tang and P.-R. Hsueh, Int. J. Antimicrob. Agents, 2020, 55, 105924 CrossRef CAS PubMed.
- D. L. Sykes, L. Holdsworth, N. Jawad, P. Gunasekera, A. H. Morice and M. G. Crooks, Lung, 2021, 199, 113–119 CrossRef CAS PubMed.
- M. Irwin, B. Lazarevic, D. Soled and A. Adesman, Curr. Opin. Pediatr., 2022, 34, 107–115 CrossRef CAS PubMed.
- P. S. Masters, Adv. Virus Res., 2006, 66, 193–292 CrossRef CAS PubMed.
- C. Bai, Q. Zhong and G. F. Gao, Sci. China: Life Sci., 2021, 1–15 CAS.
- R. Arya, S. Kumari, B. Pandey, H. Mistry, S. C. Bihani, A. Das, V. Prashar, G. D. Gupta, L. Panicker and M. Kumar, J. Mol. Biol., 2021, 433, 166725 CrossRef CAS PubMed.
- V. Borges, J. Isidro, N. S. Trovão, S. Duarte, H. Cortes-Martins, H. Martiniano, I. Gordo, R. Leite, L. Vieira, A. J. S. Lira, A. M. Sousa Fernandes, A. Estrada, A. Nunes, A. Rodrigues, A. Caldas, A. Constança, A. M. Henriques, A. M. Matos, A. Oliveira, A. P. Dias, A. Pelerito, A. R. Couto, A. Vilares, A. Albuquerque, B. Nunes, B. R. Gouveia, C. de Fátima Rodrigues, C. Feliciano, C. Roque, C. Cardoso, C. Sousa, C. Paulino, C. R. Bettencourt, C. C. Branco, C. N. dos Santos, C. Godinho, C. P. Caetano, C. Correia, C. Toscano, C. Veríssimo, D. Silva, D. P. P. da Silva, E. Costa, E. Pádua, F. Martins, F. Vale, F. Vilarinho, F. Branca, F. Caldeira, F. Lacerda, F. Rocha, G. Andrade, H. Ribeiro, H. Rodrigues, H. Jesus, H. Sousa, I. Ferreira, I. Baldaque, I. Costa, I. Gomes, I. Slobidnyk, I. Albergaria, I. Dias, I. Fernandes, I. L. de Carvalho, I. Água-Doce, J. B. Armas, J. Ramos, J. C. Sousa, J. Costa, J. Dias, J. Rodrigues, J. Sobral, J. Machado, J. Meneses, J. Alves, J. V. Constantino, L. Brum, L. Silveira, L. Zé-Zé, L. Santos, L. Freitas, L. Silva, L. Mota-Vieira, L. Lopes, L. Monteiro, M. Faria, M. Farinha, M. Vaz, M. A. Pinto, M. A. Pessanha, M. B. Tomaz, M. C. Vellés, M. da G. Maciel de Soveral, M. H. Ramos, M. I. Veiga, M. J. Gargate, M. J. Peres, M. J. Borrego, M. M. Figueiredo, M. Martins, M. Viana, M. Melim, M. B. Jorreto, M. Fevereiro, M. Pinheiro, M. Oleastro, N. Seixas, N. Ventura, N. Verdasca, O. Costa, P. Barros, P. Fonseca, P. Miguel, P. Bajanca-Lavado, P. Branquinho, P. Palminha, P. Soares, P. Valente, P. Leandro, P. Pereira, P. Cardoso, P. Pechirra, P. Ramos, R. Neves, R. Rocha, R. Rodrigues, R. Sabino, R. Sá, R. F. R. Ferreira, R. Rodrigues, R. C. Veloso, R. Cordeiro, R. Côrte-Real, R. de Sousa, R. Gralha, R. Macedo, R. Matos, R. Rodrigues, S. Paulo, S. Sousa, S. Lopo, S. M. S. Magalhães, S. Rodrigues, S. Silva, S. Ladeiro, S. Martins, S. Silva, T. Salvado, T. Luís, V. Alves, V. Manageiro, R. Guiomar, J. P. Gomes and P. network for S.-C.-2 genomics (Consortium), Commun. Med., 2022, 2, 10 CrossRef PubMed.
- D. J. Calleja, G. Lessene and D. Komander, Front. Chem., 2022, 10, 876212 CrossRef CAS PubMed.
- I. Antonopoulou, E. Sapountzaki, U. Rova and P. Christakopoulos, Comput. Struct. Biotechnol. J., 2022, 20, 1306–1344 CrossRef CAS PubMed.
- H. Jiang, P. Yang and J. Zhang, Front. Chem., 2022, 10, 822785 CrossRef CAS PubMed.
- J. E. Blanchard, N. H. Elowe, C. Huitema, P. D. Fortin, J. D. Cechetto, L. D. Eltis and E. D. Brown, Chem. Biol., 2004, 11, 1445–1453 CrossRef CAS PubMed.
- D. R. Owen, C. M. N. Allerton, A. S. Anderson, L. Aschenbrenner, M. Avery, S. Berritt, B. Boras, R. D. Cardin, A. Carlo, K. J. Coffman, A. Dantonio, L. Di, H. Eng, R. Ferre, K. S. Gajiwala, S. A. Gibson, S. E. Greasley, B. L. Hurst, E. P. Kadar, A. S. Kalgutkar, J. C. Lee, J. Lee, W. Liu, S. W. Mason, S. Noell, J. J. Novak, R. S. Obach, K. Ogilvie, N. C. Patel, M. Pettersson, D. K. Rai, M. R. Reese, M. F. Sammons, J. G. Sathish, R. S. P. Singh, C. M. Steppan, A. E. Stewart, J. B. Tuttle, L. Updyke, P. R. Verhoest, L. Wei, Q. Yang and Y. Zhu, Science, 2021, 374, 1586–1593 CrossRef CAS PubMed.
- B. Boras, R. M. Jones, B. J. Anson, D. Arenson, L. Aschenbrenner, M. A. Bakowski, N. Beutler, J. Binder, E. Chen, H. Eng, H. Hammond, J. Hammond, R. E. Haupt, R. Hoffman, E. P. Kadar, R. Kania, E. Kimoto, M. G. Kirkpatrick, L. Lanyon, E. K. Lendy, J. R. Lillis, J. Logue, S. A. Luthra, C. Ma, S. W. Mason, M. E. McGrath, S. Noell, R. S. Obach, M. N. O’ Brien, R. O'Connor, K. Ogilvie, D. Owen, M. Pettersson, M. R. Reese, T. F. Rogers, R. Rosales, M. I. Rossulek, J. G. Sathish, N. Shirai, C. Steppan, M. Ticehurst, L. W. Updyke, S. Weston, Y. Zhu, K. M. White, A. García-Sastre, J. Wang, A. K. Chatterjee, A. D. Mesecar, M. B. Frieman, A. S. Anderson and C. Allerton, Nat. Commun., 2021, 12, 6055 CrossRef CAS PubMed.
- Coronavirus (COVID-19) | Drugs. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 11 July 2022).
- C. Ma, M. D. Sacco, B. Hurst, J. A. Townsend, Y. Hu, T. Szeto, X. Zhang, B. Tarbet, M. T. Marty, Y. Chen and J. Wang, Cell Res., 2020, 30, 678–692 CrossRef CAS PubMed.
- W. Vuong, M. B. Khan, C. Fischer, E. Arutyunova, T. Lamer, J. Shields, H. A. Saffran, R. T. McKay, M. J. van Belkum, M. A. Joyce, H. S. Young, D. L. Tyrrell, J. C. Vederas and M. J. Lemieux, Nat. Commun., 2020, 11, 4282 CrossRef CAS PubMed.
- C.-N. Chen, C. P. C. Lin, K.-K. Huang, W.-C. Chen, H.-P. Hsieh, P.-H. Liang and J. T.-A. Hsu, Evidence-Based Complementary Altern. Med., 2005, 2, 607278 Search PubMed.
- C.-W. Lin, F.-J. Tsai, C.-H. Tsai, C.-C. Lai, L. Wan, T.-Y. Ho, C.-C. Hsieh and P.-D. L. Chao, Antiviral Res., 2005, 68, 36–42 CrossRef CAS PubMed.
- J. Osipiuk, S.-A. Azizi, S. Dvorkin, M. Endres, R. Jedrzejczak, K. A. Jones, S. Kang, R. S. Kathayat, Y. Kim, V. G. Lisnyak, S. L. Maki, V. Nicolaescu, C. A. Taylor, C. Tesar, Y.-A. Zhang, Z. Zhou, G. Randall, K. Michalska, S. A. Snyder, B. C. Dickinson and A. Joachimiak, Nat. Commun., 2021, 12, 743 CrossRef CAS PubMed.
- Y. M. Báez-Santos, S. E. st. John and A. D. Mesecar, Antiviral Res., 2015, 115, 21–38 CrossRef PubMed.
- R. Kulandaisamy, T. Kushwaha, A. Dalal, V. Kumar, D. Singh, K. Baswal, P. Sharma, K. Praneeth, P. Jorwal, S. R. Kayampeta, T. Sharma, S. Maddur, M. Kumar, S. Kumar, A. Polamarasetty, A. Singh, D. Sehgal, S. L. Gholap, M. B. Appaiahgari, M. R. Katika and K. K. Inampudi, Front. Microbiol., 2022, 13, 877813 CrossRef PubMed.
- H. Tan, Y. Hu, P. Jadhav, B. Tan and J. Wang, J. Med. Chem., 2022, 65, 7561–7580 CrossRef CAS PubMed.
- M. S. Matos, J. D. Anastácio and C. Nunes dos Santos, Pharmaceutics, 2021, 13, 991 CrossRef CAS PubMed.
- M. Chadwick, H. Trewin, F. Gawthrop and C. Wagstaff, Int. J. Mol. Sci., 2013, 14, 12780–12805 CrossRef PubMed.
- J. Perović, V. Tumbas Šaponjac, J. Kojić, J. Krulj, D. A. Moreno, C. García-Viguera, M. Bodroža-Solarov and N. Ilić, Food Chem., 2021, 336, 127676 CrossRef PubMed.
- M. S. Matos, J. D. Anastácio, J. W. Allwood, D. Carregosa, D. Marques, J. Sungurtas, G. J. McDougall, R. Menezes, A. A. Matias, D. Stewart and C. N. Dos Santos, Nutrients, 2020, 12, 1–20 Search PubMed.
- S. T. Häkkinen, M. Soković, L. Nohynek, A. Ćirić, M. Ivanov, D. Stojković, I. Tsitko, M. Matos, J. P. Baixinho, V. Ivasiv, N. Fernández, C. Nunes dos Santos and K.-M. Oksman-Caldentey, Pharmaceutics, 2021, 14, 941 CrossRef PubMed.
- J.-F. Liu, L. Wang, Y.-F. Wang, X. Song, L.-J. Yang and Y.-B. Zhang, Fitoterapia, 2015, 104, 41–44 CrossRef CAS PubMed.
- H. Yan, M.-Y. Ba, X.-H. Li, J.-M. Guo, X.-J. Qin, L. He, Z.-Q. Zhang, Y. Guo and H.-Y. Liu, Fitoterapia, 2016, 115, 64–68 CrossRef CAS PubMed.
- Y. Wang, B. Zhou, J. Lu, Q. Chen, H. Ti, W. Huang, J. Li, Z. Yang, Z. Jiang and X. Wang, BMC Complementary Altern. Med., 2017, 17, 25 CrossRef PubMed.
- J. P. Baixinho, J. D. Anastácio, V. Ivasiv, K. Cankar, D. Bosch, R. Menezes, M. de Roode, C. N. Dos Santos, A. A. Matias and N. Fernández, Molecules, 2021, 26, 2583 CrossRef CAS PubMed.
- R. Kiss, M. Sandor and F. A. Szalai, J. Cheminf., 2012, 4, P17 Search PubMed.
- D. W. Kneller, G. Phillips, H. M. O'Neill, R. Jedrzejczak, L. Stols, P. Langan, A. Joachimiak, L. Coates and A. Kovalevsky, Nat. Commun., 2020, 11, 3202 CrossRef CAS PubMed.
- C. A. Lipinski, Drug Discovery Today: Technol., 2004, 1, 337–341 CrossRef CAS PubMed.
- C. Guijarro-Real, M. Plazas, A. Rodríguez-Burruezo, J. Prohens and A. Fita, Foods, 2021, 10, 1503 CrossRef CAS PubMed.
- I. Nawrot-Hadzik, M. Zmudzinski, A. Matkowski, R. Preissner, M. Kęsik-Brodacka, J. Hadzik, M. Drag and R. Abel, Pharmaceuticals, 2021, 14, 742 CrossRef CAS PubMed.
- A. M. Sardanelli, C. Isgrò and L. L. Palese, Molecules, 2021, 26, 1409 CrossRef CAS PubMed.
- A. Tito, A. Colantuono, L. Pirone, E. Pedone, D. Intartaglia, G. Giamundo, I. Conte, P. Vitaglione and F. Apone, Front. Chem., 2021, 9, 638187 CrossRef CAS PubMed.
- K. Dogan, E. Erol, M. Didem Orhan, Z. Degirmenci, T. Kan, A. Gungor, B. Yasa, T. Avsar, Y. Cetin, S. Durdagi and M. Guzel, Phytochem. Anal., 2022, 33, 303–319 CrossRef CAS PubMed.
- Y.-M. Kim, F. Abas, Y.-S. Park, Y.-K. Park, K.-S. Ham, S.-G. Kang, M. Lubinska-Szczygeł, A. Ezra and S. Gorinstein, Molecules, 2021, 26, 4405 CrossRef CAS PubMed.
- L. Xu, L. Zhang, Y.-H. Li, L.-Y. Li, Z.-H. Xie and Z.-C. Tu, J. Food Biochem., 2022, e14101 CAS.
- Y. K. Bosken, T. Cholko, Y.-C. Lou, K.-P. Wu and C.-E. A. Chang, Front. Mol. Biosci., 2020, 7, 174 CrossRef CAS PubMed.
- C. M. Montone, S. E. Aita, A. Arnoldi, A. L. Capriotti, C. Cavaliere, A. Cerrato, C. Lammi, S. Piovesana, G. Ranaldi and A. Laganà, Molecules, 2021, 26, 6744 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- B. Krichel, S. Falke, R. Hilgenfeld, L. Redecke and C. Uetrecht, Biochem. J., 2020, 477, 1009–1019 CrossRef CAS PubMed.
- L.-C. Zhang, H.-L. Zhao, J. Liu, L. He, R.-L. Yu and C.-M. Kang, Future Med. Chem., 2022, 14, 393–405 CrossRef CAS PubMed.
- M. Mandal, S. K. Chowdhury, A. A. Khan, N. Baildya, T. Dutta, D. Misra and N. N. Ghosh, J. Mol. Struct., 2021, 1234, 130152 CrossRef CAS PubMed.
- H. M. Mengist, T. Dilnessa and T. Jin, Front. Chem., 2021, 9, 622898 CrossRef CAS PubMed.
- Y. W. Chen, C.-P. B. Yiu and K.-Y. Wong, F1000Research, 2020, 9, 129 CAS.
- D. D. Nguyen, K. Gao, J. Chen, R. Wang and G.-W. Wei, Chem. Sci., 2020, 11, 12036–12046 RSC.
- A. Narayanan, M. Narwal, S. A. Majowicz, C. Varricchio, S. A. Toner, C. Ballatore, A. Brancale, K. S. Murakami and J. Jose, Commun. Biol., 2022, 5, 169 CrossRef CAS PubMed.
- M. H. R. Amorim, R. M. Gil da Costa, C. Lopes and M. M. Bastos, Crit. Rev. Toxicol., 2013, 43, 559–579 CrossRef CAS PubMed.
- R. S. Joshi, S. S. Jagdale, S. B. Bansode, S. S. Shankar, M. B. Tellis, V. K. Pandya, A. Chugh, A. P. Giri and M. J. Kulkarni, J. Biomol. Struct. Dyn., 2021, 39, 3099–3114 CAS.
- A. Paço, T. Brás, J. O. Santos, P. Sampaio, A. C. Gomes and M. F. Duarte, Molecules, 2022, 27, 1142 CrossRef PubMed.
- J. R. Woods, H. Mo, A. A. Bieberich, T. Alavanja and D. A. Colby, MedChemComm, 2013, 4, 27–33 RSC.
- P. Rao, R. Patel, A. Shukla, P. Parmar, R. M. Rawal, M. Saraf and D. Goswami, Mol. Divers., 2022, 26, 309–329 CrossRef CAS PubMed.
- D. Shin, R. Mukherjee, D. Grewe, D. Bojkova, K. Baek, A. Bhattacharya, L. Schulz, M. Widera, A. R. Mehdipour, G. Tascher, P. P. Geurink, A. Wilhelm, G. J. van der Heden van Noort, H. Ovaa, S. Müller, K.-P. Knobeloch, K. Rajalingam, B. A. Schulman, J. Cinatl, G. Hummer, S. Ciesek and I. Dikic, Nature, 2020, 587, 657–662 CrossRef CAS PubMed.
- D. Li, J. Luan and L. Zhang, Biochem. Biophys. Res. Commun., 2021, 538, 72–79 CrossRef CAS PubMed.
- D. F. Veber, S. R. Johnson, H.-Y. Cheng, B. R. Smith, K. W. Ward and K. D. Kopple, J. Med. Chem., 2002, 45, 2615–2623 CrossRef CAS PubMed.
- R. Badraoui, M. Saoudi, W. S. Hamadou, S. Elkahoui, A. J. Siddiqui, J. M. Alam, A. Jamal, M. Adnan, A. M. E. Suliemen, M. M. Alreshidi, D. K. Yadav, H. Naïli and H. Ben-Nasr, Pharmaceuticals, 2022, 15, 129 CrossRef CAS PubMed.
- K. Farmanpour-Kalalagh, A. Beyraghdar Kashkooli, A. Babaei, A. Rezaei and A. R. van der Krol, Front. Plant Sci., 2022, 13, 780257 CrossRef PubMed.
- G. Li, M. Yuan, H. Li, C. Deng, Q. Wang, Y. Tang, H. Zhang, W. Yu, Q. Xu, Y. Zou, Y. Yuan, J. Guo, C. Jin, X. Guan, F. Xie and J. Song, Int. J. Antimicrob. Agents, 2021, 57, 106216 CrossRef CAS PubMed.
- R. Cao, H. Hu, Y. Li, X. Wang, M. Xu, J. Liu, H. Zhang, Y. Yan, L. Zhao, W. Li, T. Zhang, D. Xiao, X. Guo, Y. Li, J. Yang, Z. Hu, M. Wang and W. Zhong, ACS Infect. Dis., 2020, 6, 2524–2531 CrossRef CAS PubMed.
- S. S. Ebada, N. A. Al-Jawabri, F. S. Youssef, D. H. El-Kashef, T.-O. Knedel, A. Albohy, M. Korinek, T.-L. Hwang, B.-H. Chen, G.-H. Lin, C.-Y. Lin, S. M. Aldalaien, A. M. Disi, C. Janiak and P. Proksch, RSC Adv., 2020, 10, 38128–38141 RSC.
- A. M. Sayed, A. R. Khattab, A. M. AboulMagd, H. M. Hassan, M. E. Rateb, H. Zaid and U. R. Abdelmohsen, RSC Adv., 2020, 10, 19790–19802 RSC.
- J.-Y. Park, J. H. Kim, Y. M. Kim, H. J. Jeong, D. W. Kim, K. H. Park, H.-J. Kwon, S.-J. Park, W. S. Lee and Y. B. Ryu, Bioorg. Med. Chem., 2012, 20, 5928–5935 CrossRef CAS PubMed.
- Z. Chen, Q. Cui, L. Cooper, P. Zhang, H. Lee, Z. Chen, Y. Wang, X. Liu, L. Rong and R. Du, Cell Biosci., 2021, 11, 45 CrossRef CAS PubMed.
- C. Liu, Q. Zhou, Y. Li, L. V. Garner, S. P. Watkins, L. J. Carter, J. Smoot, A. C. Gregg, A. D. Daniels, S. Jervey and D. Albaiu, ACS Cent. Sci., 2020, 6, 315–331 CrossRef CAS PubMed.
- J. S. Morse, T. Lalonde, S. Xu and W. R. Liu, ChemBioChem, 2020, 21, 730–738 CrossRef CAS PubMed.
- B. Luan, T. Huynh, X. Cheng, G. Lan and H.-R. Wang, J. Proteome Res., 2020, 19, 4316–4326 CrossRef CAS PubMed.
- N. Sepay, A. Sekar, U. C. Halder, A. Alarifi and M. Afzal, J. Mol. Struct., 2021, 1228, 129433 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |