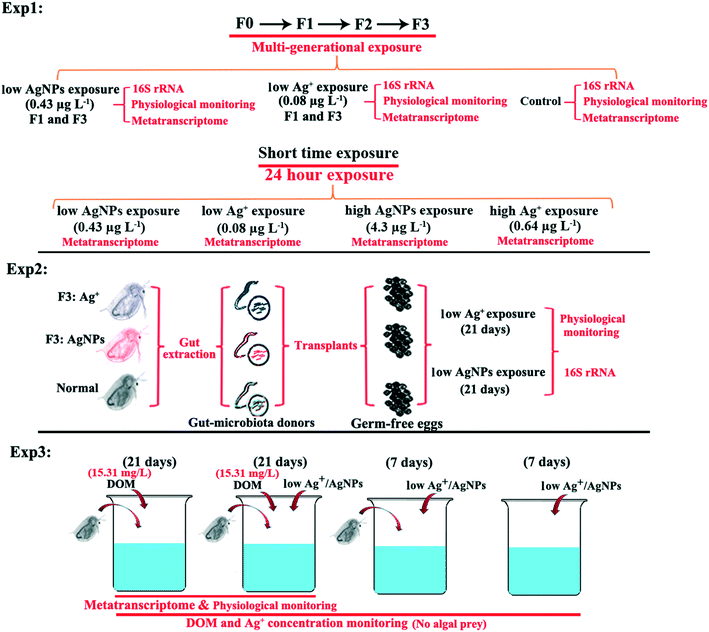
Open Access Article
Open Access ArticleGut-microbial adaptation and transformation of silver nanoparticles mediated the detoxification of Daphnia magna and their offspring†
Yingdong
Li
 ab,
Wen-Xiong
Wang
ab,
Wen-Xiong
Wang
 *c and
Hongbin
Liu
*abcd
*c and
Hongbin
Liu
*abcd
aDepartment of Ocean Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China. E-mail: liuhb@ust.hk; ylifc@connect.ust.hk
bSouthern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
cSchool of Energy and Environment, State Key Laboratory of Marine Pollution, Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory, City University of Hong Kong, Kowloon, Hong Kong, China. E-mail: wx.wang@cityu.edu.hk
dHong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory, The Hong Kong University of Science and Technology, Hong Kong, China
First published on 7th December 2021
Abstract
Despite extensive studies on the toxicity of antibacterial silver (either ionic Ag+ or nanoparticles – AgNPs) at the cellular or organism level, little is known about the differences in toxicity at the community level, especially regarding the gut microbiota. In the present study, we applied 16S rRNA sequencing, metatranscriptome sequencing and gut microbiota transplants (GMTs) to investigate the gut-microbial adaptation and transformation of different silver (Ag) forms over 4 generations of exposure of Daphnia magna to environmentally relevant concentrations of Ag+ and AgNPs. Our results demonstrated that the gut-accumulated Ag+ and AgNPs were transformed by gut microbial sulfidation, which subsequently affected the toxic symptoms of D. magna. Multi-generational exposure revealed that selection of the toxicity-adapted gut microbiota was both Ag form- and exposure time- dependent, resulting in a distinctive gut-microbial community between generations and treatments. Specifically, the expression of gut microbial genes for sulfide synthesis and organic matter degradation was simultaneously expressed when encountering Ag challenge and was positively correlated with the reduced toxic symptoms. The reciprocal GMTs further illustrated that the Ag+- and AgNP-adapted gut microbiota were unable to fully acclimate to the toxicity of other Ag forms, resulting in a dramatic community shift and aggravated toxic symptoms of recipient D. magna. Toxic differences between Ag+ and AgNPs were related to the enriched organic matter in the gut, which functioned as an electron donor for sulfidation-based detoxification of Ag+ through degradation and as an inhibitor of Ag+ release from AgNPs by surface adsorption. Our findings provided fundamental understanding about gut-microbial detoxification and transformation of Ag+ and AgNPs, which finally resulted in physiological changes of D. magna and their offspring.
Environmental significanceOur findings provided a fundamental understanding about gut-microbial detoxification and transformation of Ag+ and AgNPs, which heavily relied on the enriched organic matter in the gut and finally resulted in physiological changes of Daphnia and their offspring. In addition, this study also shows the importance of taking host–microbe interactions into account in assessing the toxic effects of silver species on microbially colonized hosts and provides a method for evaluating the time-dependent selection on detoxification microbiota. |
1. Introduction
Silver ion (Ag+) and its associated forms are widely applied in water and air purification, biomedical applications, food production, cosmetics, clothing, and numerous household products.1 As one of the most widely utilized Ag+ forms, antibacterial Ag nanoparticles (AgNPs) are discharged into the environment and generate concern about their potential toxic effects on organisms.2,3 Previous studies mainly investigated the toxicity of different Ag forms on single cells or organisms, reporting unmatched toxic effects between Ag+ and AgNPs.4,5 However, at the community level, little is known about the toxic effects of different Ag forms, especially regarding the toxicological relevance of the gut microbiota interplay in living organisms.Gut microbiota mediate the physiology, detoxification, digestion, nutrient assimilation, and immunological reactions of aquatic metazoans.6,7 Any effects on their relationship can generate great impacts on the host and cause severe environmental damage.8,9 Despite the growing awareness of the critical roles of gut microbiota in aquatic ecosystems, the toxicological relevance of gut microbiota for the host has been rarely studied,10,11 especially regarding the effects of antibacterial chemicals. Previous studies reported that AgNPs could have a more severe damage on the cell membrane compared with Ag+ due to their strong adhesion onto the cell membrane while releasing Ag+.12 Following chronic exposure, bacteria could develop resistance to AgNPs by synthesizing flagellin, a protein that induces the aggregation and deactivation of AgNPs.13 At low concentrations, Ag could covalently and rapidly bind to cyanographene to overcome bacterial resistance in a low concentration.14 Knowledge of how bacteria respond to various Ag species under different exposure conditions is still limited. Interactions between microbiota and host might play an essential role in bacterial resistance to AgNPs. For instance, our previous study demonstrated that the gut environment promoted gut microbial resistance to low concentrations of AgNPs under chronic exposure, resulting in a higher tolerance of zooplankton to AgNPs.15 Other studies also reported that the protective microbes inside zebrafish larvae displayed some degrees of resistance to the acute toxicity of AgNPs.16 However, it is still unknown whether different Ag species could trigger distinctive resistant mechanisms of bacteria under different exposure conditions.
Due to their strong homology with metazoan genomes,17,18 zooplankton Daphnia magna is an established model system in environmental toxicology research. D. magna have unique features such as sizeable gut–body ratio, simple extraction of the gut, short life cycle, and filter-feeding behavior, and are particularly suitable for studying the toxic effects of antibacterial Ag on the gut microbiota.19,20 In the present study, the gut microbiota of D. magna were subjected to sequencing after being exposed to AgNPs (0.43 μg L−1) or Ag+ (0.08 μg L−1) at environmentally relevant concentrations21,22 for two to four generations. After four generations of exposure, the gut microbiota previously exposed to Ag+ or AgNPs was transplanted to axenic D. magna, which was further exposed to both Ag species to validate the existence of different resistant mechanisms. The goal of our study was to reveal the underlying mechanisms of gut microbial resistance to Ag+ and AgNPs and the effects of different Ag species on the interactions between microbiota and host.
2. Materials and methods
2.1. Preparation of organisms and materials
The zooplankton D. magna were raised in Aachener Daphnien Medium (ADaM),23 and Chlamydomonas reinhardtii (CC1690) were cultured as the food in sterile liquid BG11 medium.24 Both were cultured in a sterile, temperature-controlled chamber at 23 ± 1 °C on a 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 h light/dark cycle under 20 μmol m−2 s−1 illumination with constant stirring and aeration. D. magna were kept at a density of one individual per 10 mL and fed with saturating amounts of C. reinhardtii (105 cells per mL) each day, while the medium was refreshed weekly. C. reinhardtii prey was centrifuged to remove the culture medium and resuspended with an appropriate amount of ADaM before being fed to D. magna. AgNPs were synthesized and characterized according to a previous study.15 Detailed information is shown in the ESI.† Detailed methods to validate the toxicity and 48 h LC50 of Ag+ and AgNPs are provided in the ESI.† Ag+ and AgNP concentrations were determined using single particle inductively coupled plasma mass spectrometry (spICP-MS, Perkin Elmer NexION 300Q), which enabled the study of Ag species at dilute (ng L−1) concentrations. After the operation optimization, the dwell time was 10 μs, and the data acquisition time was 120 s for 107Ag signals. The flow rate before the injection of samples was 0.32–0.35 mL min−1. A single 100 ng L−1 Ag dissolved calibration check standard was analyzed in spICP-MS mode after every ten runs to maintain the machine's sensitivity. If drift in the standard signal was detected, the particle sizing was adjusted accordingly for the decrease in sensitivity.25 Synthesized AgNPs were also observed by transmission electron microscopy (TEM), and the results are listed in Fig. S1.†
10 h light/dark cycle under 20 μmol m−2 s−1 illumination with constant stirring and aeration. D. magna were kept at a density of one individual per 10 mL and fed with saturating amounts of C. reinhardtii (105 cells per mL) each day, while the medium was refreshed weekly. C. reinhardtii prey was centrifuged to remove the culture medium and resuspended with an appropriate amount of ADaM before being fed to D. magna. AgNPs were synthesized and characterized according to a previous study.15 Detailed information is shown in the ESI.† Detailed methods to validate the toxicity and 48 h LC50 of Ag+ and AgNPs are provided in the ESI.† Ag+ and AgNP concentrations were determined using single particle inductively coupled plasma mass spectrometry (spICP-MS, Perkin Elmer NexION 300Q), which enabled the study of Ag species at dilute (ng L−1) concentrations. After the operation optimization, the dwell time was 10 μs, and the data acquisition time was 120 s for 107Ag signals. The flow rate before the injection of samples was 0.32–0.35 mL min−1. A single 100 ng L−1 Ag dissolved calibration check standard was analyzed in spICP-MS mode after every ten runs to maintain the machine's sensitivity. If drift in the standard signal was detected, the particle sizing was adjusted accordingly for the decrease in sensitivity.25 Synthesized AgNPs were also observed by transmission electron microscopy (TEM), and the results are listed in Fig. S1.†
2.2. Experimental designs
To measure the ingestion rate, a separate set of 150 mL PC bottles was prepared (again in triplicate for the different experimental groups) containing 100 mL ADaM and 10 D. magna with the addition of corresponding chemicals. As a control, another 3 × 150 mL PC bottles were prepared using the same concentration of C. reinhardtii but no D. magna. The ingestion rate was then determined using a method described previously.27 The same groups of D. magna were also used to measure the respiration rate using a SensorDish® Reader; details are provided in the ESI.†
2.3. Gut extraction of zooplankton
After the D. magna were exposed to AgNPs or Ag+, they were transferred into autoclaved ADaM for 24 h to evacuate any remaining food particles. Their gut was extracted using sterile dissection tweezers (Regine 5, Switzerland) in a sterile Petri dish under a stereomicroscope.15 Before the gut extraction procedure, the dissection tweezers were autoclaved, steeped in 70% ethanol, and flame-sterilized. The guts from 80 individuals in each treatment group were transferred to 1.5 mL sterile Eppendorf tubes and prepared for triplicate metatranscriptome sequencing, whereas the individually collected guts from multi-generational exposure and GMTs were collected into separate tubes for subsequent 16S rRNA amplicon sequencing. The extracted gut, which contains gut tissue of D. magna and the whole microbiome in it, was then crushed into small sections inside a 1.5 mL sterile Eppendorf tube using a blunt-pointed pestle self-made from a 200 μL pipette. The small gut chips were then further filtered through 0.22 μm polycarbonate membranes (EMD Millipore, Billerica, MA, USA) and preserved with the addition of 1 mL RNA protection solution in a 1.5 ml sterile Eppendorf tube in a −80 °C freezer until further extraction of RNA and DNA. The dissection tool rinsing water was also collected and filtered through a 0.22 μm membrane to evaluate the likelihood of any operational contamination.2.4. Gut microbiota community analysis
Total DNA was extracted from the following: the preserved filters from the dissection tool rinsing water, the randomly sampled axenic eggs and the individually sampled gut microbiota using a PureLink Genomic DNA Kit (Invitrogen, Thermo Fisher Scientific Corp., Carlsbad, CA, USA). Amplification was conducted with the 16S rRNA specific targeting primers as follows: forward primer, 341F (5′-CCTACGGGRSGCAGCAG-3′) and reverse primer, 787R (5′-CTACNRGGGTATCTAA-3′) using a previously reported PCR system.31 The PCRs were conducted in triplicate, and the products were pooled together and sequenced using a HiSeq 2500 System (Illumina, San Diego, CA, USA) with 2 × 250 bp paired-end read configurations.The amplicon sequencing results were analyzed using the QIIME2 pipeline (version 2018.4), following reported procedures for quality filtration, demultiplexing, denoising with dada2,32 using a uniform sequence number, differentiating the partial 16S rRNA gene amplicon sequence variants (ASVs), taxonomy assignment, and diversity analysis. The taxonomic assignment of ASVs was achieved using the SILVA database (release 132), and the α diversity (within samples) of the gut microbial community was determined using the “qiime diversity alpha group significance” command. A filtered ASVs table at 0.1% abundance of each sample was generated with QIIME 1.9.1, and the summarize_taxa.py script was used to treat the ASVs table into relative abundances for LDA (linear discriminant analysis) effective size (LEfSe) analysis, which was then typically used to compare taxonomic units between exposure treatments.33 In addition, Sloan's neutral community model (NCM) was applied to evaluate the relative contribution of neutral and selection processes in shaping the gut community.34
2.5. Gut microbiota associated metatranscriptome analysis
The collected filters for metatranscriptome sequencing were briefly thawed on ice and the RNA protection solution was removed as previously described.15 According to the manufacturer's protocol, the total RNA was extracted using the Totally RNA isolation kit (Ambion Inc., Austin, TX, USA). The NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) was used to prepare the sequencing library.35 The pooled RNA from each sample was barcoded and sequenced using an Illumina HiSeq 2500 sequencer (Novogene Co., Ltd., Beijing, China).In total, 21 metatranscriptome samples were sequenced in this study (including triplicates for low Ag+, high Ag+, F1 low Ag+, F3 low Ag+, DOM-Ag+, DOM-AgNPs, and DOM), which were analyzed together with our previously reported metatranscriptomic data for AgNP exposure (triplicate for normal, low AgNP, high AgNP, F1 low AgNPs and F3 low AgNPs).15 Since the control group for Ag+-related experiments was under the same conditions as our previous study, we applied the previously sequenced metatranscriptomic data for the control group in this study. The different holobiont part-affiliated short reads were separated and distinguished according to a previously reported method.36 In brief, the genome and previously published RNA-seq of D. magna were downloaded to our local server to construct a reference dataset.37 In addition, the bacterial section of the Tara Oceans meta-genomic gene catalogue (OM-RGC) and non-redundant (nr) database was extracted using the blastdbcmd program38 to construct a bacterial reference dataset. After quality control of the short reads,39 the SRC_c software (in the default setting) was used to map the sequenced short reads to either a D. magna or bacteria affiliated dataset.40 According to a previous paper,41 the D. magna and gut microbiota affiliated short reads were separately assembled, predicted for open reading frames (ORFs), and annotated with the Kyoto Encyclopedia of Genes and Genomes (KEGG), Cazymes, and non-redundant (nr) database using Diamond software with k set to 1.42 The coverage of ORFs was calculated by mapping the short reads back to the assembled transcripts using Bowtie 2.2.9 (ref. 43) and SAMtools v1.9.44 The edgeR package in R was used to calculate the differentially expressed genes (DEGs) between groups with transcripts per million (TPM) values. The DEGs were defined with the criteria of |log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (fold change)| >1 and p-value <0.05 shown at the comparisons between experimental groups.
2 (fold change)| >1 and p-value <0.05 shown at the comparisons between experimental groups.
Co-expressed networks (modules) were constructed using the functions in the weighted correlation network analysis (WGCNA) package in R.45 Generally, genes with a similar expression pattern across all the samples were clustered into different co-expression metabolic eigengenes (MEs). In each experimental group, the regression correlation analysis was performed between the module eigengene values (i.e., the average expression level of all the genes in each ME) and the recorded reproduction rate of D. magna (as an indicator for toxic effects of Ag pollutants). The results are presented as a heatmap using R (V3.6.1), and the detailed analysis procedures are listed in the ESI.†
2.6. Gene expression validation
Six universal microbial genes were selected to verify the transcriptomic results. The primers for bacterial genes are listed in Table S1.† Any DNA contamination in the extracted RNA was detected through reverse transcription (RT) control of each pair of primers. In general, HiScript® III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme Biotech, Nanjing, China) was applied for the reverse transcription of extracted RNA. Subsequently, 1 μL (47 ng) synthesized cDNA was used for qPCR with the FastStart Universal SYBR Green Master Mix Kit (Roche, Germany) in a LightCycler 384 device (Roche, Germany), under thermocycling conditions as previously described.15 The 16S rRNA genes were selected as the inner references for normalization of gene expression in the gut microbes. The relative amount of mRNA was calculated using the 2−ΔΔCt method.2.7. Statistical analyses
The Bray–Curtis dissimilarity between gut-microbial communities was computed and visualized using the vegan package in R. The differences between datasets were statistically examined using ANOSIM.3. Results
3.1. Characterization and bioaccumulation of AgNPs or Ag+ in D. magna gut
The transmission electron microscopy (TEM) results revealed that the synthesized AgNPs were spherical particles with mean ± SD 20.0 ± 0.53 nm (Fig. S1†). The zeta potential of the synthesized AgNPs is −15.6 mV, and the hydrodynamic diameter is around 24 nm. After 48 h of exposure of D. magna to AgNPs (0–20 mg L−1) or Ag+ (0–2 mg L−1), the mortality rate of D. magna was measured, and the calculated 48 h LC50 of AgNPs was 9.03 μg L−1 as compared to 0.82 μg L−1 for Ag+ (Fig. S2A†). Because of AgNP dissolution, released Ag+ after 40 h and 90 h was ∼0.1 μg L−1 for the low and high concentration treatment, respectively (Fig. S2B†). Since D. magna did not survive for more than a week at high concentrations of AgNPs (4.3 μd L−1) and Ag+ (0.64 μ( L−1), they were only exposed to these concentrations for 24 h for sequencing of the metatranscriptome. The metatranscriptomic results for acute toxicity of high concentration of Ag+ and AgNPs showed that the number of gut microbiota affiliated reads was very low (below 1%). In comparison, the gut microbiota affiliated reads were relatively higher in number at low concentrations of Ag+ and AgNPs (higher than 5%, Table S2†). The respiration rate of D. magna was severely inhibited under the chronic exposure to the low concentration of AgNPs or Ag+. In addition, there was a gradual increase in the respiration rate from the F0 exposure (with mean values of 0.344 and 0.351 μmol O2 per mg dw per h under AgNP and Ag+ exposure, respectively) to the F3 (with mean values of 0.355 and 0.358 μmol O2 per mg dw per h under AgNP and Ag+ exposure, respectively) (Fig. S3†).3.2. Analysis of metatranscriptomic sequencing data affiliated to gut microbiota
In order to reveal the metabolic shift of gut microbiota between experimental groups, metatranscriptomic sequencing for high Ag+ (24 h), low Ag+ (24 h), F1 low Ag+, F3 low Ag+, DOM, DOM-Ag+, and DOM-AgNPs was conducted in this study, which was analyzed together with our previously sequenced data (high AgNPs (24 h), low AgNPs (24 h), F1 low AgNPs, F3 low AgNPs). Approximately 71 to 140 million 150 bp paired-end short reads were obtained across 30 metatranscriptome samples (Table S3†). The proportion of D. magna associated short reads ranged from 58.14% to 90.11%, whereas that of the gut microbiota ranged from 0.1% to 19.15% across all our samples (Table S2†). Noticeably, the percentage of short reads assigned to gut microbiota was less than 1% in the high Ag and F1 low Ag samples. The assembly results showed that the median of assembled lengths (N50) of D. magna affiliated reads was between 1266 and 4785 bp, whereas that of the gut microbiota was between 511 and 2278 bp (Table S4†). Biological coefficient of variation (BCV) analysis revealed that the transcriptomic data triplicates were clustered together for each experimental group, indicating the reproducibility and reliability of the triplicates and the RNA-seq data (Fig. S4†). In addition, the strong Spearman correlation between the gene expression level in the transcriptome and the qPCR results (R2 > 0.85, p < 0.001) validated our sequencing data (Fig. S5†). Furthermore, the empty results from the RT-control of each pair of primers demonstrated the low level of DNA contamination in the extracted RNA.3.3. Selection of toxicity adapted gut microbial community through multi-generational exposure (expt. 1)
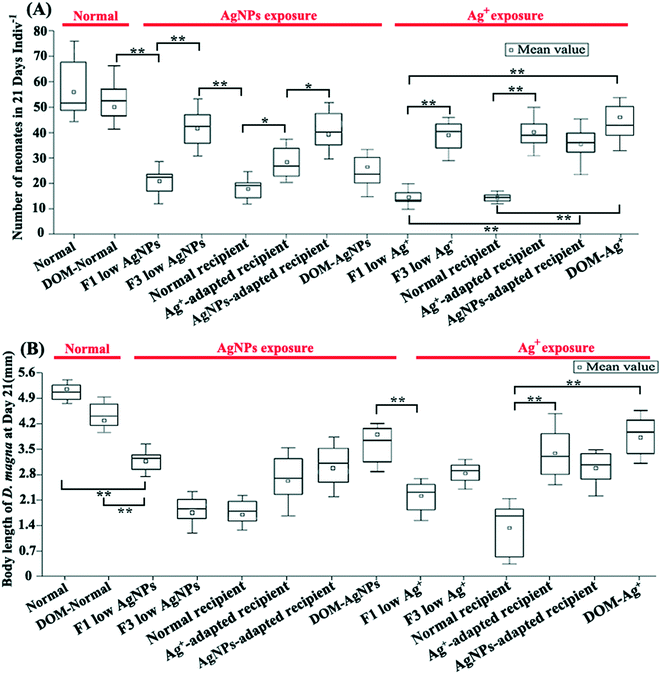
The physiological monitoring of D. magna under multi-generational exposure revealed their chronic stress adaptation to different Ag pollutants. The reproduction rate (Fig. 2A), body length (Fig. 2B), and ingestion rate (Fig. S6†) of the D. magna from F3 low Ag+ showed a marked increase compared with that from F1 low Ag+. A similar chronic stress adaptation was also found in exposures of AgNPs except the body length, which decreased in the F3 exposure, indicating a cumulative effect of AgNPs on the growth of D. magna. Overall, the reproduction rate clearly reflected the improved tolerance of D. magna to Ag+ and AgNPs under multi-generational exposure.It is generally accepted that the mechanisms of controlling the microbial community shift can be mainly divided into random (caused by drift) and selection (caused by environmental stress) processes.46 Therefore, after multi-generational exposure, it is important to quantify the contribution of these two processes in shaping the gut microbial community through neutral community model (NCM) analysis. In the results of NCM analysis, a higher R-squared value (Fig. S7†) indicated a better fit of the model for the microbial community data and illustrated the higher importance of neutral processes in shaping the community.47 In our study, although neutral processes were the dominant force in shaping the gut microbial community in all exposure groups (i.e., normal: R2 = 0.544; F1 low Ag+: R2 = 0.765; F3 low Ag+: R2 = 0.616; F1 low AgNPs: R2 = 0.794; and F3 low AgNPs: R2 = 0.629), the gut microbiota was still subjected to selection stress as the exposure time increased (Fig. S7†). In addition, it is interesting to note that after three generations of exposure, the selection stress of Ag+ for gut microbiota was much higher than that of AgNPs, indicating a stronger toxic effect of Ag+ on gut microbiota.
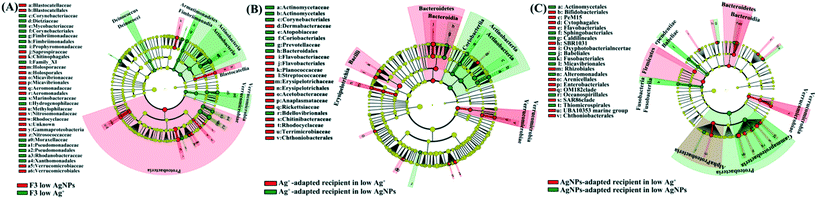
A distinctive gut-microbial composition between generations and treatments was investigated. The calculation of Bray–Curtis similarity showed that the gut microbial communities from the same treatment were grouped together, showing high similarity between individuals (Fig. S8†). Linear discriminant analysis showed that the microbial taxa characterized by anaerobic lifestyle and complex organic degradation were enriched in the F3-exposed D. magna in comparison with the F1. For example, the typical organic decomposers [including Fimbriimonadaceae,48Crocinitomicaceae,49Pseudomonadaceae,50Bacteroidetes,51 and Verrucomicrobia52 and anaerobic organisms (including Aeromonadaceae53 and Planctomycetes54)] were markedly enriched in F3 low AgNPs/Ag+ compared with F1 low AgNPs/Ag+ (Fig. S9A and B†). In addition, representative taxa for intestinal flora disorder [including Microbacteriaceae,55Enterococcaceae,56,57 and Moraxellaceae58] and strong stress resistance [including Micrococcales59 and Bacillaceae60] were enriched in the F1 low AgNPs and Ag+ (Fig. S9A and B†) compared with F3 low AgNPs and Ag+, respectively. It is noticeable that the comparison between F3 low AgNPs and F3 low Ag revealed the enrichment of microbial decomposers in F3 low Ag+, such as Deinococcus,61Fimbriimonadia,62 and Actinobacteria63 (Fig. 3A). In addition, the gut microbiota community from the control group was further compared with the other reported data,6,7 which exhibited little difference between each other (ANOSIM p > 0.1, Fig. S10†).
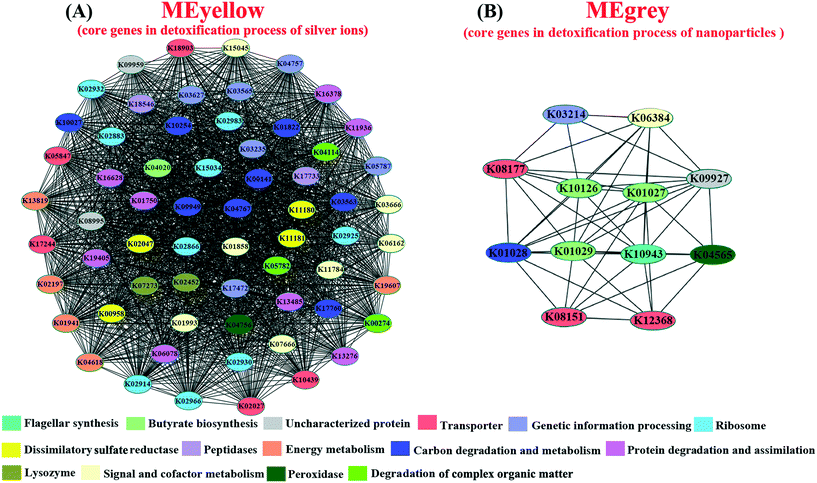
Since the reproduction rate of D. magna was highly associated with the toxic effects on gut microbiota,64 it was used as an indicator of gut microbial adaptation for different Ag pollutants. In order to assess the correlations between the functions of gut microbiota and experimental factors (the Ag pollutants in different forms and the reproduction rate of hosts), the identified microbial ORFs from multi-generational exposures (27 metatranscriptomic datasets) were first annotated with the KEGG database and then subjected to WGCNA.45 The results of co-expression clustering and its relationship with experimental factors are summarized in Table S5.† Results showed that the co-expressed gut microbial genes in MEpink were positively correlated with the exposure to Ag+ and the reproduction rate of D. magna, whereas the co-expressed genes in MEdarkgrey were positively correlated with AgNP exposure and the reproduction rate (Fig. S11B†). In order to identify the core genes within the MEpink and MEdarkgrey, the co-expression network of affiliated genes (correlation weight >4) was constructed and classified based on KEGG annotation. Within MEpink, the genes encoding for the ribosome, energy metabolism, protein degradation and assimilation, carbon degradation and metabolism, dissimilatory sulfite reductase, and degradation of complex organic matter were more crucial for the adaptation of gut microbiota under the exposure to Ag+ (Fig. 4A). In contrast, the biosynthesis of butyrate and flagellar played an important role in the microbial adaptation under the exposure to AgNPs (Fig. 4B).
3.4. Reciprocal microbiota transplants illustrate different toxicity between Ag species on gut microbiota (expt. 2)
In order to illustrate the toxic differences between different Ag species on gut microbiota, one type of Ag species adapted gut microbiota was transplanted to axenic D. magna for further exposure of both Ag species. Since the physiological traits of recipient D. magna varied significantly between groups with different reciprocal microbiota sources, our result suggested that the gut microbiota highly mediated the toxic symptoms. Under exposure of Ag+, a dramatic promotion of the traits (including reproduction rate, body length, and ingestion rate) in the recipient D. magna was detected once the gut microbial donor was pre-exposed to Ag+. A similar situation was also detected in the exposure of AgNPs when the gut microbial donor was pre-exposed to AgNPs. In contrast, the physiological traits of the recipient D. magna with the donor from Ag+-adapted gut microbiota were strongly affected by AgNPs; a similar situation was also detected in the recipient D. magna exposed to Ag+ where the AgNP-adapted gut microbiota was transplanted (Fig. 2A and B). These results suggested that Ag+ and AgNPs generated distinctive effects on gut microbiota, which was further correlated with the toxic symptoms of D. magna.The gut-microbial community of recipient D. magna was significantly shifted when the recipient was exposed to the Ag form that the gut microbiota was not adapted. Under AgNP exposure, the Coriobacteriales [indicator for the stressful gut environment65] and Prevotellaceae [indicator for unhealthy gut conditions66] were found to be enriched in the recipient D. magna inoculated with Ag+-adapted gut microbiota (Fig. 3B). In addition, the taxa with strong capability in organic degradation (such as Bacteroidetes and Firmicutes) were enriched in the recipient (inoculated with AgNP-adapted gut microbiota) exposed to Ag+ compared with the recipient under AgNP exposure (Fig. 3C). Notably, a high similarity was detected between gut-microbial donors and the recipient once they were exposed to the same Ag form (Fig. S12A and B†), indicating that once the gut microbiota had adapted to one Ag form, the recipient also appeared to adapt to this Ag form with long-term persistence.
3.5. DOM enrichment experiments validate the importance of organic compounds in microbial detoxification of Ag species (expt. 3)
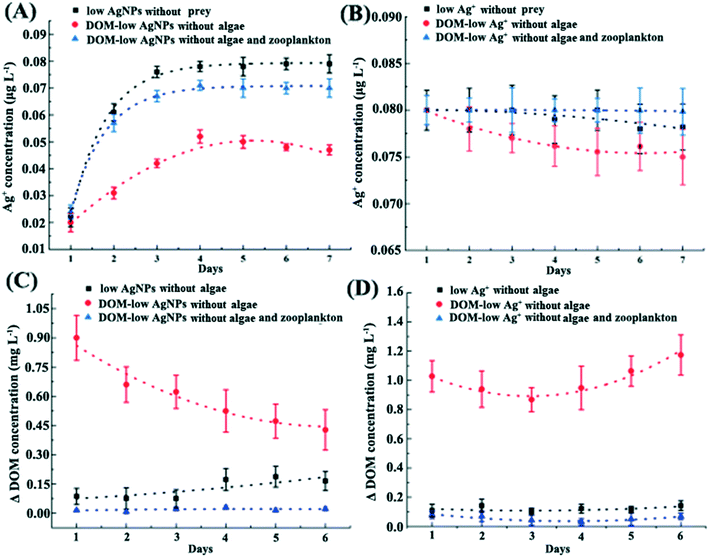
The addition of DOM dramatically reduced the toxic effects of Ag+ on D. magna with elevated reproduction rate and body length as compared with that in the F1 exposure (p < 0.01, Fig. 2). DOM addition also slightly reduced the released Ag+ in AgNP exposure, which was around 0.08 μg L−1 and 0.07 μg L−1 in the exposure to 0.43 μg L−1 AgNPs with and without the addition of DOM, respectively (Fig. 5A). In addition, DOM reduced the Ag+ concentration in both Ag+ and AgNP exposures with the existence of D. magna and massive DOM consumption (Fig. 5). The transcriptomic analysis confirmed that under Ag+ exposure, the DOM stimulated the up-regulation of gut-microbial genes encoding for organic degradation when compared with AgNP exposure and control (DOM group), indicating distinctive modulation of organic metabolism in gut microbiota under different Ag exposures (Fig. S13†). Results of differentially expressed genes between comparison groups are given in Table S6.†4. Discussion
4.1. D. magna gain fitness benefits from gut microbiota under Ag exposure
The reproduction rate and ingestion rate indicated that the tolerance of D. magna for Ag was dramatically promoted under multi-generational exposure. Further GMT experiment demonstrated that the tolerance of D. magna for Ag compound was highly correlated with gut microbiota metabolism. In a previous study, the gut microbiota of D. magna helped detoxify the cyanobacterial microcystin.7 Our finding further illustrates that the gut microbiota of D. magna promoted the tolerance of D. magna to biotoxin and metals, especially for Ag species. This may be due to the consistent selection stress for gut-microbial phylotypes that adapted to the toxicity under multi-generational exposure. In addition, the fitness benefits from gut microbiota were also found to be Ag form-dependent, which indicated that the Ag+ and AgNPs may differently affect D. magna through their distinctive effects on gut microbiota. Many studies attempted to differentiate the toxic differences between Ag+ and AgNPs. For instance, AgNPs showed strong colocalization with lipid droplets when using fluorescent signals to track their transportation inside D. magna, which triggered the maternal transfer of AgNPs since lipids are the main energy source of D. magna embryos, while Ag+ was irregularly distributed in different sites.67 Due to the strong adhesion of AgNPs to the cell membrane, a high proportion of Ag+ released from AgNPs was detected in the mitochondria, causing exhaustion of the respiratory reserve capacity and cell death.68 Also, the Ag+ displayed a diffusive cytoplasmic distribution pattern, while AgNPs were taken up mainly through endocytosis and distributed heterogeneously in the cytoplasm of algal cells.19 Our results based on multi-generational exposure and GMT experiments provided a novel insight into the toxicology of Ag+ and AgNPs and suggested that the gut microbiota may be overlooked for differentiating the toxicity of Ag+ from AgNPs.By conducting community analysis of samples from multi-generational exposure, we illustrated that different exposure times resulted in distinctive gut-microbial communities. The enriched representative microbial taxa for intestinal flora disorder and stress resistance in the F1 exposures indicated that Ag+ and AgNPs strongly affected the gut microbiota at the beginning of the exposure, which was consistent with the detected strong toxic symptoms of D. magna. Similar results were also detected in mice with dietary AgNPs (from 0 to 4600 ppb) for 28 days, where serious gut microbiota disturbance was detected.69 Differently, along with the symptomatic improvement of D. magna, the recovery of gut microbiota was evidenced by the enriched anaerobic microbial taxa in F3 exposure of this study, since gut microbiota normally contributed to the food digestion and fermentation processes.70 Similarly, a previous study also reported that the bacterial community in natural water was strongly affected but then recovered under exposure to low concentrations of Ag+ and AgNPs.71 These results illustrated that Ag pollutants' effects on gut microbiota were highly exposure time-dependent, which further mediated the toxic symptoms of D. magna.
The community analysis of samples from the GMT experiment illustrated that different Ag forms may generate distinctive effects on gut microbiota, which further mediated the toxic symptoms of D. magna. Although previous studies reported that the AgNPs and Ag+ can distinctively affect organisms in environments,72,73 our results further demonstrated that the effects of Ag on D. magna could also be triggered by the form-dependent toxicity on the gut-microbial community.
4.2. Sulfidation and organic degradation based on gut-microbial transformation and adaptation
Previous studies reported that the sulfidation- and chlorination-based transformation of Ag+ and AgNPs is the primary antidote to their toxicity.74,75 For instance, although AgNPs altered the detoxification enzymes, enhanced the liver oxidative stress, and affected the brain acetylcholinesterase activity in zebrafish, sulfidation of AgNPs resulted in significant alleviation of these toxic effects.75 The heritable reproductive toxicity of AgNPs to Caenorhabditis elegans was also relieved under sulfidized AgNPs.76 In complex microcosms, nearly 100% of Ag was bound to sulfur, which decreased the bioavailability and toxicity of Ag+ and AgNPs.77 Another study also showed that Ag2S was the main form in both AgNP- and AgNO3-treated sediments, and the proportion of Ag2S was 48–49% of the total Ag for AgNP and AgNO3 treatments.78 In addition, no acute toxicity of sulfidized AgNPs on Pseudokirchneriella subcapitata (algae), D. magna (crustacean), Danio rerio (fish), and Hydra vulgaris (cnidarian) was detected.79 Our studies demonstrated the possibility that the anaerobic gut environment of D. magna provided an ideal condition for the gut-microbial transformation of Ag+ and AgNPs through sulfidation with enriched sulfate and sulfate-reducing bacteria,80 since low oxygen is one of the requirements for microbial sulfate reduction.81 For instance, the expression of gut-microbial genes for sulfate reduction was tightly correlated with the toxic symptoms of D. magna across the metatranscriptomic datasets from Ag-treated exposures, where the highest expression of sulfate reduction and significant alleviation of toxic symptoms in D. magna were detected in the F3 generation. Considering the gut accumulation of Ag in zooplankton,82,83 the sulfidation-based gut-microbial transformation of Ag+ and AgNPs may be crucial for its detoxification. For instance, due to its strong damage on the cell membrane and electron transfer, Ag showed significant effects on phytoplankton, especially the metabolic processes for photosynthesis and nitrogen fixation.84,85 The gut-microbial sulfidation-based transformation of Ag may contribute to nutrient recycling by preventing the effects on primary production. Since the physiological status of zooplankton highly influenced the aquatic food web, the identified fitness benefits from gut-microbial detoxification could play an important role in ecological equilibrium.86 In addition, the low bioavailability of Ag2S could dramatically reduce the trophic transfer and accumulation of Ag in high trophic levels.For the first time, we showed that the enriched organic matter in the gut played a distinctive role in gut microbial adaptation under multi-generational exposure. Within the F3 exposures, the abundance of microbial organic decomposers increased and suggested that the organics may play a key role in the adaptation of gut microbiota for Ag+ and AgNPs. This finding was consistent with recent reports that surrounding organic matter decreased the toxicity of AgNPs and Ag+ to green algae and zebrafish.87,88 However, further analysis showed that the microbial organic decomposers were enriched in the exposure of F3 low Ag+ compared with exposure of F3 low AgNPs. This result indicated that organic degradation might play a more important role in gut-microbial adaptation for Ag+ than AgNPs. In addition, the distinctive role of organics in gut-microbial adaptation for different Ag was further evidenced by the decreased abundance of microbial organic decomposers in the AgNP-exposed recipient zooplankton (donors from Ag-adapted gut microbiota).
With the application of co-expression analysis (WGCNA), we revealed the underlying mechanisms of the distinctive gut-microbial adaptation for Ag+ and AgNPs. Since the reproduction rate of D. magna well represented the detoxification and adaptation of gut microbiota,6,64 this trait was used for correlation analysis with clustered co-expression genes. The analysis revealed that the gut-microbial gene expression for the organic degradation process was correlated with their gene expression for the bio-synthesis of sulfide (sulfate reduction), which may neutralize Ag+. This may be due to the organic electron donors for sulfate reduction in gut microbiota under Ag+ exposure,89,90 where the sulfate reduction process is favored by the electrons generated in the organic degradation process. Subsequently, the Ag+ in water reacts with the synthesized sulfide to generate Ag2S, which has been demonstrated to be the main strategy for bacteria to cope with the strong oxidation of Ag+.91,92 However, this co-expression pattern was not for the detoxification process of AgNPs, indicating the suppressed organic degradation for gut-microbial adaptation of AgNPs. This result was further supported by the significantly down-regulated gut-microbial genes for organic degradation in DOM-AgNP exposure compared with the expression profile in DOM-Ag+ and DOM (control). Since the organics can strongly adsorb on the surface of AgNPs to prevent the release of Ag+ from AgNPs,93,94 the suppressed gene expression for organic degradation may be a strategy of gut microbiota to reduce the released Ag+ from AgNPs. Therefore, the gut-microbial gene expression for organic degradation may be dynamically regulated by the requirement for neutralizing the released Ag+ (as organic electron donors for sulfate reduction) and preventing the release of Ag+ under AgNP exposure. In addition, this conclusion was further supported by the reduced saturation concentration of the released Ag+ from AgNPs in the DOM incubation experiment. The identified core genes for detoxification of AgNPs were consistent with the previous report, where the bacterial flagellar protein reacted with nanoparticles (NPs) to form nontoxic precipitation.13
5. Conclusion
In this study, we investigated for the first time the toxic effects of Ag+ and AgNPs on gut microbiota under multi-generational exposures and identified the potential strategy of gut-microbial adaptation as well as their implications for the host and its offspring. Our results showed that multi-generational exposures to Ag+ and AgNPs caused intensive selection pressure for gut-microbial phylotypes, resulting in toxicity-adapted phylotypes in F3 exposures. The gut microbial transplant experiment further demonstrated that gut microbiota adaptation for Ag pollutants was highly Ag form dependent. Transcriptomic analysis revealed the potentially sulfidation-based gut-microbial transformation of Ag+ with organic decomposition as the electron donor in Ag+ exposure. In contrast, the suppressed gene expression for organic degradation suggested the possible adaptation strategy of gut microbiota during AgNP exposure, such as retaining the organic matter to enhance organic adsorption on AgNPs to reduce Ag+ release. Since the gut microbiota played an important role in the environmental fitness of host and ecological processes, our study provided crucial information for the environmental risk assessment of antibacterial Ag pollutants.Data availability
Sequence data were deposited in GenBank (Sequence Read Archive) and are available under the BioProject (PRJNA540581).Author contributions
Yingdong Li and Hongbin Liu designed the experiment. Hongbin Liu and Wen-Xiong Wang provided the experimental species and materials. Yingdong Li conducted the experiment, analyzed the data, and wrote the manuscript. Hongbin Liu and Wen-Xiong Wang reviewed the manuscript.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This study was supported by the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC01), the Hong Kong Research Grants Council (grant no. CityU16103120 and T21-602/16-R), and the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0409).References
- C. Marambio-Jones and E. M. Hoek, A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment, J. Nanopart. Res., 2010, 12(5), 1531–1551 CrossRef CAS.
- R. Kaegi, B. Sinnet, S. Zuleeg, H. Hagendorfer, E. Mueller, R. Vonbank, M. Boller and M. Burkhardt, Release of silver nanoparticles from outdoor facades, Environ. Pollut., 2010, 158(9), 2900–2905 CrossRef CAS PubMed.
- N. C. Mueller and B. Nowack, Exposure modeling of engineered nanoparticles in the environment, Environ. Sci. Technol., 2008, 42(12), 4447–4453 CrossRef CAS PubMed.
- M. Milić, G. Leitinger, I. Pavičić, M. Zebić Avdičević, S. Dobrović, W. Goessler and I. Vinković Vrček, Cellular uptake and toxicity effects of silver nanoparticles in mammalian kidney cells, J. Appl. Toxicol., 2015, 35(6), 581–592 CrossRef PubMed.
- M. K. Ha, S. J. Kwon, J. S. Choi, N. T. Nguyen, J. Song, Y. Lee, Y. E. Kim, I. Shin, J. W. Nam and T. H. Yoon, Mass Cytometry and Single-Cell RNA-seq Profiling of the Heterogeneity in Human Peripheral Blood Mononuclear Cells Interacting with Silver Nanoparticles, Small, 2020, 16(21), 1907674 CrossRef CAS PubMed.
- M. Callens, H. Watanabe, Y. Kato, J. Miura and E. Decaestecker, Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia, Microbiome, 2018, 6(1), 1–12 CrossRef PubMed.
- E. Macke, M. Callens, L. De Meester and E. Decaestecker, Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria, Nat. Commun., 2017, 8(1), 1–13 CrossRef CAS PubMed.
- Z. Zhou, E. Ringø, R. Olsen and S. Song, Dietary effects of soybean products on gut microbiota and immunity of aquatic animals: a review, Aquacult. Nutr., 2018, 24(1), 644–665 CrossRef CAS.
- A. Larsen, H. Mohammed and C. Arias, Characterization of the gut microbiota of three commercially valuable warmwater fish species, J. Appl. Microbiol., 2014, 116(6), 1396–1404 CrossRef CAS PubMed.
- S. P. Claus, H. Guillou and S. Ellero-Simatos, The gut microbiota: a major player in the toxicity of environmental pollutants?, npj Biofilms Microbiomes, 2016, 2(1), 1–11 CrossRef PubMed.
- X. You, N. Xu, X. Yang and W. Sun, Pollutants affect algae-bacteria interactions: a critical review, Environ. Pollut., 2021, 116723 CrossRef CAS PubMed.
- L. Maurer and J. Meyer, A systematic review of evidence for silver nanoparticle-induced mitochondrial toxicity, Environ. Sci.: Nano, 2016, 3(2), 311–322 RSC.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek and O. Tomanec, Bacterial resistance to silver nanoparticles and how to overcome it, Nat. Nanotechnol., 2018, 13(1), 65–71 CrossRef PubMed.
- D. Panáček, L. Hochvaldová, A. Bakandritsos, T. Malina, M. Langer, J. Belza, J. Martincová, R. Večeřová, P. Lazar and K. Poláková, Silver Covalently Bound to Cyanographene Overcomes Bacterial Resistance to Silver Nanoparticles and Antibiotics, Adv. Sci., 2021, 2003090 CrossRef PubMed.
- Y. Li, N. Yan, T. Y. Wong, W.-X. Wang and H. Liu, Interaction of antibacterial silver nanoparticles and microbiota-dependent holobionts revealed by metatranscriptomic analysis, Environ. Sci.: Nano, 2019, 6(11), 3242–3255 RSC.
- B. W. Brinkmann, B. E. Koch, H. P. Spaink, W. J. Peijnenburg and M. G. Vijver, Colonizing microbiota protect zebrafish larvae against silver nanoparticle toxicity, Nanotoxicology, 2020, 14(6), 725–739 CrossRef CAS PubMed.
- D. Ebert, Ecology, epidemiology, and evolution of parasitism in Daphnia, National Library of Medicine, 2005 Search PubMed.
- A. Stollewerk, The water flea Daphnia-a'new'model system for ecology and evolution?, J. Biol., 2010, 9(2), 1–4 CrossRef PubMed.
- N. Yan and W.-X. Wang, Novel imaging of silver nanoparticle uptake by a unicellular alga and trophic transfer to Daphnia magna, Environ. Sci. Technol., 2021, 55(8), 5143–5151 CrossRef CAS PubMed.
- J. Li, M. Tang and Y. Xue, Review of the effects of silver nanoparticle exposure on gut bacteria, J. Appl. Toxicol., 2019, 39(1), 27–37 CrossRef CAS PubMed.
- A. Tsiola, C. Toncelli, S. Fodelianakis, G. Michoud, T. D. Bucheli, A. Gavriilidou, M. Kagiorgi, I. Kalantzi, K. Knauer and G. Kotoulas, Low-dose addition of silver nanoparticles stresses marine plankton communities, Environ. Sci.: Nano, 2018, 5(8), 1965–1980 RSC.
- J. T. Quik, J. J. de Klein and A. A. Koelmans, Spatially explicit fate modelling of nanomaterials in natural waters, Water Res., 2015, 80, 200–208 CrossRef CAS PubMed.
- B. Klüttgen, U. Dülmer, M. Engels and H. Ratte, ADaM, an artificial freshwater for the culture of zooplankton, Water Res., 1994, 28(3), 743–746 CrossRef.
- R. Rippka, J. Deruelles, J. B. Waterbury, M. Herdman and R. Y. Stanier, Generic assignments, strain histories and properties of pure cultures of cyanobacteria, Microbiology, 1979, 111(1), 1–61 Search PubMed.
- D. Mitrano, J. F. Ranville, A. Bednar, K. Kazor, A. S. Hering and C. P. Higgins, Tracking dissolution of silver nanoparticles at environmentally relevant concentrations in laboratory, natural, and processed waters using single particle ICP-MS (spICP-MS), Environ. Sci.: Nano, 2014, 1(3), 248–259 RSC.
- J. Liu and R. H. Hurt, Ion release kinetics and particle persistence in aqueous nano-silver colloids, Environ. Sci. Technol., 2010, 44(6), 2169–2175 CrossRef CAS PubMed.
- S. Zhang, H. Liu, P. M. Glibert, C. Guo and Y. Ke, Effects of prey of different nutrient quality on elemental nutrient budgets in Noctiluca scintillans, Sci. Rep., 2017, 7(1), 7622 CrossRef PubMed.
- Y. Kan and J. Pan, A ONE-SHOT SOLUTION TO BACTERIAL AND FUNGAL CONTAMINATION IN THE GREEN ALGA CHLAMYDOMONAS REINHARDTII CULTURE BY USING AN ANTIBIOTIC COCKTAIL 1, J. Phycol., 2010, 46(6), 1356–1358 CrossRef CAS.
- B. C. Mcmeans, A.-M. Koussoroplis, M. T. Arts and M. J. Kainz, Terrestrial dissolved organic matter supports growth and reproduction of Daphnia magna when algae are limiting, J. Plankton Res., 2015, 37(6), 1201–1209 CAS.
- J. Koprivnjak, J. Blanchette, R. Bourbonniere, T. Clair, A. Heyes, K. Lum, R. McCrea and T. Moore, The underestimation of concentrations of dissolved organic carbon in freshwaters, Water Res., 1995, 29(1), 91–94 CrossRef CAS.
- H. Liu, S. Tan, J. Xu, W. Guo, X. Xia and S. Yan Cheung, Interactive regulations by viruses and dissolved organic matter on the bacterial community, Limnol. Oceanogr., 2017, 62(S1), S364–S380 CrossRef CAS.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13(7), 581–583 CrossRef CAS PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Metagenomic biomarker discovery and explanation, Genome Biol., 2011, 12(6), 1–18 CrossRef PubMed.
- W. T. Sloan, M. Lunn, S. Woodcock, I. M. Head, S. Nee and T. P. Curtis, Quantifying the roles of immigration and chance in shaping prokaryote community structure, Environ. Microbiol., 2006, 8(4), 732–740 CrossRef PubMed.
- K. D. Hansen, S. E. Brenner and S. Dudoit, Biases in Illumina transcriptome sequencing caused by random hexamer priming, Nucleic Acids Res., 2010, 38(12), e131 CrossRef PubMed.
- A. Meng, C. Marchet, E. Corre, P. Peterlongo, A. Alberti, C. Da Silva, P. Wincker, E. Pelletier, I. Probert and J. Decelle, A de novo approach to disentangle partner identity and function in holobiont systems, Microbiome, 2018, 6(1), 105 CrossRef PubMed.
- L. Orsini, D. Gilbert, R. Podicheti, M. Jansen, J. B. Brown, O. S. Solari, K. I. Spanier, J. K. Colbourne, D. B. Rusch and E. Decaestecker, Daphnia magna transcriptome by RNA-Seq across 12 environmental stressors, Sci. Data, 2016, 3(1), 1–16 Search PubMed.
- C. Camacho, G. Coulouris, V. Avagyan, N. Ma, J. Papadopoulos, K. Bealer and T. L. Madden, BLAST+: architecture and applications, BMC Bioinf., 2009, 10(1), 421 CrossRef PubMed.
- W. Gong, H. Paerl and A. Marchetti, Eukaryotic phytoplankton community spatiotemporal dynamics as identified through gene expression within a eutrophic estuary, Environ. Microbiol., 2018, 20(3), 1095–1111 CrossRef CAS PubMed.
- C. Marchet, L. Lecompte, A. Limasset, L. Bittner and P. Peterlongo, A resource-frugal probabilistic dictionary and applications in bioinformatics, Discrete Appl. Math., 2020, 274, 92–102 CrossRef.
- Y. Li, Z. Xu and H. Liu, Nutrient-imbalanced conditions shift the interplay between zooplankton and gut microbiota, BMC Genomics, 2021, 22(1), 1–18 CrossRef PubMed.
- B. Buchfink, C. Xie and D. H. Huson, Fast and sensitive protein alignment using DIAMOND, Nat. Methods, 2015, 12(1), 59–60 CrossRef CAS PubMed.
- B. Langmead and S. L. Salzberg, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 2012, 9(4), 357 CrossRef CAS PubMed.
- H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis and R. Durbin, The sequence alignment/map format and SAMtools, Bioinformatics, 2009, 25(16), 2078–2079 CrossRef CAS PubMed.
- P. Langfelder and S. Horvath, WGCNA: an R package for weighted correlation network analysis, BMC Bioinf., 2008, 9(1), 559 CrossRef PubMed.
- S. Woodcock, C. J. Van Der Gast, T. Bell, M. Lunn, T. P. Curtis, I. M. Head and W. T. Sloan, Neutral assembly of bacterial communities, FEMS Microbiol. Ecol., 2007, 62(2), 171–180 CrossRef CAS PubMed.
- J. Chave, Neutral theory and community ecology, Ecol. Lett., 2004, 7(3), 241–253 CrossRef.
- S. Chen, M. Yan, T. Huang, H. Zhang, K. Liu, X. Huang, N. Li, Y. Miao and R. Sekar, Disentangling the drivers of microcystis decomposition: Metabolic profile and co-occurrence of bacterial community, Sci. Total Environ., 2020, 739, 140062 CrossRef CAS PubMed.
- Z. Zhao, M. Gonsior, P. Schmitt-Kopplin, Y. Zhan, R. Zhang, N. Jiao and F. Chen, Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity, ISME J., 2019, 13(10), 2551–2565 CrossRef CAS PubMed.
- A. Bains, O. Perez-Garcia, G. Lear, D. Greenwood, S. Swift, M. Middleditch, E. P. Kolodziej and N. Singhal, Induction of microbial oxidative stress as a new strategy to enhance the enzymatic degradation of organic micropollutants in synthetic wastewater, Environ. Sci. Technol., 2019, 53(16), 9553–9563 CrossRef CAS PubMed.
- B. Fernández-Gomez, M. Richter, M. Schüler, J. Pinhassi, S. G. Acinas, J. M. González and C. Pedros-Alio, Ecology of marine Bacteroidetes: a comparative genomics approach, ISME J., 2013, 7(5), 1026–1037 CrossRef PubMed.
- A. Sichert, C. H. Corzett, M. S. Schechter, F. Unfried, S. Markert, D. Becher, A. Fernandez-Guerra, M. Liebeke, T. Schweder and M. F. Polz, Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan, Nat. Microbiol., 2020, 5(8), 1026–1039 CrossRef CAS PubMed.
- R. Colwell, M. MacDonell and J. De Ley, Proposal to Recognize the Family Aeromonadaceae fam. nov, Int. J. Syst. Evol. Microbiol., 1986, 36(3), 473–477 Search PubMed.
- J. S. Sinninghe Damsté, W. I. C. Rijpstra, J. A. Geenevasen, M. Strous and M. S. M. Jetten, Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox), FEBS J., 2005, 272(16), 4270–4283 CrossRef PubMed.
- J. Plaza-Díaz, A. Gómez-Fernández, N. Chueca, M. J. D. L. Torre-Aguilar, Á. Gil, J. L. Perez-Navero, K. Flores-Rojas, P. Martín-Borreguero, P. Solis-Urra and F. J. Ruiz-Ojeda, Autism spectrum disorder (ASD) with and without mental regression is associated with changes in the fecal microbiota, Nutrients, 2019, 11(2), 337 CrossRef PubMed.
- G. M. Nava, H. J. Friedrichsen and T. S. Stappenbeck, Spatial organization of intestinal microbiota in the mouse ascending colon, ISME J., 2011, 5(4), 627–638 CrossRef CAS PubMed.
- A. Lo Presti, F. Zorzi, F. Del Chierico, A. Altomare, S. Cocca, A. Avola, F. De Biasio, A. Russo, E. Cella and S. Reddel, Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease, Front. Microbiol., 2019, 10, 1655 CrossRef PubMed.
- N. D. Vaziri, J. Wong, M. Pahl, Y. M. Piceno, J. Yuan, T. Z. DeSantis, Z. Ni, T.-H. Nguyen and G. L. Andersen, Chronic kidney disease alters intestinal microbial flora, Kidney Int., 2013, 83(2), 308–315 CrossRef PubMed.
- X. Sun, L. Zhang, J. Pei and L.-F. Huang, Regulatory relationship between quality variation and environment of Cistanche deserticola in three ecotypes based on soil microbiome analysis, Sci. Rep., 2020, 10(1), 1–12 CrossRef PubMed.
- A. M. Tarnecki, M. Wafapoor, R. N. Phillips and N. R. Rhody, Benefits of a Bacillus probiotic to larval fish survival and transport stress resistance, Sci. Rep., 2019, 9(1), 1–11 CAS.
- C. C. Lange, L. P. Wackett, K. W. Minton and M. J. Daly, Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments, Nat. Biotechnol., 1998, 16(10), 929–933 CrossRef CAS PubMed.
- W.-T. Im, Z.-Y. Hu, K.-H. Kim, S.-K. Rhee, H. Meng, S.-T. Lee and Z.-X. Quan, Description of Fimbriimonas ginsengisoli gen. nov., sp. nov. within the Fimbriimonadia class nov., of the phylum Armatimonadetes, Antonie van Leeuwenhoek, 2012, 102(2), 307–317 CrossRef CAS PubMed.
- M.-È. Lacombe-Harvey, R. Brzezinski and C. Beaulieu, Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution, Appl. Microbiol. Biotechnol., 2018, 102(17), 7219–7230 CrossRef CAS PubMed.
- S. Akbar, L. Gu, Y. Sun, Q. Zhou, L. Zhang, K. Lyu, Y. Huang and Z. Yang, Changes in the life history traits of Daphnia magna are associated with the gut microbiota composition shaped by diet and antibiotics, Sci. Total Environ., 2020, 705, 135827 CrossRef CAS PubMed.
- K. M. Bangsgaard Bendtsen, L. Krych, D. B. Sørensen, W. Pang, D. S. Nielsen and K. Josefsen, et al., Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse, PLoS One, 2012, 7, e46231 CrossRef PubMed.
- L. J. Spielman, D. L. Gibson and A. Klegeris, Unhealthy gut, unhealthy brain: The role of the intestinal microbiota in neurodegenerative diseases, Neurochem. Int., 2018, 120, 149–163 CrossRef CAS PubMed.
- N. Yan, S. M. J. Tsim, X. He, B. Z. Tang and W.-X. Wang, Direct visualization and quantification of maternal transfer of silver nanoparticles in zooplankton, Environ. Sci. Technol., 2020, 54(17), 10763–10771 CrossRef CAS PubMed.
- N. Yan, B. Z. Tang and W.-X. Wang, Intracellular trafficking of silver nanoparticles and silver ions determined their specific mitotoxicity to the zebrafish cell line, Environ. Sci.: Nano, 2021, 8(5), 1364–1375 RSC.
- S. Van Den Brûle, J. Ambroise, H. Lecloux, C. Levard, R. Soulas, P.-J. De Temmerman, M. Palmai-Pallag, E. Marbaix and D. Lison, Dietary silver nanoparticles can disturb the gut microbiota in mice, Part. Fibre Toxicol., 2015, 13(1), 1–16 CrossRef PubMed.
- H. M. Freese and B. Schink, Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna, Microb. Ecol., 2011, 62(4), 882 CrossRef PubMed.
- P. Das, C. J. Williams, R. R. Fulthorpe, M. E. Hoque, C. D. Metcalfe and M. A. Xenopoulos, Changes in bacterial community structure after exposure to silver nanoparticles in natural waters, Environ. Sci. Technol., 2012, 46(16), 9120–9128 CrossRef CAS PubMed.
- Y. Yang, J. Quensen, J. Mathieu, Q. Wang, J. Wang, M. Li, J. M. Tiedje and P. J. Alvarez, Pyrosequencing reveals higher impact of silver nanoparticles than Ag+ on the microbial community structure of activated sludge, Water Res., 2014, 48, 317–325 CrossRef CAS PubMed.
- Y. Guo, N. Cichocki, F. Schattenberg, R. Geffers, H. Harms and S. Müller, AgNPs change microbial community structures of wastewater, Front. Microbiol., 2019, 9, 3211 CrossRef PubMed.
- C. Levard, E. M. Hotze, B. P. Colman, A. L. Dale, L. Truong, X. Yang, A. J. Bone, G. E. Brown Jr, R. L. Tanguay and R. T. Di Giulio, Sulfidation of silver nanoparticles: natural antidote to their toxicity, Environ. Sci. Technol., 2013, 47(23), 13440–13448 CrossRef CAS PubMed.
- G. P. Devi, K. B. A. Ahmed, M. S. Varsha, B. Shrijha, K. S. Lal, V. Anbazhagan and R. Thiagarajan, Sulfidation of silver nanoparticle reduces its toxicity in zebrafish, Aquat. Toxicol., 2015, 158, 149–156 CrossRef CAS PubMed.
- A. Wamucho, J. M. Unrine, T. J. Kieran, T. C. Glenn, C. L. Schultz, M. Farman, C. Svendsen, D. J. Spurgeon and O. V. Tsyusko, Genomic mutations after multigenerational exposure of Caenorhabditis elegans to pristine and sulfidized silver nanoparticles, Environ. Pollut., 2019, 254, 113078 CrossRef CAS PubMed.
- H. S. Jiang, L. Yin, N. N. Ren, L. Xian, S. Zhao, W. Li and B. Gontero, The effect of chronic silver nanoparticles on aquatic system in microcosms, Environ. Pollut., 2017, 223, 395–402 CrossRef CAS PubMed.
- S. Gao, T. Fang, G. Wang, S. Bao and W. Tang, The transportation of silver nanoparticles between water and sediments, Shuisheng Shengwu Xuebao, 2015, 39(2), 375–381 CAS.
- J. S. Kang and J. W. Park, Silver Ion Release Accelerated in the Gastrovascular Cavity of Hydra vulgaris Increases the Toxicity of Silver Sulfide Nanoparticles, Environ. Toxicol. Chem., 2021, 40(6), 1662–1672 CrossRef CAS PubMed.
- W. Peng, X. Li, M. Lin, H. Gui, H. Xiang, Q. Zhao and W. Fan, Biosafety of cadmium contaminated sediments after treated by indigenous sulfate reducing bacteria: Based on biotic experiments and DGT technique, J. Hazard. Mater., 2020, 384, 121439 CrossRef CAS PubMed.
- G. Antler, A. V. Turchyn, S. Ono, O. Sivan and T. Bosak, Combined 34S, 33S and 18O isotope fractionations record different intracellular steps of microbial sulfate reduction, Geochim. Cosmochim. Acta, 2017, 203, 364–380 CrossRef CAS.
- S. Kuehr, J. Klehm, C. Stehr, M. Menzel and C. Schlechtriem, Unravelling the uptake pathway and accumulation of silver from manufactured silver nanoparticles in the freshwater amphipod Hyalella azteca using correlative microscopy, NanoImpact, 2020, 19, 100239 CrossRef.
- N. Yan, B. Z. Tang and W.-X. Wang, In vivo bioimaging of silver nanoparticle dissolution in the gut environment of zooplankton, ACS Nano, 2018, 12(12), 12212–12223 CrossRef CAS PubMed.
- C. F. McGee, The effects of silver nanoparticles on the microbial nitrogen cycle: a review of the known risks, Environ. Sci. Pollut. Res., 2020, 27, 31061–31073 CrossRef CAS PubMed.
- S.-H. Nam and Y.-J. An, Size-and shape-dependent toxicity of silver nanomaterials in green alga Chlorococcum infusionum, Ecotoxicol. Environ. Saf., 2019, 168, 388–393 CrossRef CAS PubMed.
- K. A. Ger, L. A. Hansson and M. Lürling, Understanding cyanobacteria-zooplankton interactions in a more eutrophic world, Freshwater Biol., 2014, 59(9), 1783–1798 CrossRef.
- M. Khoshnamvand, S. Ashtiani, Y. Chen and J. Liu, Impacts of organic matter on the toxicity of biosynthesized silver nanoparticles to green microalgae Chlorella vulgaris, Environ. Res., 2020, 185, 109433 CrossRef CAS PubMed.
- B. Xiao, X. Wang, J. Yang, K. Wang, Y. Zhang, B. Sun, T. Zhang and L. Zhu, Bioaccumulation kinetics and tissue distribution of silver nanoparticles in zebrafish: The mechanisms and influence of natural organic matter, Ecotoxicol. Environ. Saf., 2020, 194, 110454 CrossRef CAS PubMed.
- S. Coetser, W. Pulles, R. Heath and T. E. Cloete, Chemical characterisation of organic electron donors for sulfate reduction for potential use in acid mine drainage treatment, Biodegradation, 2006, 17(2), 67–77 CrossRef CAS PubMed.
- B. Hausmann, K.-H. Knorr, K. Schreck, S. G. Tringe, T. G. Del Rio, A. Loy and M. Pester, Consortia of low-abundance bacteria drive sulfate reduction-dependent degradation of fermentation products in peat soil microcosms, ISME J., 2016, 10(10), 2365–2375 CrossRef CAS PubMed.
- B. Reinsch, C. Levard, Z. Li, R. Ma, A. Wise, K. Gregory, G. Brown Jr and G. Lowry, Sulfidation of silver nanoparticles decreases Escherichia coli growth inhibition, Environ. Sci. Technol., 2012, 46(13), 6992–7000 CrossRef CAS PubMed.
- M. Kraas, K. Schlich, B. Knopf, F. Wege, R. Kägi, K. Terytze and K. Hund-Rinke, Long-term effects of sulfidized silver nanoparticles in sewage sludge on soil microflora, Environ. Toxicol. Chem., 2017, 36(12), 3305–3313 CrossRef CAS PubMed.
- S. M. Wirth, G. V. Lowry and R. D. Tilton, Natural organic matter alters biofilm tolerance to silver nanoparticles and dissolved silver, Environ. Sci. Technol., 2012, 46(22), 12687–12696 CrossRef CAS PubMed.
- Z. Zhang, X. Yang, M. Shen, Y. Yin and J. Liu, Sunlight-driven reduction of silver ion to silver nanoparticle by organic matter mitigates the acute toxicity of silver to Daphnia magna, J. Environ. Sci., 2015, 35, 62–68 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1en00765c |
| This journal is © The Royal Society of Chemistry 2022 |