Open Access Article
Open Access ArticleReal-time personalized health status prediction of lithium-ion batteries using deep transfer learning†
Guijun
Ma
 a,
Songpei
Xu
a,
Songpei
Xu
 b,
Benben
Jiang
c,
Cheng
Cheng
b,
Benben
Jiang
c,
Cheng
Cheng
 b,
Xin
Yang
d,
Yue
Shen
b,
Xin
Yang
d,
Yue
Shen
 e,
Tao
Yang
f,
Yunhui
Huang
e,
Tao
Yang
f,
Yunhui
Huang
 e,
Han
Ding
a and
Ye
Yuan
e,
Han
Ding
a and
Ye
Yuan
 *b
*b
aSchool of Mechanical Science and Engineering, Huazhong University of Science and Technology, Wuhan 430074, China
bSchool of Artificial Intelligence and Automation, Huazhong University of Science and Technology, Wuhan 430074, China. E-mail: yye@hust.edu.cn
cDepartment of Automation, Tsinghua University, Beijing 100084, China
dCollege of Electrical and Information Engineering, Hunan University, Changsha 410082, China
eSchool of Materials Science and Engineering, Huazhong University of Science and Technology, Wuhan 430074, China
fState Key Laboratory of Synthetical Automation for Process Industries, Northeastern University, Shenyang 110819, China
First published on 30th July 2022
Abstract
Real-time and personalized lithium-ion battery health management is conducive to safety improvement for end-users. However, personalized prognostic of the battery health status is still challenging due to diverse usage interests, dynamic operational patterns and limited historical data. We generate a comprehensive dataset consisting of 77 commercial cells (77 discharge protocols) with over 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 charge–discharge cycles—the largest dataset to our knowledge of its kind, and develop a transfer learning framework to realize real-time personalized health status prediction for unseen battery discharge protocols, at any charge–discharge cycle. Our method can achieve mean testing errors of 0.176% and 8.72% for capacity estimation and remaining useful life (RUL) prediction, respectively. Additionally, the proposed framework can leverage the knowledge from two other well-known battery datasets, with a variety of charge configurations and a different battery chemistry respectively, to reliably estimate the capacity (0.328%/0.193%) and predict the RUL (9.80%/9.90%) of our cells. This study allows end users to tailor battery consumption plans and motivates manufacturers to improve battery designs.
000 charge–discharge cycles—the largest dataset to our knowledge of its kind, and develop a transfer learning framework to realize real-time personalized health status prediction for unseen battery discharge protocols, at any charge–discharge cycle. Our method can achieve mean testing errors of 0.176% and 8.72% for capacity estimation and remaining useful life (RUL) prediction, respectively. Additionally, the proposed framework can leverage the knowledge from two other well-known battery datasets, with a variety of charge configurations and a different battery chemistry respectively, to reliably estimate the capacity (0.328%/0.193%) and predict the RUL (9.80%/9.90%) of our cells. This study allows end users to tailor battery consumption plans and motivates manufacturers to improve battery designs.
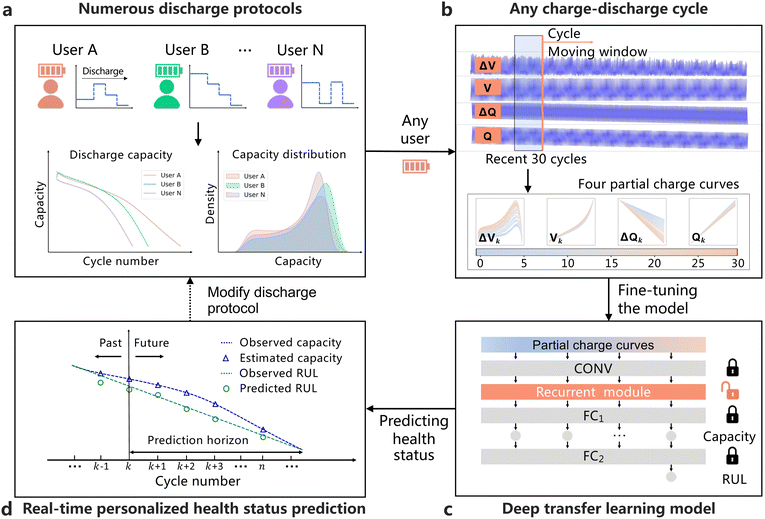
Broader contextWith the rapid development of lithium-ion batteries, battery health status management has attracted more and more attention in recent years. For safety and sustainability considerations, each end-user would like to know the current battery health status (e.g., the state of health and remaining useful life) of electronic devices (e.g., electrified vehicles). However, due to personalized battery usage preferences, building a unified and reliable health status prediction model is hindered by cell-to-cell and cycle-to-cycle discharge variabilities. To solve this realistic issue, we develop a transfer learning framework to customize personalized battery health status prediction for each end-user. Using this framework, each end-user could accurately obtain the current battery health status by inputting the cycling data in the recent 30 cycles and then tailor battery consumption plans accordingly. Our approach has been demonstrated by LFP/graphite cells and NMC/graphite cells, and experimental results showcase the effectiveness and generalizability of the proposed approach. |
Introduction
Lithium-ion batteries have experienced over 50 years of revolution and nowadays have been successfully used in cell phones, laptops, electrified vehicles, etc.1–5 In recent years, charge protocol optimization of lithium-ion batteries has attracted substantial research.6,7 In comparison, discharge protocols of lithium-ion batteries have been less extensively investigated due to person-to-person usage preference. Real-time prognostic of the battery health status (i.e., capacity and remaining useful life (RUL)) for a personalized discharge protocol would provide end-users with a safe and scheduled usage scenario8 and enhance battery health management.9 Also, battery health status prediction techniques could further facilitate the development of battery self-repairing materials10–12 and recycling technologies13–15 and guide the next generation of battery production.16–18 For example, end-users could estimate the current health status of a battery after a high-power discharge protocol and draw up a new usage plan to extend the battery RUL according to the current estimation. Likewise, manufacturers could test the real-time performance of batteries after undergoing extreme discharge to improve the design of batteries. Meanwhile, recyclers could assess the current health status of batteries to determine if they are best suited for re-use (and if so, for which applications), remanufacturing or recycling.19 Consequently, customizing personalized battery health management is an urgent requirement for end-users, manufacturers and recyclers in the field of energy technology.20–22Two significant challenges stand in the way of developing a reliable real-time personalized health status prediction strategy: variability of discharge protocols and real-time requirements.23–25 First, due to the high usage variability, as well as different battery chemistries and ambient temperatures, the same discharge protocols and degradation patterns rarely exist in the historical database. Thus, if the health status of a new discharge protocol is predicted by other historical protocols directly, there is a possibility of bias.26 Previous studies focused on either performing general prediction by extracting common high-performance features from early-cycle battery data,6,27 or proposing case-by-case prediction using at least 25% degradation data of each battery.28–33 On the one hand, general prediction approaches using common features produced conceivable prediction errors due to the neglect of the personalized degradation pattern of each battery. On the other hand, the case-by-case prediction approaches had a contradiction between the amount of data and the prediction accuracy. Second, health status prediction is required to be real-time, so that end-users are continuously informed about the current battery health condition and may schedule maintenance or replacements accordingly.34 Previous research focused on predicting the battery health status at pre-defined cycle(s),6,27–33 which does not match real-time requirements.
Recent research interest in transfer learning aims to improve the performance of specific learners by transferring the knowledge from other different but related learners.35,36 As an important branch of transfer learning, deep transfer learning requires no manual feature engineering.37 Deep transfer learning methods have been successfully applied to various applications, such as medical diagnosis,38,39 chemistry,40,41 natural language processing,42,43 and smart manufacturing.44,45 An increasing number of deep transfer learning technologies have been employed for health status prediction of lithium-ion batteries. However, these studies require at least 50% prior degradation data of the target battery to perform a curve matching from the historical database, without considering the necessity of real-time prediction along with battery degradation.46–48 There still exists a considerable opportunity to fill the gap between deep transfer learning and real-time personalized health status prediction for a wide range of usage patterns.
In this work, we design a transferable deep network to achieve personalized and real-time health status prediction of lithium-ion batteries, using partial cycling data of the recent 30 cycles at any interested cycle (Fig. 1). To this end, we construct an experimental platform containing 77 lithium-iron-phosphate (LFP)/graphite cells with cycle lives ranging from 1100 to 2700 cycles. The cells undergo different multi-stage discharge protocols to approximate usage variability, thus acquiring 146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 122 charge–discharge cycles in total. Our method achieves mean testing errors of 0.176% and 8.72% for capacity estimation and RUL prediction respectively, at all charge–discharge cycles. In addition, we also perform two further tasks to predict the health status of the testing cells in our dataset by transferring the degradation knowledge from two other datasets, which have different charge–discharge conditions and a different battery chemistry from our dataset, respectively. Mean testing errors of 0.328% (0.193%) and 9.80% (9.9%) for capacity estimation and RUL prediction are achieved in two further tasks. These results illustrate the effectiveness and generalizability of the deep transfer learning framework on health status prediction according to the personalized usage patterns.
122 charge–discharge cycles in total. Our method achieves mean testing errors of 0.176% and 8.72% for capacity estimation and RUL prediction respectively, at all charge–discharge cycles. In addition, we also perform two further tasks to predict the health status of the testing cells in our dataset by transferring the degradation knowledge from two other datasets, which have different charge–discharge conditions and a different battery chemistry from our dataset, respectively. Mean testing errors of 0.328% (0.193%) and 9.80% (9.9%) for capacity estimation and RUL prediction are achieved in two further tasks. These results illustrate the effectiveness and generalizability of the deep transfer learning framework on health status prediction according to the personalized usage patterns.
Experimental data
We developed a battery degradation experiment (Fig. S1, ESI†) with 77 cells (LFP/graphite A123 APR18650M1A, 1.1 Ah nominal capacity and 3.3 V nominal voltage; see more details in the Methods section) considering 77 different multi-stage discharge protocols, but an identical fast-charging protocol (Fig. S2 and Table S1, ESI†) in two thermostatic chambers at 30 °C. Fig. 2a shows the discharge capacity as a function of cycle number, where the colours of curves are scaled by the cycle lives of cells ranging from 1100 to 2700 cycles (average 1898 with a standard deviation of 387). The dataset contains 146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 122 discharge cycles in total, which is the largest dataset considering various discharge protocols as far as the authors know. Fig. 2b shows the joint distribution of cycle life and initial capacity for all 77 cells, where the distributions of observed cycle life and initial discharge capacity are both close to normal distributions with large standard deviations, and the colours are scaled by the quantity density. For ease of validation, the cells are divided into two parts: 55 training cells (55 discharge protocols) and 22 testing cells (22 discharge protocols, see Table S1, ESI†).
122 discharge cycles in total, which is the largest dataset considering various discharge protocols as far as the authors know. Fig. 2b shows the joint distribution of cycle life and initial capacity for all 77 cells, where the distributions of observed cycle life and initial discharge capacity are both close to normal distributions with large standard deviations, and the colours are scaled by the quantity density. For ease of validation, the cells are divided into two parts: 55 training cells (55 discharge protocols) and 22 testing cells (22 discharge protocols, see Table S1, ESI†).
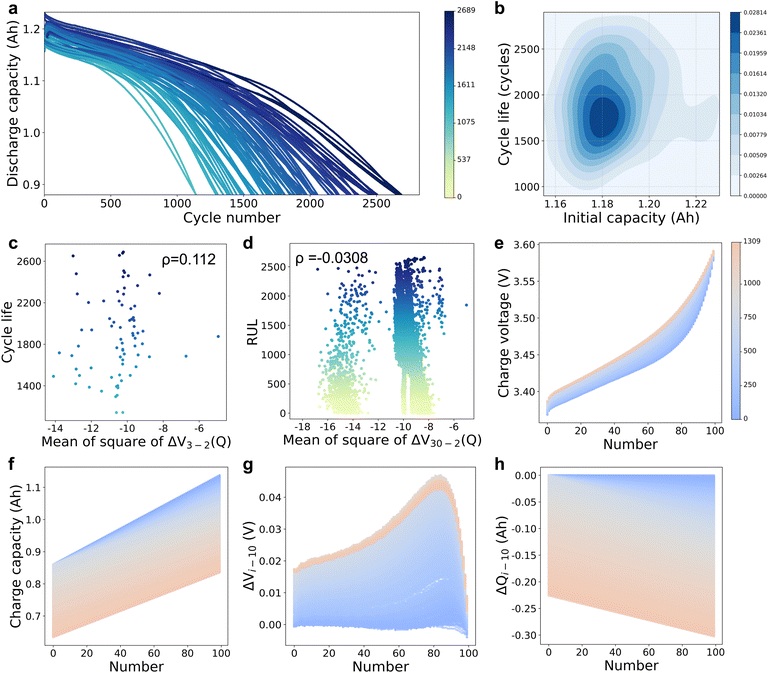 | ||
| Fig. 2 (a) Discharge capacity of 77 LFP/graphite cells with completely different discharge protocols. The colour of each curve is scaled by the battery cycle life. (b) The joint distribution of cycle life and initial capacity, where the cycle life and initial capacity both have large standard deviations. Two handcrafted features have poor predictive performance: (c) cycle life as a function of a common handcrafted feature6 (i.e., mean of square of ΔV3−2(Q); ΔV3−2(Q) is the difference of the charge voltage curves between the 3rd cycle and the 2nd cycle), (d) RUL as a function of mean of square of ΔV30−2(Q) (ΔV30−2(Q) represents the difference of the charge voltage curves between the 30th cycle and the 2nd cycle in every 30 cycles). Four important feature curves are collected from 80% SOC to the first 3.6 V for a representative cell: (e) charge voltage curves, (f) charge capacity curves, (g) difference of charge voltage curves between each cycle and the 10th cycle and (h) difference of charge capacity curves between each cycle and the 10th cycle. The feature curves are interpolated to a fixed length of 100 points and the colour of each curve is scaled by the cycle number in the representative cell. | ||
We also employ two other well-known public datasets which contain different charge–discharge configurations, ambient temperatures and battery chemistries from our dataset. Among them, the first public dataset comprises more than 100 LFP/graphite cells (see Fig. S3a, ESI† for degradation curves) from Severson et al.27 and includes 72 different charge protocols, but an identical discharge protocol, which focuses on charge variability other than discharge reflected in our dataset. The second public dataset developed by Preger et al.49 contains 22 lithium–manganese–cobalt-oxide (NMC)/graphite cells (see Fig. S3b, ESI† for degradation curves), where the NMC/graphite cells were cycled at three ambient temperatures (15 °C, 25 °C and 35 °C) with an identical charge protocol, but four different one-stage discharge protocols (0.5C, 1C, 2C and 3C). It could be found that the second public dataset and our dataset have different battery chemistries, cycling ambient temperature and discharge configurations. More details of the two public datasets are described in the Methods section. In this work, cross-scenario and cross-chemistry battery health status predictions are investigated when training data are limited. To be specific, the two public datasets are used for training two deep models, which will then be applied to our testing cells for real-time personalized health status prediction, respectively.
Deep transfer learning approach
We develop a deep transfer learning approach to perform real-time personalized battery health status predictions, by leveraging the knowledge from other different, but related battery degradation data. The deep transfer learning model is first pre-trained using multi-cycle sensory data of the training cells and will be transferred online to any charge–discharge cycle of any testing protocol. Our choice of a deep transfer learning model requires no high-performance handcrafted features that are hard to satisfy all charge–discharge cycles and discharge protocols. In addition, the deep transfer learning model is flexible to rapidly adapt to a new working condition by fine-tuning specific network layers dynamically.To alleviate the dependence on the battery charge ranges, partial charge data with a commonly-used charge range from 80% SOC to the first 3.6 V is used as input for the deep learning model that could automatically extract valuable features from charge data without complicated feature engineering. Four partial feature curves in the charging process are employed, i.e., charge voltage curve (V, Fig. 2e), charge capacity curve (Q, Fig. 2f), the difference of the charge voltage curve between each cycle and the 10th cycle (ΔVi−10 = Vi − V10, Fig. 2g) and the difference of the charge capacity curve between each cycle and the 10th cycle (ΔQi−10 = Qi − Q10, Fig. 2h). The four feature curves are fitted as a function of time and linearly interpolated to a fixed length of 100 for the convenience of model manipulations. Among the four feature curves, ΔQi−10 is inspired by Severson et al.,27 who proved that a high-performance feature (log variance of ΔQ100−10(V), i.e., the difference of the discharge capacity curves between the 100th cycle and the 10th cycle) had a strong correlation with the battery cycle lives (the absolute value of correlation coefficient is 0.92). ΔVi−10 is another important feature curve inspired by Jiang et al.,6 who proved that ΔVb−a(Q) (difference of the charge voltage curves between the bth cycle and the ath cycle) also contained high-performance features (e.g., mean of square of ΔV3−2(Q)) related to battery cycle life. However, the features proposed by previous studies are not suitable for our work due to the following three reasons: (1) the correlation is weak between the two-cycle features (e.g., mean of square of ΔV3−2(Q)) and cycle life (ρ = 0.1120, Fig. 2c; see Fig. S4, ESI† for the performance of different statistical features); (2) the correlation is weak between the two-cycle features (e.g., mean of square of ΔV30−2(Q), the 30th cycle and the 2nd cycle in every 30 cycles) and RUL (ρ = −0.0308, Fig. 2d; see Fig. S5, ESI† for the performance of different statistical features); and (3) the discharge current frequently changes during the battery discharge process (Fig. S2b, ESI†), thus the discharge feature curves in shared ranges are hard to be collected. Thus, we enrich the model input information using four charge feature curves at the current cycle combined with the previous 29 cycles (a total of 30 cycles). In particular, 10 cycles are taken from 30 cycles (sampling 1 cycle every 3 cycles in 30 cycles) for the ease of calculation. Thus, each input sample has a shape of 1 × 4 × 100 × 10 (i.e., 1 sample, 4 feature curves, 100 sampling points and 10 cycles) and its label consists of 10 battery discharge capacities at the corresponding 10 cycles and 1 RUL value at the current cycle.
As shown in Fig. 3, we design a deep learning model which consists of one convolutional module, one recurrent module and two fully connected modules. Among them, the convolutional module is used as a feature extractor to automatically extract valuable intra-cycle features from four feature curves and the recurrent module is used as an encoder to learn cycle-to-cycle knowledge. Additionally, two fully connected modules are designed for capacity estimation and RUL prediction, respectively. Two stages, the model pre-training stage and the model transfer stage, are designed in the transfer learning model. At the model pre-training stage, the deep learning model is pre-trained using input feature curves and labeled health status of training cells where each cell has thousands of charge–discharge cycles. Then, the model transfer stage aims to realize a real-time personalized health status prediction at any cycle of any new discharge protocol, through inputting feature curves in the recent 10 charge–discharge cycles (after resampling). In particular, input feature curves and labeled discharge capacities in the first 9 cycles are employed to fine-tune the recurrent module online by freezing other modules (see Note S1, ESI†). Then, the capacity and RUL at the current charge–discharge cycle are estimated and predicted online by the fine-tuned model, respectively. More computational details are described in the Methods section. With the proposed deep transfer learning model, any end-user with a personalized discharge pattern could obtain the battery health status at any charge–discharge cycle and make new usage plans accordingly.
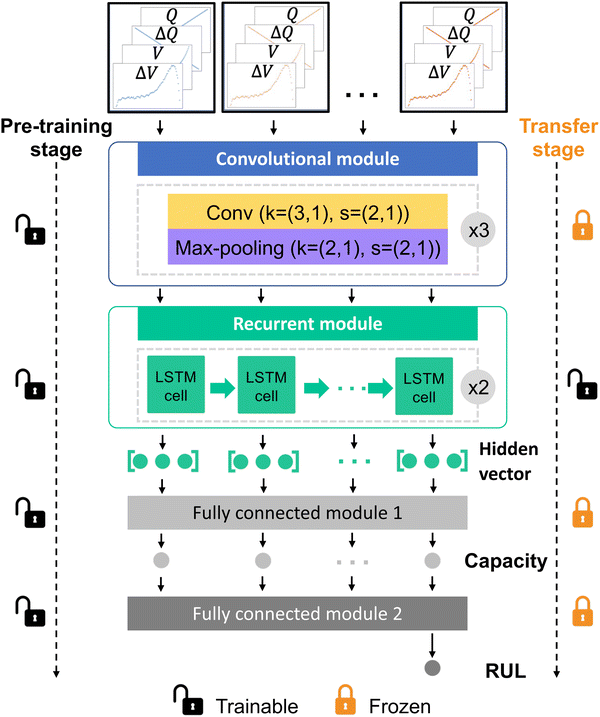 | ||
| Fig. 3 The proposed model comprises an offline model pre-training stage and an online model transfer stage. The offline model pre-training stage aims to mine the implicit relationship between four partial charge curves and health status using a deep learning model that consists of a convolutional module, a recurrent module and two fully connected modules. The convolutional module is used to extract intra-cycle features from each cycle using a stack of three convolutional layers and max-pooling layers, and the recurrent module is used to learn the cycle-to-cycle knowledge using two long short-term memory (LSTM) layers. The output of the recurrent module is then mapped to 10 capacities using the first fully connected module. The mapped capacities are finally used to predict the RUL at the current cycle using another fully connected module. The model parameters configuration and the output sizes of each layer are detailed in Table S2 (ESI†). The online model transfer stage aims to real-time predict the capacity and RUL at any cycle of any discharge protocol, by fine-tuning the recurrent module while freezing other modules using four partial charge curves and capacities at the previous 9 charge–discharge cycles. | ||
Based on the proposed deep transfer learning approach and three datasets, we investigate three different tasks for real-time and personalized prediction of 22 unseen discharge protocols via transferring degradation knowledge from (1) 55 cells with different discharge protocols in our training dataset (denoted by Task A); (2) cells with different charge–discharge configurations in the first public dataset (denoted by Task B); and (3) cells with a different battery chemistry (NMC/graphite) in the second public dataset (denoted by Task C). In all cases, the 22 unseen discharge protocols are taken from our dataset. Three evaluation metrics, i.e., root mean squared error (RMSE, see eqn (4)), coefficient of determination (R2, see eqn (5)) and mean absolute percentage error (MAPE, see eqn (6)), are employed to evaluate the prediction performance of battery health status.
Results and discussion
Performance of the deep transfer learning model
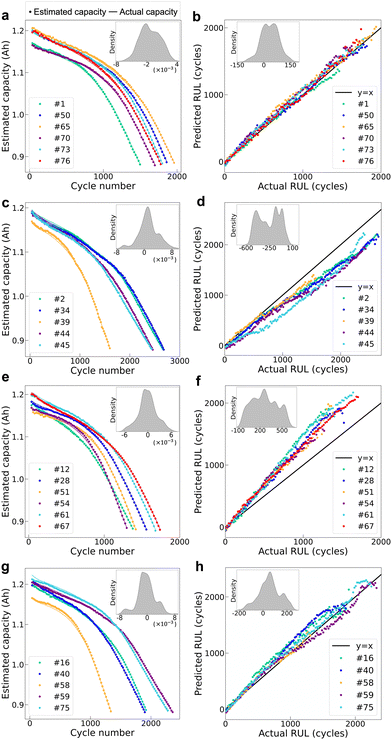
We first seek to customize personalized battery health status prediction for each new end-user based on historical data provided by other end-users, where cells for end-users have the same chemistry, but different usage protocols. To this end, we investigate a task (Task A) of battery health status prediction among discharge protocols based on our LFP/graphite battery dataset and the proposed deep transfer learning approach. For the sake of battery health status prediction at any charge–discharge cycle for new discharge protocols, we predict the battery health status at all charge–discharge cycles of 22 representative testing protocols, respectively, via transferring the health status knowledge from the other 55 protocols. Finally, the results for capacity estimation and RUL prediction of the testing protocols are illustrated as follows:| Capacity | RMSE (mA h) | R 2 | MAPE (%) |
|---|---|---|---|
| a Results for 22 testing protocols by transferring degradation knowledge from cells in our dataset. b Results for 22 testing protocols by transferring the degradation knowledge from cells in a well-known dataset27 with different charge–discharge configurations. c Results for 22 testing protocols by transferring the degradation knowledge from cells in another well-known dataset49 with a different battery chemistry. | |||
| Task Aa | 2.57 | 0.999 | 0.176 |
| Task Bb | 4.65 | 0.997 | 0.328 |
| Task Cc | 3.08 | 0.999 | 0.193 |
| RUL | RMSE (cycles) | R 2 | MAPE (%) |
|---|---|---|---|
| Task A | 186 | 0.804 | 8.72 |
| Task B | 240 | 0.770 | 9.80 |
| Task C | 216 | 0.794 | 9.90 |
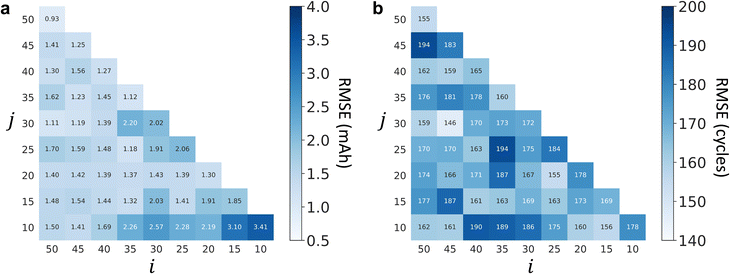
It should be noted that the cycling data in the recent 30 cycles and a sampling rate of 3 cycles are used to perform health status prediction by giving consideration to a smaller number of input cycles, smaller computation costs and smaller prediction errors. To investigate the possibility and robustness of the proposed model using different numbers of input cycles, we perform additional analysis to compare the testing performance on different numbers of input cycles after a sparse sampling. Fig. 5 shows the prediction errors using different numbers of sparse input cycles for capacity estimation and RUL prediction and the results reveal that our choice of the number of input cycles in this work is general, but not the best, and the proposed model with a smaller number of input cycles is also feasible (for instance, i = 20 and j = 10 achieve RMSEs of 2.19 mA h and 160 cycles for capacity estimation and RUL prediction, respectively); the proposed model is insensitive to the number of input cycles as well as the sampling rates (average 1.65 mA h with a standard deviation of 0.50 mA h for capacity estimation; average 171 cycles with a standard deviation of 11 cycles for RUL prediction), thus demonstrating the robustness of the proposed deep transfer learning model.
In summary, the above results illustrate the predictive ability of our approach for unseen discharge protocols, showcasing the high flexibility and effectiveness to customize personalized battery health management for each end-user.
Performance by transferring degradation knowledge from cells with different charge–discharge configurations
We then seek to investigate the possibility of customizing personalized battery health status prediction for each end-user by generalizing the operating conditions of training cells. In this case, we assume that the database has no or few historical data of training cells provided by other end-users, where the insufficient training data are unable to train a reliable deep learning model due to the overfitting phenomenon.51 To this end, we develop a task (Task B) of personalized battery health status prediction between two different datasets, including one public dataset27 and our dataset which have identical chemistry (LFP/graphite), but different charge–discharge protocols. To be specific, we attempt to predict the battery health status at all charge–discharge cycles of the same 22 representative testing protocols in our dataset, respectively, via transferring the degradation knowledge from cells in the public dataset. Table 1 (see Task B) shows the evaluation metrics for the testing protocols, where our model achieves an overall RMSE of 4.65 mA h (240 cycles), MAPE of 0.328% (9.8%) and R2 of 0.997 (0.77) for capacity estimation and RUL prediction, respectively. Moreover, we benchmark the performance of our model against the deep learning model without a model transfer stage (RMSEs of 85.3 mA h and 760 cycles for capacity estimation and RUL prediction, respectively, see Tables S5 and S6 and Fig. S6 Task B, ESI†) and the elastic net with two-cycle handcrafted features (RMSE of 607 cycles for RUL prediction, see Table S6 and Fig. S6b Task B, ESI†), revealing that the performance of our approach is significantly superior to the benchmark models and the model transfer stage is able to substantially improve the prediction accuracy. It is noted that the prediction error of our model in this task is slightly higher when compared with Task A, which is attributed to quite different charge–discharge configurations and battery testing platforms between the two datasets. Nevertheless, our model still obtains satisfactory results (Fig. S7–S10, ESI†) of personalized battery health status prediction even if the training cells and the testing cells are from cross-scenario datasets. The above results further underscore the ability of our approach for personalized battery health status prediction.Performance by transferring knowledge from cells with different chemistry
We further ask whether our model has the capability of customizing personalized battery health status prediction for each end-user based on training cells with a different battery chemistry. For this purpose, we design a task (Task C) of personalized health status prediction for LFP/graphite cells in our dataset by transferring the degradation knowledge from NMC/graphite cells in another public dataset.49 Notably, the nominal capacity, nominal voltage, ambient temperature, and the charge–discharge configuration are different between the NMC/graphite dataset and our LFP/graphite dataset, resulting in significantly different battery degradation trajectories, where the LFP/graphite cells and NMC/graphite cells have increasing and decreasing aging speeds, respectively (see Fig. 2a and Fig. S3b, ESI†). Although the cross-chemistry health status prediction task seems challenging, our model achieves an overall RMSE of 3.08 mA h (216 cycles), MAPE of 0.193% (9.9%) and R2 of 0.999 (0.794), for capacity estimation and RUL prediction, respectively (see Table 1, Fig. S11–S14, ESI†). Furthermore, our approach also significantly outperforms the benchmark models, i.e., the deep learning model without a model transfer stage (RMSEs of 92.4 mA h and 832 cycles for capacity estimation and RUL prediction, respectively, see Tables S7 and S8 and Fig. S6 Task C, ESI†) and the elastic net with two-cycle handcrafted features (RMSE of 749 cycles for RUL prediction, see Table S8 and Fig. S6b Task C, ESI†), thus highlighting the feasibility of the proposed method for the cross-chemistry battery health status prediction task. Additionally, the prediction error in this task is slightly higher than Task A, which indicates that our approach is not heavily limited by the battery chemistries. The above results further demonstrate the effectiveness and generalizability of the proposed deep transfer learning method, even under challenging cross-chemistry conditions.It is worth mentioning that the model transfer stage fine-tunes the recurrent module to adapt to the personalized battery usage conditions. A seemingly natural question is whether we can fine-tune the convolutional module at the model transfer stage. To further evaluate our proposed transfer strategy, we investigate the performance of personalized battery health status prediction by fine-tuning the convolutional module rather than the recurrent module at the model transfer stage (see Table S9, ESI†). The results for the three tasks consistently show that fine-tuning the convolutional module yields performance degradation in terms of both capacity estimation and RUL prediction, demonstrating that the cycle-to-cycle knowledge is strongly related to the battery health status and that the recurrent module could effectively learn new cycle-to-cycle knowledge after fine-tuning the model.
The above results present the capability of the proposed approach to LFP/graphite cells. Beyond LFP/graphite cells, our approach can be applied to NMC/graphite cells.49 To this end, we perform two additional experiments for personalized health status prediction of NMC/graphite cells by transferring degradation knowledge from NMC/graphite cells and LFP/graphite cells, respectively. On the one hand, we use a cross-validation strategy to predict the health status for four discharge protocols (constant-current discharge rates of 0.5C, 1C, 2C and 3C) of NMC/graphite cells, where one protocol is used for validation while the other three protocols are used to pre-train the deep model each time. We achieve overall RMSEs of 12.7 mA h and 98 cycles for capacity estimation and RUL prediction, respectively (Fig. S15, ESI†). On the other hand, we also predict the health status for the same four discharge protocols of NMC/graphite cells by transferring degradation knowledge from the 55 training LFP/graphite cells in our dataset. Overall RMSEs of 19.38 mA h and 103 cycles are achieved for capacity estimation and RUL prediction, respectively (Fig. S16, ESI†). The results further showcase the effectiveness of our approach for personalized battery health status prediction to the NMC/graphite chemistry other than LFP/graphite chemistry.
Conclusions
In summary, we have successfully customized the personalized battery health status prediction for each discharge protocol using the developed deep transfer learning approach. This approach achieves satisfactory effectiveness, generalizability and flexibility on a variety of unseen usage protocols using cycling knowledge from cells with completely different usage protocols, charge–discharge configurations, and battery chemistries. Using the proposed approach, the battery management system could rapidly adapt to new operating conditions and predict the battery health status for every usage preference, thus end-users can take immediate action to ensure safety or draw up new usage plans to extend battery RUL. This approach could extend to other discharge scenarios, such as constant-current discharging and stochastic discharging, as well as to fast-charging scenarios, such as multi-step charging, pulsed charging and constant-power charging. Furthermore, the proposed approach could also be scalable to quasi-solid-state batteries, solid-state batteries and post-lithium-ion batteries52 (e.g., lithium–sulfur batteries, sodium-ion batteries, etc.).While the approach has achieved satisfactory performance in this work, there are still some limitations. First, the approach works when cycling data in a fixed charge range are available, but the stochastic charge process (random charge ranges) which approximates real application is not investigated. Then, the approach requires the recent 30 cycles to predict the cell health status, with the result that the health status in the first several cycles cannot be predicted. Also, our approach relies on the quality of cycling data in recent cycles, where some singular measurement data will certainly influence the prediction results. In addition, our work simplifies the operating conditions in real applications, where more practical operating scenarios such as different temperatures, depth of discharges and discharge powers are not included in our dataset, thus further investigations are still required. Nevertheless, this approach obtains considerable generalizability in a variety of usage scenarios and has the potential to be extended to real applications.
Methods
Data generation
Three battery datasets are used in this work. Among them, the first dataset and the second dataset comprise LFP/graphite A123 APR18650M1A cells, which have a nominal capacity of 1.1 Ah and a nominal voltage of 3.3 V. The third dataset contains NMC/graphite LG 18650HG2 cells (3 Ah nominal capacity and 3.6 V nominal voltage) that have a different battery chemistry compared with the other two datasets. The first dataset developed by our laboratory contains 77 cells with various discharge protocols, which is mainly used to support this work; the second dataset and the third dataset are used to discuss a scenario when the training cells are scarce so that other datasets with a different battery chemistry or different charge–discharge configurations will be employed to pre-train the model; the third dataset is also used to demonstrate the effectiveness and generalizability of proposed approach on a different battery chemistry.All cells in the first dataset were tested in an 80-channel CT-4008 Neware battery tester at a constant temperature of 30 °C (Fig. S1, ESI†). The cells were cycled with an identical fast-charging protocol, but completely different multi-stage discharge protocols (Fig. S2 and Table S1, ESI†), resulting in different cycle lives ranging from 1100 to 2700 cycles. The charge process consisted of a constant-current (CC) charge of 5C from 0% to 80% state of charge (SOC), a CC charge of 1C from 80% SOC to 3.6 V and a constant–voltage (CV) charge until 100% SOC with a current cutoff of C/20. The discharge stage comprised four CC discharge steps, where the first three steps were intended to discharge from 100% to 60% SOC, from 60% to 40% SOC and from 40% to 20% SOC, respectively; the last step was intended to discharge from 20% to 0% SOC using a CC of 1C with a voltage cutoff of 2 V. For example, discharge protocol with a 5C–4C–3C–1C protocol consisted of a 5C discharge from 100% to 60% SOC, a 4C discharge from 60% to 40% SOC, a 3C discharge from 40% to 20% SOC and a 1C discharge from 20% to 0% SOC. It should be noted that 1C is 1.1 A in this dataset, or the current is required to fully (dis)charge the nominal capacity (1.1 A h) in 1 h. The rest time between two arbitrary steps was 30 s. Voltage, current and capacity were collected along with battery cycling until the maximum capacity first reached 80% of nominal capacity (i.e., failure threshold). A total of 80 cells were cycled with 80 different discharge protocols and 3 of 80 cells were excluded from the dataset due to sudden faults before degradation failure. In this work, 55 of 77 cells are used to pre-train a benchmark deep learning model and the other 22 cells are used to validate the effectiveness of the proposed method.
The second dataset is a famous public dataset, where cells in the dataset were cycled with 72 different fast-charging protocols, but with an identical discharge protocol at a constant temperature of 30 °C. The cells were charged with one-step or two-step CC protocols from 0% to 80% SOC, a uniform CC of 1C charge step to 3.6 V and a uniform CV charge with a current cutoff of C/50. Then, the cells were discharged with a uniform CC discharge of 4C to 2 V. The cells have cycle lives ranging from 150 to 2,300 cycles, and more details of the dataset could be found in the study proposed by Severson et al.27 In this work, all cells in the dataset are used to pre-train a deep learning model, then the knowledge would be transferred to predict the health status of 22 testing cells in the first dataset.
The third dataset developed by Preger et al.49 is also a public dataset. A total of 22 NMC/graphite cells were cycled with an identical charge protocol, but with four different discharge protocols (0.5C, 1C, 2C and 3C) at three different ambient temperatures (15 °C, 25 °C and 35 °C). In each charge–discharge cycle, the cells were charged with the same CC charge rate of 0.5C from 0% SOC to 4.2 V, followed by a CV charge. Then, the cells were CC discharged with four different discharge protocols from 100% SOC to a voltage cutoff of 2 V. The cells have cycle lives ranging from 380 cycles to 1321 cycles, and more details of the dataset could be found in the study proposed by Preger et al.49 In the personalized health status prediction experiment for LFP/graphite cells by transferring degradation knowledge from NMC/graphite cells, we pre-train a deep learning model using all the 22 NMC/graphite cells and then transfer it to our 22 testing LFP/graphite cells for personalized health status prediction. In the personalized health status prediction experiment for NMC/graphite cells by virtue of other NMC/graphite cells, we use a cross-validation strategy to predict the health status by four discharge protocols (four NMC/graphite cells), where one protocol is used for validation, while the other three protocols are used to pre-train the deep model each time. Additionally, in the personalized health status prediction experiment for NMC/graphite cells by virtue of LFP/graphite cells, the 55 training LFP/graphite cells in our dataset are used to pre-train the deep model, which would then be deployed to the same four NMC/graphite cells.
Each battery could last for thousands of cycles before failure, which would waste much data storage space if voltage/current sensors continuously collect cycling data. Compared with all-time cycling data, partial cycling data is more practical and economical in real applications, i.e., voltage/current sensors only collect partial data corresponding to a specific range of SOC. To reduce the dependence on the data sampling time, charge voltage and charge capacity collected from a commonly-used range (80% SOC to the first 3.6 V (4.2 V for NMC/graphite cells)) are utilized as the model input in this work. To unify the length of sampling data, all charge curves from 80% SOC to the first 3.6 V are fitted as a function of sampling time and evaluated at 100 linearly spaced points. These uniformly sized samples enable direct model input and data manipulations like voltage/capacity subtraction.
Model development
This section introduces the proposed deep transfer learning model (see Table S2, ESI† for more details about model parameters). In essence, given a sample xi ∈ R1×k×s×p at the ith cycle that includes cycling data from the (i − p + 1)th cycle to the ith cycle, s time steps at each cycle and k feature channels, the deep transfer learning model takes the data xi as input and outputs p discharge capacities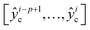 as well as one RUL
as well as one RUL 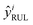 , which is represented by the following two equations
, which is represented by the following two equations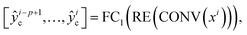 | (1) |
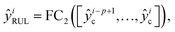 | (2) |
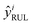 according to the estimated discharge capacities.
according to the estimated discharge capacities.
At the model training stage, M training samples (each sample has p actual discharge capacities 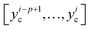 and one RUL yiRUL) are used to compute the mean squared error (MSE) loss between model outputs and actual measurements
and one RUL yiRUL) are used to compute the mean squared error (MSE) loss between model outputs and actual measurements
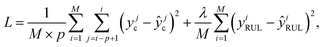 | (3) |
At the fine-tuning stage, the capacity and RUL at the current cycle are required to be estimated and predicted using the pre-trained model. To implement personalized prediction for the testing cells, random d actual discharge capacities in the previous p−1 cycles are used to fine-tune the model weights in RE, and the other p−1−d actual capacities are used to validate the performance after fine-tuning. Only weights in RE are updated because RULs are determined by multi-cycle knowledge. We also find that the model weights in FC2 are general for all discharge protocols, so the pre-trained weights in FC2 are also frozen while fine-tuning the model.
Once the estimated capacities and predicted RULs are obtained, three standard metrics, i.e., RMSE, R2 and MAPE, are chosen to evaluate the model performance. RMSE for capacity estimation or RUL prediction is defined as
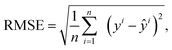 | (4) |
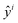 is the estimated capacity or predicted RUL at the ith cycle and yi is the corresponding observed capacity or RUL. n is the total number of cycles in a single cell. R2 is defined as
is the estimated capacity or predicted RUL at the ith cycle and yi is the corresponding observed capacity or RUL. n is the total number of cycles in a single cell. R2 is defined as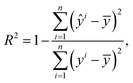 | (5) |
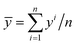 ; other variables are defined as above. In addition, MAPE is defined as
; other variables are defined as above. In addition, MAPE is defined as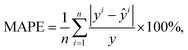 | (6) |
The operating system for algorithm implementation is Ubuntu 18.04; NVIDIA GeForce RTX 3090 GPU with 24 GB memory is used to accelerate the model training; CUDA 11.1 is deployed for the GPU-accelerated applications; Python 3.8.8 is used to process data and analyse data, where the pandas, Numpy and scikit-learn packages are employed to load, manipulate and process data; the PyTorch framework is used to construct the deep learning model.
Data and code availability
The developed dataset with 77 LFP/graphite cells, including 77 discharge protocols, has been deposited at the Mendeley Data53 at https://doi.org/10.17632/nsc7hnsg4s.2; the first public dataset27 with 72 fast-charging protocols is available at https://data.matr.io/1/projects/5c48dd2bc625d700019f3204; and the second public dataset49 with 22 NMC/graphite cells is available at https://www.batteryarchive.org/list.html. The code for personalized battery health status prediction is available at the GitHub54 at https://doi.org/10.5281/zenodo.6827566.Author contributions
Conceptualization, Y. Y. and G. M.; data curation, G. M., S. X. and Y. Y.; formal analysis, G. M., Y. S. and X. Y.; funding acquisition, Y. Y.; methodology, G. M. and S. X.; project administration, Y. H., H. D. and Y. Y.; software, G. M., S. X. and Y. Y.; supervision, Y. Y.; validation, G. M. and B. J.; visualization, G. M., S. X. and C. C.; writing – original draft, G.M., C. C., B. J., T. Y. and Y. Y.; and writing – review & editing, G. M., S. X., C. C. and Y. Y.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the National Key R&D Program of China under Grant 2018YFB1701202, and in part by the National Natural Science Foundation of China under Grant 92167201.References
- J. B. Goodenough and Y. Kim, Chem. Mater., 2010, 22, 587–603 CrossRef CAS.
- D. L. Wood, J. Li and S. J. An, Joule, 2019, 3, 2884–2888 CrossRef.
- Z. Deng, Z. Huang, Y. Shen, Y. Huang, H. Ding, A. Luscombe, M. Johnson, J. E. Harlow, R. Gauthier and J. R. Dahn, Joule, 2020, 4, 2017–2029 CrossRef CAS.
- G. Harper, R. Sommerville, E. Kendrick, L. Driscoll, P. Slater, R. Stolkin, A. Walton, P. Christensen, O. Heidrich, S. Lambert, A. Abbott, K. Ryder, L. Gaines and P. Anderson, Nature, 2019, 575, 75–86 CrossRef CAS PubMed.
- Y. Liu, Y. Zhu and Y. Cui, Nat. Energy, 2019, 4, 540–550 CrossRef.
- B. Jiang, W. E. Gent, F. Mohr, S. Das, M. D. Berliner, M. Forsuelo, H. Zhao, P. M. Attia, A. Grover, P. K. Herring, M. Z. Bazant, S. J. Harris, S. Ermon, W. C. Chueh and R. D. Braatz, Joule, 2021, 5, 3187–3203 CrossRef CAS.
- P. M. Attia, A. Grover, N. Jin, K. A. Severson, T. M. Markov, Y. H. Liao, M. H. Chen, B. Cheong, N. Perkins, Z. Yang, P. K. Herring, M. Aykol, S. J. Harris, R. D. Braatz, S. Ermon and W. C. Chueh, Nature, 2020, 578, 397–402 CrossRef CAS PubMed.
- P. Tagade, K. S. Hariharan, S. Ramachandran, A. Khandelwal, A. Naha, S. M. Kolake and S. H. Han, J. Power Sources, 2020, 445, 227281 CrossRef CAS.
- S. M. Rezvanizaniani, Z. Liu, Y. Chen and J. Lee, J. Power Sources, 2014, 256, 110–124 CrossRef CAS.
- R. Narayan, C. Laberty-Robert, J. Pelta, J. M. Tarascon and R. Dominko, Adv. Energy Mater., 2022, 12, 202102652 Search PubMed.
- F. Elizalde, J. Amici, S. Trano, G. Vozzolo, R. Aguirresarobe, D. Versaci, S. Bodoardo, D. Mecerreyes, H. Sardon and F. Bella, J. Mater. Chem. A, 2022, 10, 12588–12596 RSC.
- Z. Li, J. Fu, S. Zheng, D. Li and X. Guo, Small, 2022, 18, 2200891 CrossRef CAS PubMed.
- J. Neumann, M. Petranikova, M. Meeus, J. D. Gamarra, R. Younesi, M. Winter and S. Nowak, Adv. Energy Mater., 2022, 12, 2102917 CrossRef CAS.
- Z. Qin, Y. Xie, X. Meng, D. Qian, D. Mao, X. Ma, C. Shan, J. Chen, L. Wan and Y. Huang, Energy Storage Mater., 2022, 49, 360–369 CrossRef.
- J. Wang, Q. Zhang, J. Sheng, Z. Liang, J. Ma, Y. Chen, G. Zhou and H.-M. Cheng, Natl. Sci. Rev., 2022, 9, nwac097 CrossRef.
- M. A. A. M. Abdah, M. Mokhtar, L. T. Khoon, K. Sopian, N. A. Dzulkurnain, A. Ahmad, Y. Sulaiman, F. Bella and M. S. Su’ait, Energy Rep., 2021, 7, 8677–8687 CrossRef.
- T. Wang, J. Duan, B. Zhang, W. Luo, X. Ji, H. Xu, Y. Huang, L. Huang, Z. Song, J. Wen, C. Wang, Y. Huang and J. B. Goodenough, Energy Environ. Sci., 2022, 15, 1325–1333 RSC.
- A. Mannodi-Kanakkithodi and M. K. Y. Chan, Energy Environ. Sci., 2022, 15, 1930–1949 RSC.
- G. Harper, R. Sommerville, E. Kendrick, L. Driscoll, P. Slater, R. Stolkin, A. Walton, P. Christensen, O. Heidrich, S. Lambert, A. Abbott, K. Ryder, L. Gaines and P. Anderson, Nature, 2019, 575, 75–86 CrossRef CAS PubMed.
- M. Huang, Batteries, 2019, 5, 1–16 CrossRef.
- H. M. Chung, S. Maharjan, Y. Zhang and F. Eliassen, IEEE Trans. Intell. Transp. Syst., 2021, 22, 7760–7771 Search PubMed.
- Z. He, X. Shen, Y. Sun, S. Zhao, B. Fan and C. Pan, J. Energy Storage, 2021, 41, 102867 CrossRef.
- M. Alinejad, O. Rezaei, A. Kazemi and S. Bagheri, J. Energy Storage, 2021, 35, 102245 CrossRef.
- S. Khaleghi, Y. Firouz, J. Van Mierlo and P. Van den Bossche, Appl. Energy, 2019, 255, 113813 CrossRef.
- R. Xiong, Y. Zhang, J. Wang, H. He, S. Peng and M. Pecht, IEEE Trans. Veh. Technol., 2019, 68, 4110–4121 Search PubMed.
- Y. Yuan, G. Ma, C. Cheng, B. Zhou, H. Zhao, H. T. Zhang and H. Ding, Natl. Sci. Rev., 2020, 7, 418–429 CrossRef PubMed.
- K. A. Severson, P. M. Attia, N. Jin, N. Perkins, B. Jiang, Z. Yang, M. H. Chen, M. Aykol, P. K. Herring, D. Fraggedakis, M. Z. Bazant, S. J. Harris, W. C. Chueh and R. D. Braatz, Nat. Energy, 2019, 4, 383–391 CrossRef.
- Y. Zhang, R. Xiong, H. He and M. G. Pecht, IEEE Trans. Veh. Technol., 2018, 67, 5695–5705 Search PubMed.
- Z. Xue, Y. Zhang, C. Cheng and G. Ma, Neurocomputing, 2020, 376, 95–102 CrossRef.
- Q. Miao, L. Xie, H. Cui, W. Liang and M. Pecht, Microelectron. Reliab., 2013, 53, 805–810 CrossRef CAS.
- Y. Zhang, R. Xiong, H. He and M. G. Pecht, IEEE Trans. Ind. Electron., 2019, 66, 1585–1597 Search PubMed.
- G. Ma, Y. Zhang, C. Cheng, B. Zhou, P. Hu and Y. Yuan, Appl. Energy, 2019, 253, 113626 CrossRef.
- Y. Chang, H. Fang and Y. Zhang, Appl. Energy, 2017, 206, 1564–1578 CrossRef.
- Y. Li, K. Liu, A. M. Foley, A. Zülke, M. Berecibar, E. Nanini-Maury, J. Van Mierlo and H. E. Hoster, Renewable Sustainable Energy Rev., 2019, 113, 109254 CrossRef.
- F. Zhuang, Z. Qi, K. Duan, D. Xi, Y. Zhu, H. Zhu, H. Xiong and Q. He, Proc. IEEE, 2021, 109, 43–76 Search PubMed.
- S. J. Pan and Q. Yang, IEEE Trans. Knowl. Data Eng., 2010, 22, 1345–1359 Search PubMed.
- C. Tan, F. Sun, T. Kong, W. Zhang, C. Yang and C. Liu, Int. Conf. Artif. Neural Networks, 2018, 11141, 270–279 Search PubMed.
- R. K. Sevakula, V. Singh, N. K. Verma, C. Kumar and Y. Cui, IEEE/ACM Trans. Comput. Biol. Bioinf., 2019, 16, 2089–2100 Search PubMed.
- D. S. Kermany, M. Goldbaum, W. Cai, C. C. S. Valentim, H. Liang, S. L. Baxter, A. McKeown, G. Yang, X. Wu, F. Yan, J. Dong, M. K. Prasadha, J. Pei, M. Ting, J. Zhu, C. Li, S. Hewett, J. Dong, I. Ziyar, A. Shi, R. Zhang, L. Zheng, R. Hou, W. Shi, X. Fu, Y. Duan, V. A. N. Huu, C. Wen, E. D. Zhang, C. L. Zhang, O. Li, X. Wang, M. A. Singer, X. Sun, J. Xu, A. Tafreshi, M. A. Lewis, H. Xia and K. Zhang, Cell, 2018, 172, 1122–1131 CrossRef CAS PubMed.
- G. Pesciullesi, P. Schwaller, T. Laino and J. L. Reymond, Nat. Commun., 2020, 11, 4874 CrossRef CAS PubMed.
- J. S. Smith, B. T. Nebgen, R. Zubatyuk, N. Lubbers, C. Devereux, K. Barros, S. Tretiak, O. Isayev and A. E. Roitberg, Nat. Commun., 2019, 10, 2903 CrossRef PubMed.
- J. G. Makin, D. A. Moses and E. F. Chang, Nat. Neurosci., 2020, 23, 575–582 CrossRef CAS PubMed.
- B. McCann, J. Bradbury, C. Xiong and R. Socher, Adv. Neural. Inf. Process. Syst., 2017, 30, 6295–6306 Search PubMed.
- C. Sun, M. Ma, Z. Zhao, S. Tian, R. Yan and X. Chen, IEEE Trans. Ind. Inform., 2019, 15, 2416–2425 Search PubMed.
- B. Yang, Y. Lei, F. Jia and S. Xing, Mech. Syst. Signal Process., 2019, 122, 692–706 CrossRef.
- J. Ma, P. Shang, X. Zou, N. Ma, Y. Ding, J. Sun, Y. Cheng, L. Tao, C. Lu, Y. Su, J. Chong, H. Jin and Y. Lin, Appl. Energy, 2021, 282, 116167 CrossRef.
- Y. Che, Z. Deng, X. Lin and L. Hu, IEEE Trans. Veh. Technol., 2021, 70, 1269–1277 Search PubMed.
- J. Ma, S. Xu, P. Shang, Y. ding, W. Qin, Y. Cheng, C. Lu, Y. Su, J. Chong, H. Jin and Y. Lin, Appl. Energy, 2020, 262, 114490 CrossRef CAS.
- Y. Preger, H. M. Barkholtz, A. Fresquez, D. L. Campbell, B. W. Juba, J. Romàn-Kustas, S. R. Ferreira and B. Chalamala, J. Electrochem. Soc., 2020, 167, 120532 CrossRef CAS.
- H. Zou and T. Hastie, J. R. Stat. Soc. Ser. B Stat. Methodol., 2005, 67, 301–320 CrossRef.
- M. Olson, A. J. Wyner and R. Berk, Adv. Neural. Inf. Process. Syst., 2018, 30, 3619–3628 Search PubMed.
- F. Duffner, N. Kronemeyer, J. Tübke, J. Leker, M. Winter and R. Schmuch, Nat. Energy, 2021, 6, 123–134 CrossRef CAS.
- Y. Yuan, G. Ma and S. Xu, Mendeley Data, 2022 DOI:10.17632/nsc7hnsg4s.2.
- Y. Yuan, G. Ma and S. Xu, GitHub, 2022 DOI:10.5281/zenodo.6827566.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ee01676a |
| This journal is © The Royal Society of Chemistry 2022 |