 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dual mode of voltammetric studies on Cu(II) complexes of His2 peptides: phosphate and peptide sequence recognition†
Aleksandra
Tobolska
 *ab,
Klaudia
Głowacz
*ab,
Klaudia
Głowacz
 a,
Patrycja
Ciosek-Skibińska
a,
Patrycja
Ciosek-Skibińska
 a,
Wojciech
Bal
a,
Wojciech
Bal
 c,
Wojciech
Wróblewski
c,
Wojciech
Wróblewski
 a and
Nina E.
Wezynfeld
a and
Nina E.
Wezynfeld
 *a
*a
aChair of Medical Biotechnology, Faculty of Chemistry, Warsaw University of Technology, Noakowskiego 3, 00-664 Warsaw, Poland. E-mail: aleksandra.tobolska@pw.edu.pl; nina.wezynfeld@pw.edu.pl
bFaculty of Chemistry, University of Warsaw, Pasteura 1, 02-093 Warsaw, Poland
cInstitute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
First published on 10th November 2022
Abstract
Copper(II) complexes of peptides with a histidine residue at the second position (His2 peptides) provide a unique profile of electrochemical behavior, offering signals of both Cu(II) reduction and Cu(II) oxidation. Furthermore, their structures with vacant positions in the equatorial coordination plane could facilitate interactions with other biomolecules. In this work, we designed a library of His2 peptides based on the sequence of Aβ5–9 (RHDSG), an amyloid beta peptide derivative. The changes in the Aβ5–9 sequence highly affect the Cu(II) oxidation signals, altered further by anionic species. As a result, Cu(II) complexes of Arg1 peptides without Asp residues were chosen as the most promising peptide-based molecular receptors for phosphates. The voltammetric data on Cu(II) oxidation for binary Cu(II)–His2 peptide complexes and ternary Cu(II)–His2 peptide/phosphate systems were also tested for His2 peptide recognition. We achieved a highly promising identification of subtle modifications in the peptide sequence. Thus, we introduce voltammetric measurement as a potential novel tool for peptide sequence recognition.
Introduction
Amyloid β (Aβ) peptides are at the center of Alzheimer's disease (AD) research. They are the main constituents of amyloid plaques. Several forms of Aβ with different lengths and modifications have been found in vivo, including Aβ1–x and N-terminal truncated Aβp3–x, Aβ4–x, Aβ5–x, Aβp11–x, and Aβ11–x (predominant x values 40 and 42).1–3 Previous studies demonstrated that each of them binds to Cu(II) ions, although the N-terminal modifications significantly impact their affinity and Cu(II)-binding sites.4–6 The structures and properties of Cu(II)–Aβ complexes depend mostly on the localization of His residues in the peptide sequence.The diversity of Cu(II)–Aβ complexes also results in their various redox properties, which could be crucial for the understanding of the elevated oxidative stress level in AD.7–9 For example, Cu(II) complexes of two major Aβ forms in the brain, Aβ1–x and Aβ4–x, show contrasting electrochemical properties. The heterogeneous Cu(II) binding site in Aβ1–x, involving the N-terminus and His6, His13, and His14, could be reduced at pH 7.4, easily entering the Cu(II)/Cu(I) cycle,10,11 which fuels the generation of reactive oxygen species.12 The electrochemical Cu(II) oxidation has not been reported for this species at this pH. In contrast, Cu(II) in complex with Aβ4–x, where His3 plays a dominant role in the metal ion coordination, could be oxidized to Cu(III),5,11,13,14 but it is highly resistant to the reduction to Cu(I).15 Interestingly, another form, Aβ5–x, which contains His2, binds to Cu(II) in such a way that it enables both Cu(II) reduction and Cu(II) oxidation. The coordination mode of the Cu(II)–Aβ5–x complex comprised three nitrogen atoms (3N) from the N-terminal amine, the first amide, and the imidazole ring of His2 (Nam, N−, and Nim) with a vacant fourth equatorial site.4 The structure of the 3N complex of Cu(II)–Aβ5–x is provided in Fig. 1. The vacant position is occupied mostly by loosely bound water molecules, which could be a gate for interactions with other biomolecules under physiological conditions and could be utilized in the design of biosensors.4,16,17
Anions are molecules of great concern to human and environmental health.18,19 Among them, phosphate anions deserve special attention. Monitoring phosphate levels, e.g., in blood, allows the diagnosis of bone, kidney, and thyroid diseases.20 The recognition of phosphates is particularly challenging because they are highly hydrated.21 Therefore, the development of new methods for determining these anions is an important scientific goal. We recently demonstrated that Cu(II) complexes of Aβ5–9 could interact with phosphates, extensively altering the potential of Cu(II)/Cu(III) oxidation.4,16,17,22 Moreover, the studied peptide also possesses Arg (R) and Asp (D), residues that are commonly applied in designing selective molecular receptors towards anions.19,23 Thus, their presence in the peptide sequence could provide additional interactions such as electrostatic forces and/or hydrogen bonding.
Taking into account the distinctive properties of Cu(II)–Aβ5–9, we designed a library of nine His2 peptides consisting of Aβ5–9 (Arg–His–Asp–Ser–Gly) and its eight analogs (Table 1), whose binary Cu(II) complexes represent potential phosphate-binding receptors. We exchanged the first and the third amino acid residues for Arg and Asp, potentially contributing to intramolecular interactions, or Gly (G) as the smallest building block of peptides, without the side chain. For simplicity, all oligopeptides will be described with the first three residues of their sequences, as provided in Table 1. We performed initial UV-vis spectroscopic studies on the formation of Cu(II)–peptide complexes. Then, we employed Cyclic Voltammetry (CV) and Differential Pulse Voltammetry (DPV) to check the electrochemical properties of binary Cu(II) complexes of His2 peptides as well as to compare the response of the complexes toward the selected anions. In the last part of the paper, we have proposed another approach. Using the same data set, i.e., voltammetric curves registered for binary (Cu(II)–His2 peptides) and ternary (Cu(II)–His2 peptides/phosphates) systems, we discriminated the His2 peptide sequences by Principal Component Analysis (PCA). Thus, we demonstrated that the analysis of the Cu(II) oxidation process in the metallocomplex provides a dual application: the recognition of phosphate species and the identification of His2 peptide sequences.
| His2 peptide sequence | Abbreviation |
|---|---|
| The maternal sequence | |
| Arg–His–Asp–Ser–Gly–NH2 (Aβ5–9) | RHD |
| Analogs | |
| Arg–His–Gly–Ser–Gly–NH2 | RHG |
| Arg–His–Arg–Ser–Gly–NH2 | RHR |
| Asp–His–Asp–Ser–Gly–NH2 | DHD |
| Asp–His–Arg–Ser–Gly–NH2 | DHR |
| Asp–His–Gly–Ser–Gly–NH2 | DHG |
| Gly–His–Gly–Ser–Gly–NH2 | GHG |
| Gly–His–Asp–Ser–Gly–NH2 | GHD |
| Gly–His–Arg–Ser–Gly–NH2 | GHR |
Results and discussion
Influence of the His2 peptide sequence on Cu(II) coordination – spectroscopic studies
We started with spectroscopic studies on Cu(II) coordination with the His2 peptides, whose sequences are compiled in Table 1. As they differ primarily by the acid–base properties of their first and third residues, we performed pH-metric titrations of solutions containing Cu(II) ions and the peptides. The UV-vis spectra are provided in Fig. S2,† pH dependence of the formation of Cu(II) complexes of His2 peptides is given in Fig. 2, and the detailed spectroscopic parameters related to 3N and 3N + OH− complexes are shown in Table 2 and Table S1,† respectively. | ||
| Fig. 2 The pH dependence of absorbance at wavelengths associated with the formation of the two main species of Cu(II) complexes of the His2 peptides, 3N complex (open symbols) and 3N + OH− (full symbols), based on the spectra from Fig. S2.† The solid lines represent fits to the Hill equation and the pK values calculated for 3N species of each peptide are given in the corresponding graph, while their detailed values are given in Table 2. | ||
| 3N complex | pH 7.4 | pK ± SEa | |
|---|---|---|---|
| λ max (nm) | ε (M−1 cm−1) | ||
| a SE stands for the standard error and represents the statistical error of pK determinations. | |||
| Cu(II)–RHR | 603 | 58 | 3.53 ± 0.02 |
| Cu(II)–DHD | 604 | 59 | 3.62 ± 0.02 |
| Cu(II)–DHG | 603 | 60 | 3.63 ± 0.05 |
| Cu(II)–DHR | 602 | 59 | 3.68 ± 0.02 |
| Cu(II)–RHG | 603 | 61 | 3.75 ± 0.02 |
| Cu(II)–RHD | 604 | 69 | 3.77 ± 0.02 |
| (Cu(II)–Aβ5–9) | |||
| Cu(II)–GHR | 608 | 58 | 3.86 ± 0.02 |
| Cu(II)–GHD | 606 | 59 | 3.94 ± 0.02 |
| Cu(II)–GHG | 608 | 63 | 4.03 ± 0.03 |
In general, during all titrations, two d–d bands for Cu(II)–peptide complexes were observed. The first one, around 605 nm, appeared at pH above 3 (see Fig. 2, open symbols) and is characteristic of the 3N [Nam, Nim, N−] square planar Cu(II) complex of the His2 peptides.4,16,24–26 What is important for this work is the presence of the 3N complex as the main Cu(II) coordination mode for all the studied complexes at physiological pH 7.4. The second d–d band around 520 nm was noticed at pH higher than 9 (see Fig. 2, full symbols) and corresponded to the formation of the 3N + OH− [Nam, Nim, N−, OH−] complex due to the deprotonation of a water molecule.
Despite these similarities, we found significant differences between the peptides in the formation of both 3N and 3N + OH− species. For example, the 3N complex appears at higher pH values for the peptides possessing the Gly residue at the first position (Gly1), Fig. 2. This is also reflected in the highest pK values calculated for those complexes, pK 3.9–4.0. In this work, pK is defined as the pH value at which half of the total copper amount is in the form of the given species (here, 3N complex). The pK values were obtained by fitting the spectroscopic data for 3N complexes provided in Fig. 2 (open symbols) to the Hill equation (see the ESI,† eqn (S1) for the fitting description and Table 2 for the detailed pK values). As such, the higher pK values for 3N complexes of Gly1 peptides are associated with the lower stability of these complexes. The pK values calculated for 3N Cu(II) complexes of the remaining His2 peptides were in the range of 3.5–3.8, with the lowest value for RHR. In this case, Arg residues likely decrease the deprotonation pKa values of Cu(II) binding groups, such as the Arg1 amine, peptide bond Arg1–His2, and His2 imidazole, which occur in the proximity of the Arg residues in the designed peptides. This, in consequence, facilitates the formation of Cu(II) complexes by Arg-containing peptides and increases their stability. The maximum of d–d bands of 3N Gly1 complexes occurred at slightly longer wavelengths (608–609 nm) than those for the 3N complexes of other peptides (Table 2). But, no systematic dependence on the peptide sequence was found for their molar extinction coefficients ranging from 58 to 69 M−1 cm−1.
The formation of the second observed species, 3N + OH−, was likely not accomplished at the highest measured pH, 11.7, as the endpoints of the titration curves for these complexes were not established (Fig. 2, solid symbols). Moreover, in the case of complexes with Asp3 peptides, the d–d band still shifted towards lower wavelengths at around pH 11.7, as given in Fig. S3.† However, we did not continue the measurements due to the non-Nernstian response of the employed glass electrode at higher pH values. During the 3N + OH− species analysis, we focused on spectroscopic parameters at the highest measured pH 11.7 given in Table S1,† which mostly resemble those of 3N + OH− Cu(II) complexes. In addition, we followed the pH dependence of A475, shifted to lower wavelengths from the maximum of the d–d band of 3N + OH−, as the signal derived exclusively from this species, without the contribution of the adjacent band of 3N species, Fig. 2 (solid symbols). At pH 11.7, the less pronounced blue shift (Table S1†) and the lowest A475 (Fig. 2, entries DHD, GHD and RHD) were noticed for the Asp3 peptides. In contrast, the most pronounced blue shift (Table S1†) and the highest A475 (Fig. 2, entries DHR, GHR and RHR) were observed for the Cu(II) complexes of Arg3 peptides.
To conclude, the first residue of His2 peptides primarily affects the 3N Cu(II) complex formation, with a destabilizing role of Gly1. On the other hand, the third residue of His2 peptides mainly impacts the formation of the following 3N + OH− complex, which Asp3 could weaken.
Influence of the His2 peptide sequence on the electrochemical response of the Cu(II) complexes
In our previous study on Cu(II) complexes with Aβ5–9 (RHD),4,16 we showed that the 3N chelate exhibited two redox couples: the Cu(II)/Cu(I) reduction at −0.45 V vs. Ag/AgCl, and the Cu(II)/Cu(III) oxidation around 1.20 V vs. Ag/AgCl at pH 7.4. We proved that those signals are characteristic of Cu(II) bound to His2 peptides and do not appear in the control measurements of peptide/KNO3 or Cu(II)/KNO3 mixtures.4The cyclic voltammograms of the 3N complexes of the His2 peptides obtained in this work and shown in Fig. S4† demonstrated similar electrochemical behaviour for all the studied analogs, but with considerable differences in the position of the peaks. Analyzing the negative range of potentials, we observed Cu(II) reduction in the potential range from −0.55 V to −0.70 V, and, in the reverse scan, Cu(I) oxidation from around −0.08 V to 0.12 V (Fig. S4A†). This significant separation between the cathodic and anodic peaks suggested a slow electron transfer, which likely results from the differences between Cu(II) and Cu(I) complex structures and their reorganization upon the redox reactions.4,27–29 A more detailed analysis revealed some dependence between the positions of the peaks of the Cu(II)/Cu(I) cycle and the His2 peptide sequence (Fig. S5†). The lowest potential of Cu(II) reduction was registered for Asp1 peptides, whereas the highest Cu(I) oxidation potential was noticed for peptide complexes containing Arg1 or/and Arg3 residues. However, in general, the differences between the peptide complexes for the Cu(II)/Cu(I) cycle were minor, with considerable variability among replicates.
A more pronounced effect of the peptide sequence was observed for the Cu(II)/Cu(III) oxidation (Fig. S4B†). In all cases, the processes were irreversible (lack of the cathodic signal in the reverse CV scan). Considering the character of the studied reactions and limitations of cyclic voltammetry, e.g., lower sensitivity due to the large capacitive current contribution, we chose the pulse technique (DPV) for further studies. The DPV results obtained for the binary systems (Cu(II)–His2 peptides) are shown in Fig. 3, and the potential values are given in Table 3.
 | ||
Fig. 3 DPV curves obtained for the Cu(II) complexes of His2 peptides (0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 ratio) recorded in 100 mM KNO3, pH 7.4. 1.0 ratio) recorded in 100 mM KNO3, pH 7.4. | ||
| His2 peptide | E Cu(II)/Cu(III) (V) vs. Ag/AgCl ± SDa | ||||
|---|---|---|---|---|---|
| Binary system | Ternary system Cu(II)–His2 peptide/anion | ||||
| Cu(II)–His2 peptide | H2PO4−/HPO42− | CH3COO− | SO42− | Cl− | |
| a E Cu(II)/Cu(III) values calculated as the mean of three independent repetitions together with the standard deviation (SD) for binary and ternary Cu(II)–His2 peptide/phosphate systems. The values for other ternary systems are given for comparison and are based on at least two independent repetitions. | |||||
| RHG | 1.180 ± 0.005 | 0.987 ± 0.015 | 1.096 ± 0.003 | 1.184 ± 0.003 | 1.180 ± 0.001 |
| RHR | 1.191 ± 0.003 | 0.996 ± 0.005 | 1.107 ± 0.003 | 1.175 ± 0.001 | 1.179 ± 0.001 |
| DHR | 1.204 ± 0.005 | 1.040 ± 0.003 | 1.120 ± 0.006 | 1.194 ± 0.003 | 1.196 ± 0.001 |
| DHG | 1.187 ± 0.003 | 1.048 ± 0.003 | 1.100 ± 0.001 | 1.180 ± 0.001 | 1.184 ± 0.001 |
| RHD16 | 1.204 ± 0.005 | 1.056 ± 0.006 | 1.120 ± 0.004 | 1.196 ± 0.004 | 1.204 ± 0.002 |
| (Aβ5–9) | |||||
| DHD | 1.203 ± 0.003 | 1.088 ± 0.007 | 1.119 ± 0.003 | 1.194 ± 0.004 | 1.199 ± 0.002 |
| GHG | 1.245 ± 0.003 | 1.087 ± 0.003 | 1.178 ± 0.003 | 1.237 ± 0.003 | 1.237 ± 0.001 |
| GHR | 1.253 ± 0.003 | 1.112 ± 0.003 | 1.191 ± 0.003 | 1.231 ± 0.003 | 1.239 ± 0.003 |
| GHD | 1.231 ± 0.004 | 1.113 ± 0.003 | 1.181 ± 0.003 | 1.227 ± 0.003 | 1.231 ± 0.001 |
The presence of Gly1 increased the Cu(II) oxidation potential (ECu(II)/Cu(III)) by 0.03–0.05 V (Fig. 3 and Table 3) compared to the complex of the maternal RHD sequence of Aβ5–9 (ECu(II)/Cu(III) = 1.20 V). Conversely, the lowest ECu(II)/Cu(III) of 1.18 V was determined for Cu(II)–RHG. Such a decrease in the Cu(II) oxidation potential for the Arg1 peptide could be caused by electrostatic interactions with δ-donor groups, which stabilize the Cu(III) state. This is in line with the results reported for the Cu(II) complexes of His3 peptides containing Lys residues.30 The Cu(II) oxidation potential values in the presence of other His2 peptide complexes were similar, in the range of 1.19–1.20 V (Table 3). Additionally, we studied the possible impact of the charge of the complex on its redox properties. A more negative complex charge could favor the electron release and decrease of the potential value of metal center oxidation. However, such an effect was not observed. For example, the [Cu(II)–DHD]+ complex underwent Cu(II)/Cu(III) oxidation at a potential similar to that of the most positively charged [Cu(II)–RHR]5+ chelate.
Another factor that could impact the Cu(II) oxidation is the size of the first and the third residues. Indeed, we noticed some tendencies for peptides containing Gly, the smallest residue in the studied peptides. However, those effects were contrasting for the first and the third positions. While Gly3 facilitates Cu(II) oxidation (the lowest ECu(II)/Cu(III) for RHG and DHG, Table 3), Gly1 hinders this process (the highest ECu(II)/Cu(III) for GHR, GHG, and GHD, Table 3). Interestingly, the GHG peptide belongs to the second group, whose complexes exhibit the highest oxidation potential values indicating that the effects of Gly1 prevail over those of Gly3. On the other hand, the Cu(II) oxidation in the Cu(II) complex of FHFSKNR–NH2 (FHF), the peptide with bulky Phe1 and Phe3 residues, was reported at around 1.25 V vs. Ag/AgCl, a very similar ECu(II)/Cu(III) to that observed by us for the GHG complex.17 Other data refer to complexes of His2 peptides, which contain a redox-active Trp residue. Its oxidation potential peak is very close to the Cu(II) oxidation peak of Cu(II)–His2 peptides.17,31 In consequence, the analysis of such data is challenging. However, the observed order of these peptides toward higher oxidation potentials, FHW ≈ WHF < WHW < FHF,17 agrees with the general conclusion of our studies that there is no single specific feature of His2 peptides which defines the susceptibility to Cu(II) oxidation of their complexes. Instead, the particular positions of the acidic/basic/small/bulk residues and their combination in the proximity of His2 seem vital for this process.
Effect of anions on Cu(II) oxidation in complexes of the His2 peptides
Next, we introduced anions into the above-described Cu(II)–His2 peptide systems. Our recent studies showed that phosphate only slightly impacts the UV-vis spectra of 3N complexes. In contrast, the electrochemical signals were much more affected by anion addition.16,22 We have found that DPV is a particularly promising technique in the studies on weak but physiologically relevant interactions between metal–peptide complexes and anions.16,22 Therefore, we also chose DPV in this work to check how the amino acid sequence of the His2 peptides affects the phosphate recognition by their Cu(II) complexes. DPV measurements were performed at pH 7.4, which ensures 3N coordination mode with a vacancy in the equatorial site for the Cu(II) complexes of all the studied His2 peptides as demonstrated by the above-described spectroscopic studies. The DPV curves of the Cu(II)–His2 peptide complexes in the presence of environmentally and biologically relevant anions are depicted in the ESI,† Fig. S6. The Cu(II) oxidation potential values for the binary Cu(II)–peptide and ternary Cu(II)–peptide/anion systems are provided in Table 3. The column chart presenting the differences in the Cu(II) oxidation potential values between the ternary and the binary systems (ΔE) is given in Fig. 4.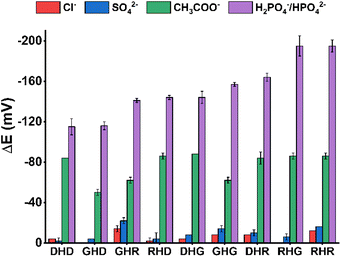 | ||
| Fig. 4 Differences between the Cu(II) oxidation potential values in ternary Cu(II)–His2 peptide/anion and binary Cu(II)–His2 peptide systems (ΔE) calculated on the basis of the results shown in Table 3, Fig. S6,† and the literature data.16 | ||
Chloride and sulfate anions only slightly affected the redox properties of all the studied Cu(II)–His2 peptide complexes. It is in line with our previous observations.16 Distinct changes in the voltammetric curves were observed in the presence of acetate or phosphate anions, i.e., the oxidation peak corresponding to the Cu(II)/Cu(III) process for ternary Cu(II)–His2 peptide/phosphate or Cu(II)–His2 peptide/acetate systems occurred at lower potentials than that of the respective binary Cu(II)–His2 peptide complex. Furthermore, the voltammetric signals obtained for chelates in the mixture of all the studied anions (Fig. S6,† dotted violet lines) resembled those recorded for the ternary Cu(II)–His2 peptide/phosphate system (Fig. S6,† solid violet lines). The only exception is Cu(II)–DHD, with very similar responses to phosphates and acetates, and in consequence, to the mixture of anions (Fig. S6†). Nevertheless, these results demonstrated the high selectivity of electrochemical phosphate recognition by Cu(II) complexes of most of the investigated His2 peptides.
The greatest shift of the Cu(II)/Cu(III) peak (the highest ΔE value) was observed after the phosphate addition to the Cu(II) complexes of RHG and RHR, the Arg1 peptides without the Asp residue (Fig. 4, the detailed values are listed in Table 3). This result can be explained by the favorable high positive charge of those chelates, [Cu(II) − RHG]4+ and [Cu(II) − RHR]5+, and the presence of the Arg1 residue in the closest neighborhood to the groups directly engaged in the Cu(II) coordination. In contrast, minor changes of the Cu(II) oxidation signal in the presence of anionic species were noticed for GHD, but particularly for DHD (Fig. 4). As mentioned, the Cu(II)/Cu(III) oxidation peaks for Cu(II)–DHD appeared relatively close to each other in the presence of acetate and phosphate anions (see Table 3). This revealed a worse selectivity of Cu(II)–DHD towards phosphates, likely caused by the lower complex charge [Cu(II) − DHD]+ provided by deprotonated Asp residues, and thus weaker electrostatic interactions between the Cu(II)–DHD and the anions.
Our studies are limited to the His2 peptides, in which the first and the third positions are occupied by three representative residues, Arg, Asp, and Gly. However, one could expect that the selectivity of Cu(II) complexes of peptides containing other negatively charged residues, such as Glu, would be similar to those containing Asp, which is insufficient for a good phosphate receptor. Alternatively, Lys-containing peptides could provide advantageous features like those observed for positively-charged Arg analogs. Furthermore, elongation of the aliphatic side chain (as for Ile/Leu) and incorporation of uncharged polar residues (such as Ser/Thr/Asn/Gln) could have an effect as well; the former likely by the stabilization of the Cu(II)–peptide complex structure and as a steric hindrance during the ternary interaction; whereas the latter by the formation of hydrogen bonds between the side chain and the phosphate molecule.32 Therefore, in our next projects, we would also like to check other sequences based on the above consideration to improve further the receptor properties of Cu(II)–His2 peptides towards phosphates.
Chemometric modeling of voltammetric data for peptide recognition
The results presented above demonstrated that the changes in the His2 peptide sequence affect the Cu(II) oxidation potential of both systems, the binary Cu(II)–His2 peptide and the ternary Cu(II)–His2 peptide/phosphate. Therefore, we combined these values for all the studied complexes in Fig. 5. This analysis provided a clear formation of three groups (clusters) of His2 peptides, depending on their sequence. The first group contained the Gly1 peptides, and it was characterized by the highest values of Cu(II) oxidation potential in both systems. The second group gathered the peptides with Asp residues but without Gly1. The third cluster consists of the Arg1 peptides without Asp, exhibiting the lowest Cu(II) oxidation potential. Accordingly, we showed that the electrochemical properties of Cu(II) complexes of His2 peptides could be employed not only for the recognition of phosphate species but also to distinguish the types of residues in the proximity of His2 in the peptide sequence.Since the obtained voltammetric data carry much more information than the Cu(II) oxidation potential, we decided to employ chemometric modeling in the follow-up studies on the voltammetric detection systems for identifying His2 peptides. The application of chemometric methods for voltammetric data processing allowed us to track signal changes observed for various peptide complexes in the entire voltammograms (i.e., intensity, signal location, and shape of the observed signals). First, we applied Principal Component Analysis (PCA), aiming to reduce the dimensionality of large data sets by determining new variables (PCs, Principal Components), which are a linear combination of original variables and order the variance contained in the data. Thus, one could assess the initial patterns in the data by analyzing the so-called score plot. Such visualization on a two- or three-dimensional graph leads to grouping objects with similar properties. To determine which variables of the original data are the most significant for the discrimination of the samples, one could also use loading plots, where the higher the absolute value of loading, the higher the contribution of the original variable in the given PC.33,34
Another chemometric model used for sample (dis)similarity evaluation is Hierarchical Cluster Analysis (HCA). In this method, the most similar objects are linked together on the dendrogram, and the length of its branches is a measure of (dis)similarity between samples, i.e., the shorter the branches in the dendrogram linking the specified objects, the higher the similarity of the objects.35
We tested three different sets of DPV data registered for (i) the binary Cu(II)–His2 peptide complex; (ii) the ternary Cu(II)–His2 peptide/phosphate system; (iii) the data fusion of Cu(II)–His2 peptide + Cu(II)–His2 peptide/phosphate. In all cases, the information contained in the first two PCs allowed us to assess the possibility of distinguishing His2 peptides based on the interactions of the peptide with Cu(II) and of the Cu(II)–peptide complex with phosphate anions. The resulting score plots are depicted in Fig. 6, the corresponding loading plots are presented in Fig. S7 and S8,† and the hierarchical cluster analysis is given in Fig. S9.†
First, we examined the ability of His2 peptide recognition when employing only the data registered for their binary Cu(II) complexes (Fig. 6A). The samples representing each His2 peptide formed separated clusters, with little overlap of RHR and RHD, as well as DHR and DHD (also confirmed by HCA, see Fig. S9A†). The cluster positions in the PCA space primarily depend on the first residue of His2 peptides. The most easily recognized ones were Gly1 analogs, characterized by positive scores on PC1. At the same time, peptides containing Asp1 and Arg1 residues exhibited negative PC1 scores. As provided in Fig. S7A,† the highest value of the PC1 loading was noticed around the maximum of the Cu(II) oxidation peak. As such, the variation in ECu(II)/Cu(III) values contributes the most to the peptide grouping against PC1. Interestingly, the distinction between Arg1 and Asp1 peptides with similar ECu(II)/Cu(III) values is provided by the information included in PC2, with PC2 > 0 for Arg1 and PC2 < 0 for Asp1 peptides. A similar pattern was observed for Gly1 peptides with Arg3 (PC2 > 0) or Asp3 (PC2 < 0). In those cases, the differences in the shape of voltammetric curves, including the position of inflection points, likely separated those groups (see loadings on PC2 in Fig. S7B†).
In the next step, we checked how the presence of phosphate anions impacts the peptide recognition based on the oxidation signals of the Cu(II) complexes. As presented in Fig. 6B, in this case, Arg1 peptides without the Asp residue were the most easily discerned, exhibiting PC1 < −0.5. Once again, PC1 carries information related to the position of the Cu(II) oxidation signal, as suggested by the loadings on PC1 in Fig. S7C,† but with a higher contribution of other peak attributes than that noticed in the case of the binary system (for comparison see Fig. S7A†). The presence of Asp in the sequence, either in the first or third position, seems to be related to a higher degree of cluster overlap in the case of the ternary system. The RHD peptide clustered in the close proximity of DHR, whereas DHD groups alongside GHD close to GHG (also confirmed by HCA, see Fig. S9B†).
Since the results presented above suggested that both approaches likely provided additional complementary information (better discrimination of different peptides), we finally performed a fusion of DPV data obtained for Cu(II)–His2 peptide and Cu(II)–His2 peptide/phosphate systems. In line with the previous cases, the position of the Cu(II) oxidation peaks impacts mostly PC1, as confirmed by loadings in Fig. S8A.† More changes were noticed in the loadings on PC2 (Fig. S8B†), suggesting that other parts of the voltammetric curves affect the grouping of the peptides against PC2 for data fusion compared to that for binary and ternary systems separately. By analyzing Fig. 6C, it is evident that this approach resulted in a better separation of the His2 peptides (a smaller degree of cluster overlap). Each investigated peptide was the most easily recognized compared to the previous cases. Only objects representing RHD and DHR overlapped, which is interesting considering that they contain the same residues in the proximity of His2, but at different positions. Similar conclusions were derived from the HCA given in Fig. S9C.† However, employing a different approach for the HCA analysis, the clustering based on current values, allowed us to discriminate the RHD and DHR sequences (see Fig. S9D†).
Thus, we showed that the analysis of voltammetric data for Cu(II) complexes of His2 peptides provides a solid foundation for the development of voltammetric detection systems employing chemometric analysis for the recognition of peptide sequences. As described at the end of previous sections, one could expect that additional features of side chains could prevail for other peptides in the binary and ternary systems. For example, the introduction of the redox-active residues (i.e. Tyr, Trp, and Met) would generate an additional signal on the voltammetric curves, more bulky side chains could alter the stability of binary complexes, and the exchange of negatively charged Asp residues to uncharged polar Asn residues could change the electrochemical response to phosphates. Moreover, different patterns of the electrochemical signals can be obtained by tuning the structure/environment of the coordination site, leading to the discrimination of various sets of samples. One could also utilize the fusion of data from different experimental conditions and/or variously modified electrodes to considerably improve the peptide sequence identification by means of an electronic tongue-sensing strategy.
Conclusions
The results obtained in this work showed the high potential of voltammetric methods in both recognition of phosphate species using a novel class of peptide-based receptors (Cu(II) complexes of His2 peptides) and the discrimination of His2 peptide sequences based on the Cu(II) oxidation signals of their complexes.We demonstrated that the type of His2 adjacent residue influenced the formation of Cu(II) complexes studied by UV-vis spectroscopy, as well as their redox activity investigated by voltammetric techniques, CV and DPV. The most notable changes among the His2 peptides were observed in the oxidation signals of their Cu(II) complexes. Due to the typically high Cu(II) oxidation values for His2 peptide complexes at pH 7.4, this reaction is believed to be of negligible importance for biological processes. However, we believe that our research will draw attention to its high potential as a source of analytical signal in various sensing applications.
The Cu(II) oxidation potential varied among the Cu(II)–His2 peptide chelates upon the addition of the anions. The most promising results were obtained for the Cu(II) complexes of the Arg1 analogs without the Asp residue, which exhibit the highest selectivity towards phosphates. Moreover, the analysis of voltammetric data, including chemometric modeling of the DPV curves of the copper complexes with and without phosphate anions, showed that these measurements could also be employed to recognize His2 peptide sequences. The results presented in this article for the first time disclose that such similar peptide analogs can be distinguished using voltammetry coupled with chemometrics.
The variety of peptide sequences is, however, much wider than that of His2 peptides. Even the family of Aβ peptides provides a vast diversity of redox properties of their copper complexes, which could be applied to discriminate peptides or other biomolecules. Besides this, one could employ other redox active metal centers and change the pattern of the obtained results. Therefore, taking into account the sensitivity of the electrochemical methods to minor changes in the environment of the redox center, we believe that our work gives a fresh perspective on studies on metal–peptide complexes and their potential applications.
Data availability
The data presented in this study are available on request from the corresponding authors.Author contributions
N. E. W., A. T., W. B., and W. W. formulated the concepts of the study. The experimental work was performed by A. T. The analyses of spectroscopic and electrochemical data were performed by A. T. and N. E. W. K. G. formulated the concept of peptide discrimination by chemometric analysis. The chemometric analysis was performed by K. G. and P. C. S. The original draft was written by A. T., K. G., and N. E. W. All authors participated in the review and editing of the paper and have given approval regarding the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financed by the National Science Centre in Poland under the project PRELUDIUM (No. 2021/41/N/ST4/00956) and implemented as part of the Operational Program Knowledge Education Development 2014–2020 (Project No. POWR.03.02.00-00-I007/16-00) co-financed by the European Social Fund (A. T.). P. C. S. acknowledges the financial support from the National Science Centre, Poland, in the frame of SONATA BIS project No. UMO-2018/30/E/ST4/00481. K. G. acknowledges the financial support from the IDUB project (Scholarship Plus programme). We gratefully acknowledge Paulina Szczerba and Dawid Płonka from Prof. Wojciech Bal's group (Institute of Biochemistry and Biophysics, Polish Academy of Sciences) for performing the synthesis and purification of the peptides. A. T. would like to thank Prof. Sławomir Sęk (Faculty of Chemistry, Biological and Chemical Research Centre, University of Warsaw) for his support and helpful discussions.Notes and references
- N. C. Wildburger, T. J. Esparza, R. D. Leduc, R. T. Fellers, P. M. Thomas, N. J. Cairns, N. L. Kelleher, R. J. Bateman and D. L. Brody, Sci. Rep., 2017, 7, 9520 CrossRef PubMed.
- E. Portelius, N. Bogdanovic, M. K. Gustavsson, I. Volkmann, G. Brinkmalm, H. Zetterberg, B. Winblad and K. Blennow, Acta Neuropathol., 2010, 120, 185–193 CrossRef CAS PubMed.
- E. Stefaniak and W. Bal, Inorg. Chem., 2019, 58, 13561–13577 CrossRef CAS PubMed.
- N. E. Wezynfeld, A. Tobolska, M. Mital, U. E. Wawrzyniak, M. Z. Wiloch, D. Płonka, K. Bossak-Ahmad, W. Wróblewski and W. Bal, Inorg. Chem., 2020, 59, 14000–14011 CrossRef CAS PubMed.
- M. Mital, N. E. Wezynfeld, T. Frączyk, M. Z. Wiloch, U. E. Wawrzyniak, A. Bonna, C. Tumpach, K. J. Barnham, C. L. Haigh, W. Bal and S. C. Drew, Angew. Chem., Int. Ed., 2015, 54, 10460–10464 CrossRef CAS PubMed.
- K. Bossak-Ahmad, M. Mital, D. Płonka, S. C. Drew and W. Bal, Inorg. Chem., 2019, 58, 932–943 CrossRef CAS PubMed.
- E. Stefaniak, D. Płonka, P. Szczerba, N. E. Wezynfeld and W. Bal, Inorg. Chem., 2020, 59, 4186–4190 CrossRef CAS PubMed.
- M. Z. Wiloch and M. Jönsson-Niedziółka, J. Electroanal. Chem., 2022, 922, 116746 CrossRef CAS.
- M. Z. Wiloch, N. Baran and M. Jönsson-Niedziółka, ChemElectroChem, 2022, 9, e202200623 CrossRef CAS.
- C. Hureau, V. Balland, Y. Coppel, P. L. Solari, E. Fonda and P. Faller, J. Biol. Inorg. Chem., 2009, 14, 995–1000 CrossRef CAS PubMed.
- M. Z. Wiloch, U. E. Wawrzyniak, I. Ufnalska, A. Bonna, W. Bal, S. C. Drew and W. Wróblewski, J. Electrochem. Soc., 2016, 163, G196–G199 CrossRef CAS.
- E. Atrián-Blasco, M. Del Barrio, P. Faller and C. Hureau, Anal. Chem., 2018, 90, 5909–5915 CrossRef PubMed.
- C. Hureau, H. Eury, R. Guillot, C. Bijani, S. Sayen, P. L. Solari, E. Guillon, P. Faller and P. Dorlet, Chem. – Eur. J., 2011, 17, 10151–10160 CrossRef CAS PubMed.
- K. P. Neupane, A. R. Aldous and J. A. Kritzer, J. Inorg. Biochem., 2014, 139, 65–76 CrossRef CAS PubMed.
- C. Esmieu, G. Ferrand, V. Borghesani and C. Hureau, Chem. – Eur. J., 2021, 27, 1777–1786 CrossRef CAS PubMed.
- A. Tobolska, N. E. Wezynfeld, U. E. Wawrzyniak, W. Bal and W. Wróblewski, Dalton Trans., 2021, 50, 2726–2730 RSC.
- I. Ufnalska, U. E. Wawrzyniak, K. Bossak-Ahmad, W. Bal and W. Wróblewski, J. Electroanal. Chem., 2020, 862, 114003 CrossRef CAS.
- M. J. Langton, C. J. Serpell and P. D. Beer, Angew. Chem., Int. Ed., 2016, 55, 1974–1987 CrossRef CAS PubMed.
- P. D. Beer and P. A. Gale, Angew. Chem., Int. Ed., 2001, 40, 486–516 CrossRef CAS PubMed.
- M. Christov and H. Jüppner, Best Pract. Res., Clin. Endocrinol. Metab., 2018, 32, 685–706 CrossRef CAS PubMed.
- M. V. Ramakrishnam Raju, S. M. Harris and V. C. Pierre, Chem. Soc. Rev., 2020, 49, 1090–1108 RSC.
- A. Tobolska, N. E. Wezynfeld, U. E. Wawrzyniak, W. Bal and W. Wróblewski, Inorg. Chem., 2021, 60, 19448–19456 CrossRef CAS PubMed.
- S. Kubik, C. Reyheller and S. Stüwe, J. Incl. Phenom. Macrocycl. Chem., 2005, 52, 137–187 CrossRef CAS.
- K. Bossak, M. Mital, J. Poznański, A. Bonna, S. Drew and W. Bal, Inorg. Chem., 2016, 55, 7829–7831 CrossRef CAS PubMed.
- K. Bossak-Ahmad, M. D. Wiśniewska, W. Bal, S. C. Drew and T. Frączyk, Int. J. Mol. Sci., 2020, 21, 6190 CrossRef CAS PubMed.
- R. Kotuniak, T. Frączyk, P. Skrobecki, D. Płonka and W. Bal, Inorg. Chem., 2018, 57, 15507–15516 CrossRef CAS PubMed.
- C. Hureau, H. Eury, R. Guillot, C. Bijani, S. Sayen, P.-L. Solari, E. Guillon, P. Faller and P. Dorlet, Chem. – Eur. J., 2011, 17, 10151–10160 CrossRef CAS PubMed.
- I. Ufnalska, S. C. Drew, I. Zhukov, K. Szutkowski, U. E. Wawrzyniak, W. Wróblewski, T. Frączyk and W. Bal, Inorg. Chem., 2021, 60, 18048–18057 CrossRef CAS PubMed.
- S. Carlotto, A. Bonna, K. Bossak-Ahmad, W. Bal, M. Porchia, M. Casarin and F. Tisato, Metallomics, 2019, 11, 1800–1804 CrossRef CAS PubMed.
- Y. Jin, M. A. Lewis, N. H. Gokhale, E. C. Long and J. A. Cowan, J. Am. Chem. Soc., 2007, 129, 8353–8361 CrossRef CAS PubMed.
- S. Mena, A. Mirats, A. B. Caballero, G. Guirado, L. A. Barrios, S. J. Teat, L. Rodriguez-Santiago, M. Sodupe and P. Gamez, Chem. – Eur. J., 2018, 24, 5153–5162 CrossRef CAS PubMed.
- P. Gonzalez, K. Bossak, E. Stefaniak, C. Hureau, L. Raibaut, W. Bal and P. Faller, Chem. – Eur. J., 2018, 24, 8029–8041 CrossRef CAS PubMed.
- R. Bro and A. K. Smilde, Anal. Methods, 2014, 6, 2812–2831 RSC.
- M. Wang, X. Cetó and M. del Valle, Biosens. Bioelectron., 2022, 198, 113807 CrossRef CAS PubMed.
- K. Drab and M. Daszykowski, J. AOAC Int., 2014, 97, 29–38 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods; UV-vis spectra of titrations with NaOH, spectral parameters of the 3N + OH− species; CV curves of Cu(II)–His2 complexes; DPV curves of Cu(II)–His2 complexes in the presence of anions; loading plots; HCA diagrams. See DOI: https://doi.org/10.1039/d2dt03078k |
| This journal is © The Royal Society of Chemistry 2022 |



