Open Access Article
Open Access ArticleRole of hydrazone substituents in determining the nuclearity and antibacterial activity of Zn(II) complexes with pyrazolone-based hydrazones†
Fabio
Marchetti
 *a,
Riccardo
Pettinari
*a,
Riccardo
Pettinari
 b,
Federico
Verdicchio
b,
Federico
Verdicchio
 b,
Alessia
Tombesi
b,
Alessia
Tombesi
 b,
Stefania
Scuri‡
b,
Sonila
Xhafa
a,
Laura
Olivieri
a,
Claudio
Pettinari
b,
Stefania
Scuri‡
b,
Sonila
Xhafa
a,
Laura
Olivieri
a,
Claudio
Pettinari
 b,
Duane
Choquesillo-Lazarte
b,
Duane
Choquesillo-Lazarte
 c,
Amalia
García-García
d,
Antonio
Rodríguez-Diéguez
c,
Amalia
García-García
d,
Antonio
Rodríguez-Diéguez
 d and
Agustín
Galindo
d and
Agustín
Galindo
 e
e
aChIP Research Center, School of Science and Technology, University of Camerino, via Madonna delle Carceri, 62032 Camerino MC, Italy. E-mail: fabio.marchetti@unicam.it
bChIP Research Center, School of Pharmacy, University of Camerino, via Madonna delle Carceri, 62032 Camerino MC, Italy
cLaboratorio de Estudios Cristalográficos, IACT, CSIC-UGR, Av. Las Palmeras n°4, 18100 Granada, Spain
dDepartment of Inorganic Chemistry, Faculty of Sciences, University of Granada, Av. Fuentenueva S/N, 18071 Granada, Spain
eDepartamento de Química Inorgánica, Facultad de Química, Universidad de Sevilla, Aptdo 1203, 41071 Sevilla, Spain
First published on 24th August 2022
Abstract
Hydrazones and their metal derivatives are very important compounds in medicinal chemistry due to their reported variety of biological activities, such as antibacterial, antifungal and anticancer action. Five hydrazone-pyrazolone ligands H2Ln (n = 1–5) were prepared and fully characterized and their tautomerism was investigated in the solid state and solution. Five zinc(II) complexes 1–5 of composition [Zn(HLn)2] (n = 1 and 2), [Zn(HLn)2(H2O)2] (n = 3 and 5) and [Zn(HL4)2]n were synthesized and characterized by elemental analysis, IR, 1H, 19F, 13C, and 15N NMR spectroscopy, and ESI mass spectrometry. In addition, the structures of two ligands and three complexes were determined by single-crystal X-ray diffraction. The ligands H2L2 and H2L4 exist both in the NH,NH tautomeric form. Complexes 1 and 2 are mononuclear compounds, while complex 4 is a one-dimensional coordination compound. Density functional theory (DFT) calculations were carried out on proligands, their anions and all zinc complexes, confirming the experimental results, supporting IR and NMR assignments and giving proofs of the mononuclear diaqua structure of complexes 3 and 5. The antibacterial activity of the free ligands and the Zn(II) complexes was established against Escherichia coli and Staphylococcus aureus, and a strong efficiency has been found for Zn(II) complexes, particularly for the polynuclear 4 and the mononuclear diaqua complex 5, the latter containing a ligand with aliphatic and fluorinated substituents able to compromise the permeability of and disrupt the bacterial cell membrane.
Introduction
Pyrazolone fragment is an important structural motif in many compounds displaying a rich biological – antibacterial, antitumor, antisepsis – activity.1–5 Edaravone (3-methyl-1-phenyl-5-pyrazolone) represents one of the most employed precursors in the synthesis of pyrazolone-based ligands. Edaravone itself displays relevant biological features, being used for brain disorders6,7 and myocardial ischemia,8 but also shows antipyretic and analgesic activity.9,10 Some analogues of edaravone are known to possess anticancer activity.11 Pyrazolone-based ligands have attracted increasing interest for their coordination chemistry toward many metal ions.12–15 Many of their metal complexes, containing for example titanium,16–18 ruthenium,19–25 platinum,26 tin27–29 and silver,30–32 have shown to possess intriguing anticancer and antibacterial activity. Although there have been numerous reports on condensation products of salicylal hydrazone and 4-acyl-5-pyrazolones and their metal complexes,33–37 fewer studies on condensation products of substituted phenyl-hydrazones and 4-acyl-5-pyrazolones have been reported. All these pyrazolones are highly coloured and have been used extensively in the dyestuffs industry.38 We are interested in the coordination chemistry of pyrazolone-base ligands and on Zn(II) complexes from a long time,39–41 and recently we expanded our studies to heteroleptic complexes containing a N,N-chelating ligand (the 4,4′-dinonyl-2,2′-bipyridine) and 4-acyl-5-pyrazolones,42 which were screened for their in vitro effect on cell proliferation of three human prostate cell lines, showing strong cytotoxic activity against all cell lines affecting key molecules such as p-AKT and p21 waf, involved in the cell proliferation and/or arrest. Moreover, other heteroleptic Zn(II) complexes containing hexacatenar 2,2′-bipyridine or 1,10-phenanthroline ligands, exhibit green luminescence, with a general and reversible red-shift related to the increase of temperature.43 Here we report the synthesis of novel hydrazone-pyrazolone ligands with different functionalities in the hydrazone fragment and their zinc(II) complexes, together with an extensive investigation of their structural and antibacterial properties.Results and discussion
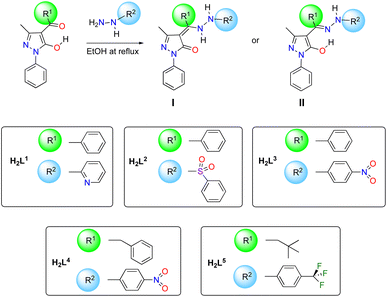
Synthesis of proligands and spectroscopic characterization
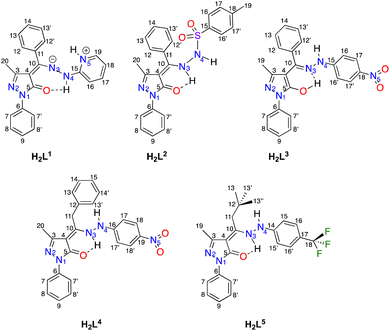
Proligands H2Ln (n = 1–5) have been prepared by reaction between previously reported 4-acyl-5-pyrazolones and monosubstituted hydrazones, following the procedure shown in Scheme 1. H2L1 and H2L3 were previously reported by us and fully characterized in combination with (arene)Ru(II) acceptors,44 whereas proligands H2L2, H2L4 and H2L5 are new. They are sharp melting solid substances, soluble in DMSO, DMF, chlorinated solvents, acetone, acetonitrile, and alcohols and are slightly soluble in diethyl ether. These molecules can in principle exist in two tautomeric forms (I and II in Scheme 1), depending on the electronic features of R1 and R2 substituents and on the solvent of crystallization.45 In detail, H2L1 is known to exists in the solid state as uncommon zwitterion while H2L3 adopts a N–H,O–H tautomeric form in both solid state and chlorinated solution (Fig. 1).44The new proligands H2L2, H2L4 and H2L5 exist in the N–H,N–H form, tautomer I (Fig. 1). In the solid state, this was confirmed by the X-ray structures of H2L2 and H2L4 (discussed below) and the IR data of H2L5. Characteristic bands were identified in IR spectra of proligands, which were assigned also by comparison with computed IR spectra (see DFT studies below). In detail, they are the asymmetric and symmetric stretching modes at 1301 and 1165 cm−1 of the sulfonyl SO2 in H2L2,46,47 the asymmetric and symmetric stretching modes at 1490–1530 and 1325–1331 cm−1 of nitro NO2 in H2L3 and H2L4,48,49 as well as bands at 1312, 1163, 1112 and 1065 cm−1 due to asymmetric and symmetric stretching vibrations of CF3 group in H2L5,50 while the typical ν(N–N) mode for all proligands is identified in the range 1040–1120 cm−1.44,511H and 13C chemical shifts were assigned based on the 1H–1H, and one-bond and long-range 1H–13C couplings, seen in the {1H,1H}-COSY, {1H,13C}-HSQC, and {1H,13C}-HMBC (see ESI†). Moreover, indirect 15N NMR chemical shifts were assigned based on the {1H,15N}-HSQC and {1H,15N}-HMBC for both proligands and complexes 1–5 (see Table 1 and Fig. 1 for C and N atoms numeration). The proton NMR spectra of proligands H2L4 and H2L5 in deuterochloroform displays broad resonances at 12.40–12.83 and 6.50–6.80 ppm, due to the two types of NH groups, in accordance with the prevalence in solution of N–H,N–H tautomer I. Although for H2L2 only one NH resonance was observed at 6.81 ppm, the N–H,N–H tautomeric form observed in the X-ray is assumed to be preserved also in solution (see DFT studies below), even if in the {1H,15N}-HSQC NMR only the N4–H direct coupling was displayed.
| Ligands | N1 | N2 | N3 | N4 | N5 |
|---|---|---|---|---|---|
| H2L1 | n.o. | n.o. | n.o. | n.o. | n.o. |
| H2L2 | 189.8 | 271.7 | n.o. | 117.3 | — |
| H2L3 | 190.1 | 271.6 | n.o. | 117.0 | 369.9 |
| H2L4 | n.o. | 288.9 | 140.6 | 98.9 | 379.4 |
| H2L5 | 191.4 | 285.8 | 143.8 | 99.7 | — |
| Complexes | |||||
| 1 | n.o. | n.o. | n.o. | 132.9 | n.o. |
| 2 | 192.9 | 276.4 | n.o. | 151.1 | — |
| 3 | 197.5 | n.o. | n.o. | 125.2 | n.o. |
| 4 | n.o. | n.o. | n.o. | n.o. | n.o. |
| 5 | n.o. | 274.1 | n.o. | 120.0 | — |
Synthesis of complexes and spectroscopic characterization
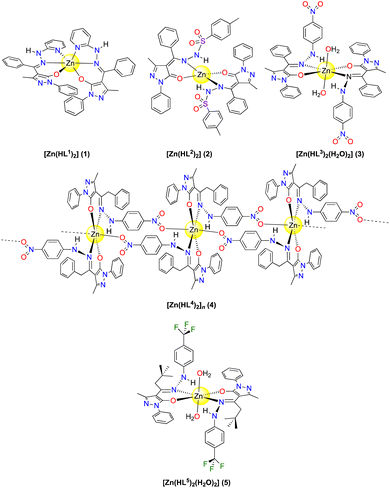
Complexes 1–5 were synthesised by interaction of Zn(O2CCH3)2·2H2O with the proligands H2Ln, in methanol at room temperature. They are sharp melting solids soluble in most organic solvents, and in the case of 2, 3 and 5 also quite soluble in methanol, while not soluble at all in water and ethers. All analytical and spectral data are in accordance with the formulation of complexes proposed in Fig. 2: while 1 and 2 are mononuclear anhydrous [Zn(HL1)2] and [Zn(HL2)2] species, complexes 3 and 5 were isolated as diaqua [Zn(HLn)2(H2O)2] compounds, with two water molecules in the zinc coordination sphere, and complex 4 is a monodimensional coordination polymer with formula [Zn(HL4)2]n (see below). While complex 3 is quite soluble in methanol and is isolated as a precipitate from methanol only after few hours of reaction, 4 is not soluble at all in methanol, from which it immediately precipitates. Melting point of 4 is higher than that of 3 of about 20 °C, in accordance with a different solid state structure. Conductivity measurements in acetonitrile indicate that 1–5 are stable and do not dissociate in solution at room temperature. The IR spectra of 1–5 provide valuable information about the nature of the functional group coordinated to the zinc atom. However, the spectra of proligands are quite complex, especially in the 1500–1650 cm−1 region from the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and more than one C
C, and more than one C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N in their structure, and definitive assignments are not straightforward. Here, we tentatively assign some band on the basis of previous literature on similar chemical systems and DFT calculations. The ν(C
N in their structure, and definitive assignments are not straightforward. Here, we tentatively assign some band on the basis of previous literature on similar chemical systems and DFT calculations. The ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) band at 1641 cm−1 for the free proligand H2L1 is shifted upon coordination, which suggests involvement of the keto group of a pyrazolone ring in coordination.44 The strong bands falling between 1626 and 1600 cm−1 in the spectra of the other free proligands, which are characteristic of the ν(>C
O) band at 1641 cm−1 for the free proligand H2L1 is shifted upon coordination, which suggests involvement of the keto group of a pyrazolone ring in coordination.44 The strong bands falling between 1626 and 1600 cm−1 in the spectra of the other free proligands, which are characteristic of the ν(>C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) azomethine group, are shifted to lower wavenumbers in the spectra of the complexes, implying the coordination of the ligands through azomethine nitrogen.52–54 Additionally, a medium and broad band at ca. 3300–3400 cm−1 in the spectra of dihydrate complexes 3 and 5 confirms the presence of water in the zinc environment. By contrast, in the IR of 4 a medium and sharp band was found at 3275 cm−1, assigned to ν(N–H) (see Fig. S47 in ESI†). Finally, the formation of new band in the low frequency regions 520–580 and 430–470 cm−1 is attributed to ν(M–N) and ν(M–O), respectively.
N–) azomethine group, are shifted to lower wavenumbers in the spectra of the complexes, implying the coordination of the ligands through azomethine nitrogen.52–54 Additionally, a medium and broad band at ca. 3300–3400 cm−1 in the spectra of dihydrate complexes 3 and 5 confirms the presence of water in the zinc environment. By contrast, in the IR of 4 a medium and sharp band was found at 3275 cm−1, assigned to ν(N–H) (see Fig. S47 in ESI†). Finally, the formation of new band in the low frequency regions 520–580 and 430–470 cm−1 is attributed to ν(M–N) and ν(M–O), respectively.
The electrospray ionization (ESI) mass spectra of 1–5 in positive ion mode, recorded in acetonitrile, show the typical isotopic patterns expected for Zn and display main peaks that correspond to the species [Zn(HL′)]+ arising from dissociation of one ligand and/or [Zn(HL′)(H2L′)]+ due to protonation of one ligand in the zinc complex or also to [Zn(HL′)2 + Na]+. 1H and 13C chemical shifts in the NMR spectra complexes 1–5 display the expected shift of those resonances due to proton and carbon atoms close to N and O donor atoms involved in bonding to zinc. In the case of polynuclear complex 4, we assume that in chloroform the weak intermolecular Zn–O(nitro) interactions are easily broken and mononuclear species are present in solution. The disappearance of resonances at 12.40–12.83 ppm but the persistence of those at 6.50–6.80 ppm, confirm deprotonation of proligands and presence of the hydrazide N–H upon coordination of the ligands to zinc.
Unfortunately, no 15N NMR chemical shift was observed for complex 4 and only some indirect 15N NMR chemical shifts of 1–3 and 5 were detected (Table 1), but not those of N3 atoms directly coordinated to zinc (and also N5 for complex 1). However, those of vicinal N4 were clearly identified to lower fields for complexes 2, 3 and 5, in accordance with a deshielding upon coordination. Complexes 1–5 are stable in DMSO-d6 and do not undergo any dissociation, as established by NMR studies (Fig. S46 in ESI†).
X-ray structural characterization
The structures of proligands H2L2 and H2L4, together with complexes 1, 2, and 4, were determined by single-crystal X-ray diffraction. Selected bond distances and angles are shown in Tables 2 and 4. The crystallographic data are given in Tables S1 and S2.† Although both proligands show a similar scaffold based on pyrazolone, hydrazone and phenyl fragments, their molecular structure are different since H2L2 crystallizes in monoclinic C2/c space group and H2L4 does in P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Fig. 3). The structures of proligands are nonplanar since the phenyl ring is turned with respect to the pyrazolone plane by 26.5(3)° and 17.8(3)° in H2L2 and H2L4, respectively. The torsion angles in hydrazone group are −92.6° for H2L2 (S–N–N–C) and −126.9° for H2L4 (C–N–N–C). The hydrazone N–N bond distances for all measured compounds are in the range of 1.397(3)–1.432(4) Å, being slightly higher for metal complexes. Proligand molecules are stabilized within the crystal through hydrogen bonds between two molecules. In both proligands the intermolecular interactions occur between the hydrogens of hydrazone (N3 and N4) and the oxygen of the pyrazolone (O1) (Table 3). Regarding the coordination compounds, complex 1 is based on ligand (HL1)−, 2 is based on (HL2)− and crystallization methanol molecules, and 4 is formed by the coordination of (HL4)− ligand (Fig. 4). 1, with general formula [Zn(HL1)2], crystallizes in the triclinic space group P
(Fig. 3). The structures of proligands are nonplanar since the phenyl ring is turned with respect to the pyrazolone plane by 26.5(3)° and 17.8(3)° in H2L2 and H2L4, respectively. The torsion angles in hydrazone group are −92.6° for H2L2 (S–N–N–C) and −126.9° for H2L4 (C–N–N–C). The hydrazone N–N bond distances for all measured compounds are in the range of 1.397(3)–1.432(4) Å, being slightly higher for metal complexes. Proligand molecules are stabilized within the crystal through hydrogen bonds between two molecules. In both proligands the intermolecular interactions occur between the hydrogens of hydrazone (N3 and N4) and the oxygen of the pyrazolone (O1) (Table 3). Regarding the coordination compounds, complex 1 is based on ligand (HL1)−, 2 is based on (HL2)− and crystallization methanol molecules, and 4 is formed by the coordination of (HL4)− ligand (Fig. 4). 1, with general formula [Zn(HL1)2], crystallizes in the triclinic space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . The asymmetric unit consists in one Zn(II) ion and two monoanionic (HL1)− ligands.
. The asymmetric unit consists in one Zn(II) ion and two monoanionic (HL1)− ligands.
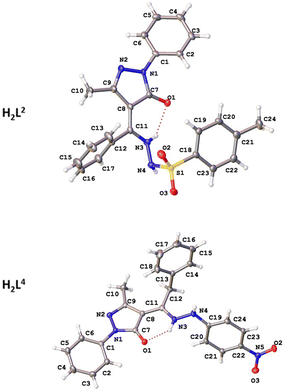 | ||
| Fig. 3 Molecular structures of ligands H2L2 and H2L4. Thermal ellipsoids are presented at 50% probability. | ||
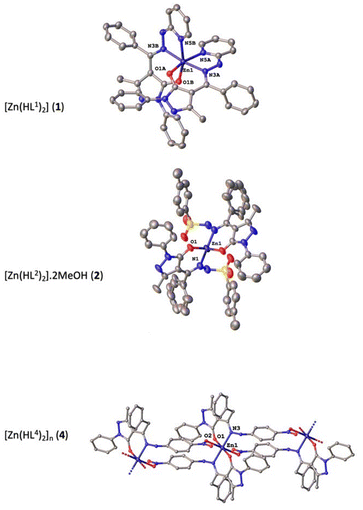 | ||
| Fig. 4 Molecular structures of the Zn(II) complexes 1, 2, and 4. Thermal ellipsoids are presented at 50% probability. Hydrogen atoms omitted for clarity. | ||
| Bond | Distance (Å) | Bond | Angle (°) |
|---|---|---|---|
| H2L2 | |||
| C1–N1 | 1.418(3) | C2–C1–N1 | 119.4(2) |
| C7–N1 | 1.369(3) | O1–C7–N1 | 125.5(2) |
| C7–O1 | 1.254(4) | C8–C11–C12 | 123.1(2) |
| C8–C11 | 1.382(4) | N3–C11–C8 | 117.6(2) |
| C9–N2 | 1.307(3) | C7–N1–N2 | 112.07(19) |
| N1–N2 | 1.403(3) | C11–N3–N4 | 122.3(2) |
| C11–N3 | 1.332(3) | N3–N4–S1 | 113.66(17) |
| N3–N4 | 1.397(3) | C19–C18–S1 | 119.7(2) |
| N4–S1 | 1.677(2) | N4–S1–C18 | 108.10(12) |
| C18–S1 | 1.755(3) | ||
| H2L4 | |||
| C1–N1 | 1.424(2) | C2–C1–N1 | 120.52(15) |
| C7–N1 | 1.371(2) | O1–C7–N1 | 125.90(15) |
| C7–O1 | 1.258(2) | C8–C11–C12 | 124.43(15) |
| C8–C11 | 1.397(2) | N3–C11–C8 | 117.81(16) |
| C9–N2 | 1.313(2) | C7–N1–N2 | 112.09(13) |
| N1–N2 | 1.4007(19) | C11–N3–N4 | 120.83(15) |
| C11–N3 | 1.332(2) | ||
| N3–N4 | 1.389(2) | ||
| C22–N5 | 1.456(2) | ||
| D–H⋯A | D–H | H⋯A | D⋯A | Angle |
|---|---|---|---|---|
| H2L2 | ||||
| N3–H3A⋯O1 | 0.88 | 2.05 | 2.706(3) | 130.4 |
| N3–H3A⋯O1 | 0.88 | 2.24 | 2.833(3) | 124.1 |
| N4–H4A⋯O1 | 0.81(3) | 2.38(3) | 2.925(3) | 125(3) |
| H2L4 | ||||
| C2–H2⋯O1 | 0.93 | 2.32 | 2.913(2) | 121.4 |
| N4–H4A⋯O1 | 0.87(2) | 2.01(2) | 2.8564(19) | 162(2) |
| N3–H3A⋯O1 | 0.89(2) | 1.94(2) | 2.6662(19) | 138.0(19) |
| Bond | Distance (Å) | Bond | Angle (°) |
|---|---|---|---|
| Complex 1 | |||
| Zn1–N3A | 2.151(2) | N3B–Zn1–N3A | 171.98(9) |
| Zn1–N3B | 2.143(2) | N5A–Zn1–N3A | 76.33(9) |
| Zn1–N5A | 2.132(2) | N5A–Zn1–N3B | 97.47(9) |
| Zn1–N5B | 2.142(2) | N5A–Zn1–N5B | 92.74(9) |
| Zn1–O1A | 2.051(2) | N5B–Zn1–N3A | 98.99(9) |
| Zn1–O1B | 2.049(2) | N5B–Zn1–N3B | 76.00(9) |
| O1A–Zn1–N3A | 87.22(8) | ||
| 01A–Zn1–N3B | 98.96(9) | ||
| O1A–Zn1–N5A | 163.54(8) | ||
| O1A–Zn1–N5B | 90.30(9) | ||
| O1B–Zn1–N3A | 98.05(9) | ||
| O1B–Zn1–N3B | 87.25(9) | ||
| O1B–Zn1–N5A | 92.94(9) | ||
| O1B–Zn1–N5B | 162.88(8) | ||
| O1B–Zn1–O1A | 88.80(8) | ||
| Complex 2 | |||
| Zn1–O1 | 1.958(3) | O1–Zn1–O1 | 97.6(2) |
| Zn1–N1 | 1.998(4) | O1–Zn1–N1 | 119.19(11) |
| O1–Zn1–N1 | 96.95(12) | ||
| O1–Zn1–N1 | 96.94(12) | ||
| O1–Zn1–N1 | 119.19(11) | ||
| N1–Zn1–N1 | 125.0(2) | ||
| Complex 4 | |||
| Zn1–N3 | 2.069(3) | N3–Zn1–N3 | 180.0 |
| Zn1–O1 | 2.004(3) | N3–Zn1–O2 | 89.09(12) |
| Zn1–O2 | 2.342(3) | N3–Zn1–O2 | 90.91(12) |
| O1–Zn1–N3 | 88.44(12) | ||
| O1–Zn1–N3 | 91.56(12) | ||
| O1–Zn1–O1 | 180.0 | ||
| O1–Zn1–O2 | 84.67(11) | ||
| O1–Zn1–O2 | 95.33(11) | ||
| O2–Zn1–O2 | 180.0 | ||
The metal centre exhibits a distorted octahedral coordination environment, ZnO2N4. Zn–O bond distances are in the 2.049(2)–2.051(2) Å range, while Zn–N bond distances range from 2.132(2) to 2.151(2) Å. Each ligand is coordinated by the oxygen of pyrazolone, one nitrogen of hydrazone and the nitrogen of pyridine rings. The structure of 1 consists of a monomer where neither hydrogen bonds nor van der Waals interactions exist.
Complex 2 consists of a monomer as well that crystallizes in the monoclinic C2/c space group with general formula [Zn(HL2)2]·2MeOH. The asymmetric unit contains half of a Zn(II) ion and one (HL2)− molecule.
Each Zn(II) ion shows a tetrahedral ZnO2N2 geometry formed by two oxygen atoms from pyrazolone and two nitrogen atoms from hydrazone of two different monoanionic (HL2)− ligands. The Zn–O bond distance is 1.958(3) Å whereas the Zn–N bond length is 1.998(4) Å. Due to the disorder within the voids, the crystallization methanol molecules were removed from the structure. Finally, complex 4 crystallizes in the triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group and the structure consists of a monodimensional coordination polymer with general formula [Zn(HL4)2]n. The asymmetric unit shows half of zinc(II) ion and one monoanionic (HL4)− ligand. Each central atom is coordinated to four different ligand molecules by showing an octahedral ZnO4N2 geometry formed by two oxygen atoms of pyrazolone ring, two oxygen atoms from nitro group and two nitrogen atoms pertaining to hydrazone moiety. The oxygens of pyrazolone occupy the basal plane of the octahedron together with nitrogen bonds, with Zn–Obasal bond distance of 2.004(3) Å and Zn–N bond length of 2.069(3) Å.
space group and the structure consists of a monodimensional coordination polymer with general formula [Zn(HL4)2]n. The asymmetric unit shows half of zinc(II) ion and one monoanionic (HL4)− ligand. Each central atom is coordinated to four different ligand molecules by showing an octahedral ZnO4N2 geometry formed by two oxygen atoms of pyrazolone ring, two oxygen atoms from nitro group and two nitrogen atoms pertaining to hydrazone moiety. The oxygens of pyrazolone occupy the basal plane of the octahedron together with nitrogen bonds, with Zn–Obasal bond distance of 2.004(3) Å and Zn–N bond length of 2.069(3) Å.
On the other hand, the oxygen atoms pertaining to nitro groups are situated at the apical position with a Zn–Oapical bond distance value of 2.342(3) Å. Each proligand molecule coordinates to two Zn(II) ions. This disposition of the ligands creates chains that propagate along b axis. Within chains stacking interaction between nitrophenyl moieties occurs, with a centroid-to-centroid distance of 4.155 Å. Only one intramolecular hydrogen bond helps to stabilize the structure between the nitrogen of hydrazone (N4) and the oxygen from pyrazolone (O1) of two different ligand molecules, with a N4–H4A⋯O1 distance of 2.786(4) Å.
Theoretical studies
Tautomers I and II of proligands H2L2, H2L4 and H2L5 (Scheme 1) were investigated using DFT at the B3LYP/6-311G** level of theory, while theoretical studies on proligands H2L1 and H2L3 were reported in the past.44 For all the proligands tautomer I showed to be the most stable in gas phase and in chloroform solution, even if the energy differences are small and comprised in the range of ca. 0.6–5.7 kcal mol−1 (see Table S2† for details). The comparison between the computed and experimental 1H, 13C and 15N NMR spectral data showed a better fit for tautomer I, proving to be the observed species in solution (for instance, see Fig. S48† for H2L5). In particular, the values for 1H and 13C are very similar for both tautomers (R2 = 0.98–0.99, range of correlation values for the NMR comparison), while R2 values from 15N NMR are entirely different, in favour of the NH,NH tautomer I. The calculated structural parameter of tautomer I for proligands H2L2 and H2L4 agree well with those observed from the X-ray diffraction results (Fig. S49†). In particular, the experimental C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond distances of 1.254(4) and 1.258(2) Å (for H2L2 and H2L4, respectively) is well reproduced by calculations with values of 1.241 and 1.242 Å for tautomer I vs. 1.321 and 1.326 Å for tautomer II, confirming the proposed assignment. Finally, the calculated infrared spectra of tautomer I fit well with the experimental ones (for example for H2L5, see Fig. S50†), furtherly supporting the IR assignments previously discussed (Table S3†). To analyse the coordination capabilities of proligands, their anionic forms were optimized as well. For all [HLn]− anions the charge localization reflects the deprotonation of tautomer I, showing a shorter C
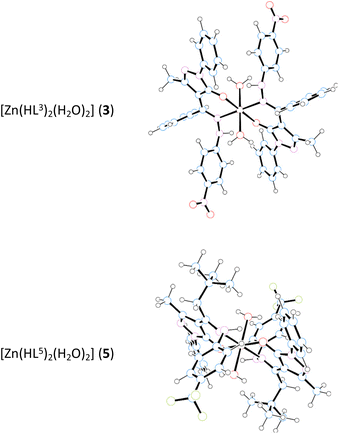
O bond distances of 1.254(4) and 1.258(2) Å (for H2L2 and H2L4, respectively) is well reproduced by calculations with values of 1.241 and 1.242 Å for tautomer I vs. 1.321 and 1.326 Å for tautomer II, confirming the proposed assignment. Finally, the calculated infrared spectra of tautomer I fit well with the experimental ones (for example for H2L5, see Fig. S50†), furtherly supporting the IR assignments previously discussed (Table S3†). To analyse the coordination capabilities of proligands, their anionic forms were optimized as well. For all [HLn]− anions the charge localization reflects the deprotonation of tautomer I, showing a shorter C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond than C–N bond (∼1.23 Å vs. ∼1.30 Å). The MOs mostly involved in the coordination to the metal centre for anions [HL2]−, [HL3]−, [HL4]− and [HL5]− are shown to be HOMO−2 and HOMO−8/−9/−10 (depending on the species). These being respectively the in-phase and out-of-phase contributions of the σ-type, which are involved in the donation to the zinc centre. Other minor contributions are present with orbitals localized on the N or O atoms. Concerning [HL1]−, additional contributions were obviously found due to its O,N,N-tridentate hapticity (see selected MOs of [HL1]−, [HL2]− and [HL4]− in Fig. S51†). The optimized structures of the anions show some differences with respect to the structure they assume upon coordination. The most noticeable difference is the conformation of the phenyl moiety (on R2 group in Scheme 1): in the optimized anion the phenyl ring is positioned on the same plane of O and N atoms, while in the complex it orientates perpendicularly. Secondly, in the complex, ligand HL2 directs its SO2 group towards the metal centre and in a similar fashion ligand HL4 directs its NO2 group to perform an intermolecular interaction. For these reasons we carried out single-point calculations of the ligands with the geometries found in the optimization of their respective complexes. No significant differences were found and the results, which were also collected in Fig. S51,† do not require further discussion. Complexes 1–3 and 5 were also analysed by DFT using the 6-311G* basis set. Complexes 1, [Zn(HL1)2], and 2, [Zn(HL2)2], are well reproduced by the calculations (Table S4†) and their optimized structures are shown in Fig. S52.†1 shows an excellent match between the calculated and the experimental data from the X-ray, while in 2 the main discrepancy is the interaction between Zn centre and the O atom of the SO2 group, 2.277 Å, which is shorter than the experimental value of 2.770 Å. In both cases, computed IR and NMR spectra of the optimized 1 and 2 are fully consistent with the experimental data (R2 correlation values of 0.9908 and 9904, respectively, for the NMR comparison). The optimized structures of complexes 3, [Zn(HL3)2(H2O)2], and 5, [Zn(HL5)2(H2O)2] (Fig. 5), display octahedral geometries with two water molecules coordinated in trans position. The coordinated ligands [HL3]− and [HL5]− show a bonding localization, which is reminiscent to the tautomer I, with different bond lengths of C
O bond than C–N bond (∼1.23 Å vs. ∼1.30 Å). The MOs mostly involved in the coordination to the metal centre for anions [HL2]−, [HL3]−, [HL4]− and [HL5]− are shown to be HOMO−2 and HOMO−8/−9/−10 (depending on the species). These being respectively the in-phase and out-of-phase contributions of the σ-type, which are involved in the donation to the zinc centre. Other minor contributions are present with orbitals localized on the N or O atoms. Concerning [HL1]−, additional contributions were obviously found due to its O,N,N-tridentate hapticity (see selected MOs of [HL1]−, [HL2]− and [HL4]− in Fig. S51†). The optimized structures of the anions show some differences with respect to the structure they assume upon coordination. The most noticeable difference is the conformation of the phenyl moiety (on R2 group in Scheme 1): in the optimized anion the phenyl ring is positioned on the same plane of O and N atoms, while in the complex it orientates perpendicularly. Secondly, in the complex, ligand HL2 directs its SO2 group towards the metal centre and in a similar fashion ligand HL4 directs its NO2 group to perform an intermolecular interaction. For these reasons we carried out single-point calculations of the ligands with the geometries found in the optimization of their respective complexes. No significant differences were found and the results, which were also collected in Fig. S51,† do not require further discussion. Complexes 1–3 and 5 were also analysed by DFT using the 6-311G* basis set. Complexes 1, [Zn(HL1)2], and 2, [Zn(HL2)2], are well reproduced by the calculations (Table S4†) and their optimized structures are shown in Fig. S52.†1 shows an excellent match between the calculated and the experimental data from the X-ray, while in 2 the main discrepancy is the interaction between Zn centre and the O atom of the SO2 group, 2.277 Å, which is shorter than the experimental value of 2.770 Å. In both cases, computed IR and NMR spectra of the optimized 1 and 2 are fully consistent with the experimental data (R2 correlation values of 0.9908 and 9904, respectively, for the NMR comparison). The optimized structures of complexes 3, [Zn(HL3)2(H2O)2], and 5, [Zn(HL5)2(H2O)2] (Fig. 5), display octahedral geometries with two water molecules coordinated in trans position. The coordinated ligands [HL3]− and [HL5]− show a bonding localization, which is reminiscent to the tautomer I, with different bond lengths of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (ca. 1.27 Å) and
O (ca. 1.27 Å) and 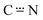 (ca. 1.32 Å).
(ca. 1.32 Å).
To rationalize the octahedral geometry in these compounds, the structures without the coordinated water ligands were additionally optimized. For [Zn(HL3)2], two structures with square planar, 3′sqp, and distorted tetrahedral, 3′Td, geometries were located, while for [Zn(HL5)2] only the distorted tetrahedral structure was found, 5′Td. In both complexes, 3 and 5, the dissociation of water molecules is not energetically favoured (ΔG values of ca. 6 kcal mol−1 for the dissociation toward the tetrahedral geometry), confirming the proposed octahedral structures. Furthermore, calculated NMR spectra of [Zn(HLn)2(H2O)2] species are well correlated to the experimental NMR with R2 values of 0.9954 and 0.9942, for 3 and 5, respectively (Fig. S53†). As already discussed in the X-ray structural characterization, complex 4, [Zn(HL4)2]n, arranges in a monodimensional polymeric structure. To gain information about the polymerization process, the geometries of the monomer [Zn(HL4)2] with a square planar disposition and that of the dimer, [Zn(HL4)2]2, were optimized. The dimerization process is exergonic with a ΔG value of −7.9 kcal mol−1, suggesting that the polymerization process to 4, [Zn(HL4)2]n, is accessible. Also, the trimer [Zn(HL4)2]3 was optimized, although frequency calculations were not carried out due to the high number of atoms and the limits of time computation facilities. In any case, ΔE value of −53.2 kcal mol−1 again suggests a very favourable polymerization process. The trimer is an excellent simulation of the polymeric 4. In fact, focusing the attention on the central Zin centre with the ZnO4N2 environment the computed structural parameters fit very well with experimental. Even the stacking interaction between nitrophenyl rings is well reproduced (see Table S4†). Finally, the breakup of the polymeric nature of 4, [Zn(HL4)2]n, was analysed in the presence of water to yield the monomer [Zn(HL4)2(H2O)2]. In particular, the hydration of dimer [Zn(HL4)2]2 to the [Zn(HL4)2(H2O)2] species is exergonic (ΔG = −13.4 kcal mol−1), while the hydration of trimer [Zn(HL4)2]3 is analogously favourable (ΔE = −56.7 kcal mol−1).
Antibacterial activity
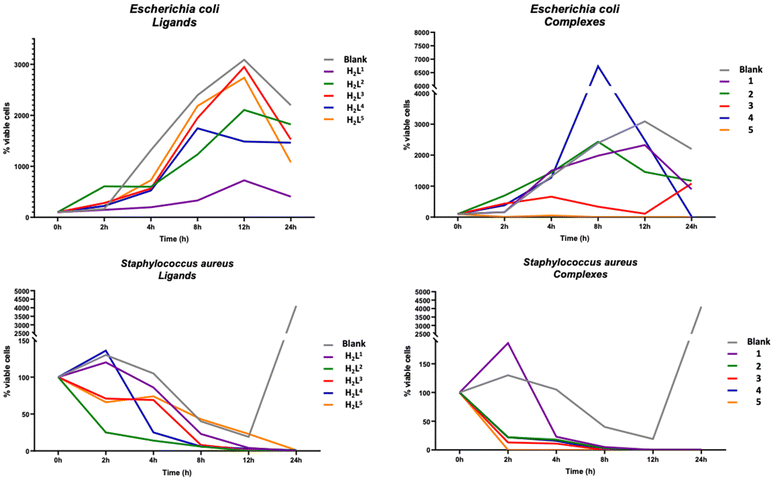
To evaluate the antibacterial activity of the ligands H2L1–H2L5 and of the Zn(II) complexes 1–5, Gram-positive S. aureus and Gram-negative E. coli were selected as models. In the following discussion, bactericidal activity is defined as the reduction of bacterial growth ≥99.9% within 18–24 h of inoculation, while bacteriostatic activity is associated with a reduction of bacterial growth maintained stable between 90 and 99%, within 18 and 24 h from inoculation.55,56 The number of viable cells was used to calculate the growth rate which numerically corresponds to the inverse of the killing rate, a parameter used to define whether a substance exhibits a bactericidal or bacteriostatic action.31 The results (Fig. 6) showed that all tested ligands and complexes did not achieve stable antibacterial activity against E. coli within 24 h, except 4 and 5. In particular, complex 4 needs 24 hours to achieve good performance, while 5 achieves 93% reduction of viable cells within the first 2 hours of exposure, then increasing to 99.99% after 24 hours. By contrast, all tested ligands and complexes showed good bacterial killing properties against S. aureus. In detail, ligands are able to inhibit the growth of bacterial cells within 24 hours while compounds 1–5 showed good performance with over 80% of bacterial death in the first 4 hours. Going deeper into the analysis of the results, it can be seen that the ligands H2L3 and H2L4, whose structures differ only in the acyl fragment, with a phenyl and a benzyl respectively, show a different efficiency as antibacterial agents, with H2L4 more active than H2L3 against both bacterial strains tested. Consistent with these results, complex 4, which differs from 3 being polynuclear rather than mononuclear, is more powerful than 3, especially against E. coli. Furthermore, 5 showed a bactericidal performance after the first two hours of inoculation and it was able to maintain the performance achieved during all treatment periods. In general, the performances of all compounds are more homogeneous towards S. aureus, both in terms of efficiency and time. A growth inhibition between 95% and 99% of S. aureus was achieved by compounds 1–5 within the first 8 hours of treatment, showing clear bacteriostatic activity. Furthermore, 1 and 2 showed bactericidal activity within 12 hours of treatment, 3 and 4 within the first 8 hours, and 5 within the first 2 hours, with 99.99% growth inhibition which remains stable even 24 hours after the test. Significantly, a different behaviour was observed towards E. coli, since 99.9% inhibition of bacterial growth was observed only by complex 5 after 8 hours, which was maintained even 24 hours after treatment, showing both bacteriostatic and bactericidal activity. Generally, other Schiff base complexes reported in the literature57–59 display an enhanced bactericidal effect than the free ligands, according to the Overtone's concept60 and the chelation theory.61Reactive oxygen species (ROS) detection assay
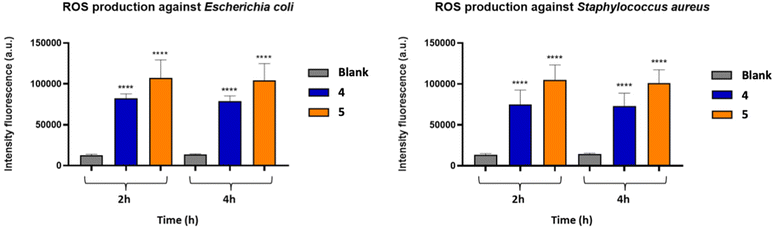
In order to understand a possible mechanism of action of the Zn(II) complexes, the most efficient 4 and 5 were further investigated through the ROS assay against E. coli and S. aureus. To detect the amount of ROS generated, the 2′,7′-dichlorofluorescein diacetate (DCFDA) was used as a reagent. This reactive is deacetylated by cellular esterase to a non-fluorescent compound, which is subsequently oxidized by ROS to 2′,7′-dichlorofluorescein (DCF). DCF is highly fluorescent and is detected by excitation/emission fluorescence spectroscopy at 485 nm/535 nm. The increased generation of free radicals leads to greater stress for the bacterial cell and the increase in the fluorescence intensity in the first hours (2 h, 4 h) of treatment (Fig. 7) confirmed the generation of ROS in both bacterial strains after incubation with compounds 4 and 5. The fluorescence signal of the untreated bacterial suspension (blank) was negligible, in agreement with a generation of free radicals in both E. coli and S. aureus cells.Propidium iodide (PI) – viability assay
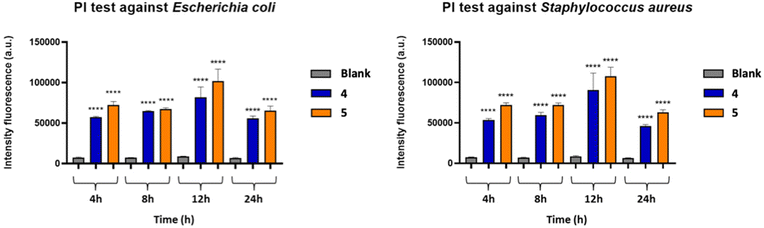
The viability test was performed to assess membrane damage of E. coli and S. aureus bacterial cells caused by 4 and 5, using the fluorescent PI dye. PI binds DNA and freely penetrates the cell membranes of dead or dying cells, but not the viable cells, allowing the evaluation of the damage in terms of permeabilization. The quantification of the damage on the bacterial cell, following the treatment with 4 and 5, was performed at different time intervals (4 h, 8 h, 12 h, 24 h) (Fig. 8). The emission of red fluorescence was determined with respect to the untreated bacterial suspension (blank). In general, it is possible to observe a moderate increase in fluorescence intensity within the first 12 hours, which is then maintained higher with respect to the blank even after 24 hours of treatment by both compounds against both bacterial strains, with a superior efficiency of complex 5 with respect to 4, thus indicating the ability of Zn(II) complexes to modify the permeability of the cell membrane by disorganizing it and leading to the death of the bacterium.Confocal laser scanning microscopy (CLSM) study
To further determine the overall viability of the bacterial cells the CLSM was performed. According to the LIVE/DEAD BacLight Bacterial Viability Kits, the SYTO 9 stain generally labels all bacteria in a population, while propidium iodide penetrates only bacteria with damaged membranes, causing a reduction in the SYTO 9 stain fluorescence when both dyes are present. Thus, with an appropriate mixture of both stains, bacteria with intact cell membranes stain fluorescent green, whereas bacteria with damaged membranes stain fluorescent red. The images obtained under the confocal microscope show in Fig. 9a and c, respectively, the untreated E. coli and S. aureus cells, which clearly appear to produce green fluorescence. In Fig. 9b and d, corresponding respectively to the E. coli cells treated with complex 4 and the S. aureus cells treated with complex 5, it is possible to observe a large number of bacterial cells marked with red fluorescent material. These results revealed that after treatment with complexes 4 and 5, the cell membrane of both bacterial strains is severely damaged, thus confirming the results obtained from previous assays.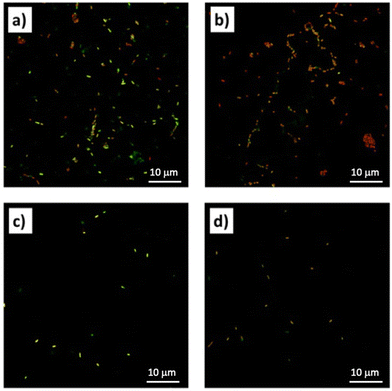 | ||
| Fig. 9 Confocal laser scanning microscopy (CLSM) images of (a) E. coli control, (b) E. coli treated with complex 4; (c) S. aureus control; (d) S. aureus treated with complex 5. | ||
Scanning electron microscopy (SEM) study
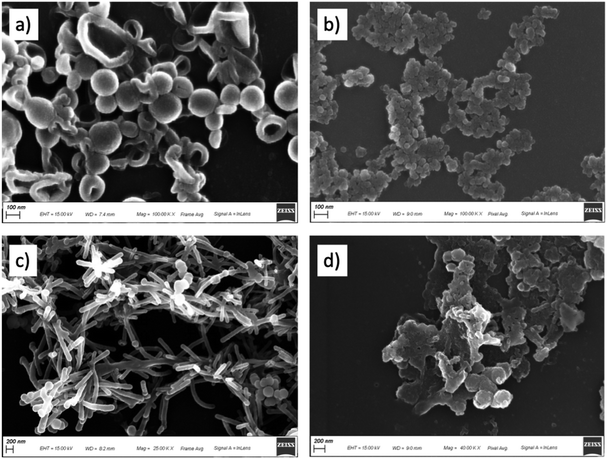
The SEM images show the morphological alterations of the bacterial cells following the treatments with complexes 4 and 5. In Fig. 10a it is possible to observe the control sample of the S. aureus, having a round appearance with a rigid cell wall. After treatment with complex 5 (Fig. 10b), the cells are extremely smaller and have vesicular structures. Fig. 10c shows the control sample of E. coli, having the classic morphology with a rod shape and smooth surface. After treatment with complex 4 (Fig. 10d), the cells completely lose their regular surface layer, resulting in deformation and agglomeration. The SEM results clearly reveal the damage to the membrane permeability of bacterial cells thus confirming the results of the previous tests carried out.In conclusion, all zinc(II) complexes are able to improve the antimicrobial activity toward E. coli within 12 hours and against S. aureus even within 8 hours, with complexes 4 and 5 displaying the best performances, the latter being able to completely inhibit the growth of both bacterial lines within 2 h after contact. Apart from the general greater efficacy of Zn(II) complexes against S. aureus compared to E. coli, clearly attributable to the different cell wall between these bacteria, with that of the Gram negative much thicker and more resistant, it is possible to hypothesize the effect of the different nuclearity of zinc complexes and also the ligands’ structures in these distinctive activities, in fact complex 4 is a polynuclear compound where nitro groups of the ligand (HL4) interact with zinc atoms of vicinal units, while in complex 5 the ligand (HL5) is the only here reported to possess an aliphatic group (a neopentyl) instead of an aromatic one on the R1 position and a trifluoromethyl on the hydrazone fragment, both improving the lipophilic behaviour of 5, and its excellent bacterial killing ability can be related to a superior facility in crossing and disrupting the cell membrane of bacteria. The viability assay seems to confirm this hypothesis, and the results of ROS detection tests give a proof to the formation of highly reactive radicals associated with a breakdown of membrane function and cellular destruction, further confirmed by CLSM and SEM studies.
Experimental
Materials and methods
All reagents were purchased (Aldrich) and used without further purification. All solvents were purified by conventional methods and stored under nitrogen. All reactions and manipulations for the syntheses of proligands H2Ln and their interactions with zinc acetate dihydrate were carried out in the air. The samples for microanalyses were dried in vacuo to constant weight (20 °C, ca. 0.1 Torr). Elemental analyses (C, H, N) were performed in-house with a Fisons Instruments 1108 CHNS-O Elemental Analyser. Melting points are uncorrected and were recorded on a STMP3 Stuart scientific instrument and on a capillary apparatus. IR spectra were recorded from 4000 to 200 cm−1 with a PerkinElmer Spectrum 100 FT-IR instrument. 1H, 19F, and 13C NMR spectra were recorded with a 500 Bruker AscendTM (500 MHz for 1H, 470.6 for 19F, 125 MHz for 13C) instrument operating at room temperature relative to TMS. Positive ESI-Mass Spectra were obtained with a Series 1100 MSI detector HP spectrometer, using acetonitrile as mobile phase. Solutions for electrospray ionization mass spectrometry (ESI-MS) were prepared using reagent grade methanol and obtained data (masses and intensities) were compared to those calculated by using the IsoPro isotopic abundance simulator version 2.1. Peaks containing zinc(II) ions were identified as the highest peak of an isotopic cluster.Synthesis of proligands
The proligands H2L1 and H2L3 were synthesized and characterized as previously reported,44 whereas H2L2, H2L4 and H2L5 are new, and their synthesis and characterization is reported below.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1611, 1582 ν(C
O), 1611, 1582 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1538 ν(C
C), 1538 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1535 ν(C–N), 1301 νas(SO2), 1165vs νs(SO2), 1089 m ν(N–N). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.61 (s, 3H, H20), 2.19 (s, 3H, H19), 6.82 (d, 2H, H17,17′), 7.20 (sbr, 1H, N4–H), 7.25 (t, 1H, H9), 7.36 (d, 2H, H12,12′), 7.45 (t, 2H, H8,8′), 7.58 (m, 3H, H13,13′, H14), 7.90 (d, 2H, H7,7′), 8.15 (d, 2H, H16,16′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 14.5 (C20), 30.9 (C19), 99.6 (C4), 111.6 (C17,17′), 120.4 (C7,7′), 126.1 (C9), 126.2 (C16,16′), 127.7 (C12,12′), 129.1 (C8,8′), 129.6 (C13,13′), 131.9 (C14), 137.5 (C6), 141.0 (C15), 147.5 (C3), 150.1 (C18), 158.7 (C10), 159.3 (C5), C11 not observed. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 117.3 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 189.8 (N1), 217.7 (N2), N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 447 [35] [H3L2]+; 469 [100] [H2L2 + Na]+; 485 [30] [H2L2 + K]+; 916 [15] [2H2L2 + Na]+.
N), 1535 ν(C–N), 1301 νas(SO2), 1165vs νs(SO2), 1089 m ν(N–N). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.61 (s, 3H, H20), 2.19 (s, 3H, H19), 6.82 (d, 2H, H17,17′), 7.20 (sbr, 1H, N4–H), 7.25 (t, 1H, H9), 7.36 (d, 2H, H12,12′), 7.45 (t, 2H, H8,8′), 7.58 (m, 3H, H13,13′, H14), 7.90 (d, 2H, H7,7′), 8.15 (d, 2H, H16,16′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 14.5 (C20), 30.9 (C19), 99.6 (C4), 111.6 (C17,17′), 120.4 (C7,7′), 126.1 (C9), 126.2 (C16,16′), 127.7 (C12,12′), 129.1 (C8,8′), 129.6 (C13,13′), 131.9 (C14), 137.5 (C6), 141.0 (C15), 147.5 (C3), 150.1 (C18), 158.7 (C10), 159.3 (C5), C11 not observed. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 117.3 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 189.8 (N1), 217.7 (N2), N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 447 [35] [H3L2]+; 469 [100] [H2L2 + Na]+; 485 [30] [H2L2 + K]+; 916 [15] [2H2L2 + Na]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1593s, 1584s ν(C
O), 1593s, 1584s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1530 m ν(C
C), 1530 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and νas(NO2), 1376 m ν(C–N), 1325s νs(NO2), 1108s ν(N–N). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 2.41 (s, 3H, C3–CH3), 4.11 (s, 2H, H11), 6.49 (s, 1H, N4–H), 6.63 (d, 2H, H17,17′), 7.14 (d, 2H, H13,13′), 7.24 (m, 4H, H14,14′, H15, H9), 7.43 (t, 2H, H8,8′), 7.98 (d, 2H, H7,7′), 8.04 (d, 2H, H18,18′), 12.40 (sbr, 1H, O–H). 13C NMR (CDCl3, 125 MHz, 298 K): δC 16.9 (C3–CH3), 33.8 (C11), 100.9 (C4), 112.0 (C17,17′), 119.7 (C7,7′), 125.2 (C9), 126.0 (C18,18′), 127.8 (C15), 128.1 (C13,13′), 129.1 (C8,8′), 129.5 (C14,14′), 134.3 (C12), 138.7 (C6), 141.9 (C19), 147.2 (C3), 151.5 (C16), 165.4 (C10), 167.1 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 98.9 (N4), 140.6 (N3). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 288.9 (N2), 98.9 (N4), 140.6 (N3), 379.4 (NO2), N1 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 428 [100] [H3L4]+; 450 [30] [H2L4 + Na]+; 491 [15] [H2L4 + MeCN + Na]+; 878 [10] [2H2L4 + Na]+.
N) and νas(NO2), 1376 m ν(C–N), 1325s νs(NO2), 1108s ν(N–N). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 2.41 (s, 3H, C3–CH3), 4.11 (s, 2H, H11), 6.49 (s, 1H, N4–H), 6.63 (d, 2H, H17,17′), 7.14 (d, 2H, H13,13′), 7.24 (m, 4H, H14,14′, H15, H9), 7.43 (t, 2H, H8,8′), 7.98 (d, 2H, H7,7′), 8.04 (d, 2H, H18,18′), 12.40 (sbr, 1H, O–H). 13C NMR (CDCl3, 125 MHz, 298 K): δC 16.9 (C3–CH3), 33.8 (C11), 100.9 (C4), 112.0 (C17,17′), 119.7 (C7,7′), 125.2 (C9), 126.0 (C18,18′), 127.8 (C15), 128.1 (C13,13′), 129.1 (C8,8′), 129.5 (C14,14′), 134.3 (C12), 138.7 (C6), 141.9 (C19), 147.2 (C3), 151.5 (C16), 165.4 (C10), 167.1 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 98.9 (N4), 140.6 (N3). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 288.9 (N2), 98.9 (N4), 140.6 (N3), 379.4 (NO2), N1 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 428 [100] [H3L4]+; 450 [30] [H2L4 + Na]+; 491 [15] [H2L4 + MeCN + Na]+; 878 [10] [2H2L4 + Na]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1577s ν(C
O), 1577s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1524 m ν(C
C), 1524 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1312vs νs(CF3), 1163, 1112 νas(CF3), 1120vs ν(N–N), 1065vs νs(CF3). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.11 (s, 9H, H13,13′,13′′), 2.49 (s, 3H, H19), 2.83 (s, 2H, H11), 6.58 (s br, 1H, N4–H), 6.79 (d, 2H, H15,15′), 7.22 (t, 1H, H9), 7.45 (t, 2H, H8,8′), 7.50 (d, 2H, H16,16′), 8.01 (d, 2H, H7,7′), 12.83 (s br, 1H, O–H). 13C NMR (CDCl3, 125 MHz, 298 K): δC 17.6 (C19), 30.6 (C13,13′,13′′), 34.7 (C12), 39.0 (C11), 100.4 (C4), 113.0 (C15,15′), 119.6 (C7,7′), 124.0q (1JC–F = 271.3 Hz, C17), 124.2q (1JC–F = 271.3 Hz, C18), 125.2 (C9), 126.9q (3JC–F = 3.9 Hz, C16,16′), 128.9 (C8,8′), 138.0 (C6), 147.0 (C3), 148.9 (C14), 164.9 (C10), 169.6 (C5). 19F NMR (CDCl3): δ, −61.77. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 143.8 (N3), 99.7 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 191.4 (N1), 285.8 (N2), 143.8 (N3), 99.7 (N4). ESI-MS (+) CH3CN (m/z, relative intensity %): 431 [95] [H3L5]+; 453 [100] [H2L5 + Na]+; 469 [15] [H2L5 + K]+; 494 [15] [H2L5 + MeCN + Na]+; 884 [20] [2H2L5 + Na]+.
N), 1312vs νs(CF3), 1163, 1112 νas(CF3), 1120vs ν(N–N), 1065vs νs(CF3). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.11 (s, 9H, H13,13′,13′′), 2.49 (s, 3H, H19), 2.83 (s, 2H, H11), 6.58 (s br, 1H, N4–H), 6.79 (d, 2H, H15,15′), 7.22 (t, 1H, H9), 7.45 (t, 2H, H8,8′), 7.50 (d, 2H, H16,16′), 8.01 (d, 2H, H7,7′), 12.83 (s br, 1H, O–H). 13C NMR (CDCl3, 125 MHz, 298 K): δC 17.6 (C19), 30.6 (C13,13′,13′′), 34.7 (C12), 39.0 (C11), 100.4 (C4), 113.0 (C15,15′), 119.6 (C7,7′), 124.0q (1JC–F = 271.3 Hz, C17), 124.2q (1JC–F = 271.3 Hz, C18), 125.2 (C9), 126.9q (3JC–F = 3.9 Hz, C16,16′), 128.9 (C8,8′), 138.0 (C6), 147.0 (C3), 148.9 (C14), 164.9 (C10), 169.6 (C5). 19F NMR (CDCl3): δ, −61.77. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 143.8 (N3), 99.7 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 191.4 (N1), 285.8 (N2), 143.8 (N3), 99.7 (N4). ESI-MS (+) CH3CN (m/z, relative intensity %): 431 [95] [H3L5]+; 453 [100] [H2L5 + Na]+; 469 [15] [H2L5 + K]+; 494 [15] [H2L5 + MeCN + Na]+; 884 [20] [2H2L5 + Na]+.
Synthesis of Zn(II) complexes 1–5
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1595vs ν(C
N), 1595vs ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1567s ν(C
N), 1567s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1536s ν(C
C), 1536s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1509 m ν(C
C), 1509 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1092 m ν(N–N), 410 m ν(Zn–N), 364s ν(Zn–N), 321s ν(Zn–O). 1H NMR (CDCl3): δ, 1.50 (s, 6H, H20), 6.31 (d, 2H, H16), 6.58 (t, 2H, H18), 7.01 (t, 2H, H17), 7.21 (t, 4H, H8,8′), 7.34 (t, 2H, H14), 7.49 (s, 2H, N4–H), 7.58 (d, 4H, H12,12′), 7.70 (t, 4H, H13,13′), 7.80 (d, 4H, H7,7′), 7.90 (d, 2H, H19). 13C NMR (CDCl3): δ, 15.3 (C20), 96.6 (C4), 108.7 (C16), 114.9 (C17), 120.6 (C7,7′), 123.7 (C9), 128.2 m (C8,8′,12,12′), 129.8 m (C13,13′,14), 133.3 (C11), 134.6 (C6), 138.5 (C18), 139.8 (C3), 144.9 (C19), 152.0 (C15), 154.6 (C10), 162.2 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 132.9 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N1, N2, N3 and N5 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 432 [20] [Zn(HL1)]+; 801 [80] [Zn(HL1)(H2L1)]+; 823 [70] [Zn(HL1)2 + Na+].
O), 1092 m ν(N–N), 410 m ν(Zn–N), 364s ν(Zn–N), 321s ν(Zn–O). 1H NMR (CDCl3): δ, 1.50 (s, 6H, H20), 6.31 (d, 2H, H16), 6.58 (t, 2H, H18), 7.01 (t, 2H, H17), 7.21 (t, 4H, H8,8′), 7.34 (t, 2H, H14), 7.49 (s, 2H, N4–H), 7.58 (d, 4H, H12,12′), 7.70 (t, 4H, H13,13′), 7.80 (d, 4H, H7,7′), 7.90 (d, 2H, H19). 13C NMR (CDCl3): δ, 15.3 (C20), 96.6 (C4), 108.7 (C16), 114.9 (C17), 120.6 (C7,7′), 123.7 (C9), 128.2 m (C8,8′,12,12′), 129.8 m (C13,13′,14), 133.3 (C11), 134.6 (C6), 138.5 (C18), 139.8 (C3), 144.9 (C19), 152.0 (C15), 154.6 (C10), 162.2 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 132.9 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N1, N2, N3 and N5 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 432 [20] [Zn(HL1)]+; 801 [80] [Zn(HL1)(H2L1)]+; 823 [70] [Zn(HL1)2 + Na+].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1562s ν(C
N), 1562s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1528 m ν(C
O), 1528 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1522 ν(C
C), 1522 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1481vs νas(SO2), 1475 ν(C–N), 1164vs νs(SO2), 1079 m ν(N–N), 551s ν(Zn–N), 430 m ν(Zn–O). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.22 (s, 3H, H20), 2.45 (s, 3H, H19), 6.61 (sbr, 1H, N4–H), 6.78 (d, 2H, H17,17′), 7.19 (d, 2H, H12,12′), 7.24 (t, 1H, H9), 7.42 (t, 2H, H8,8′), 7.45 (m, 3H, H13,13′, H14), 7.83 (d, 2H, H7,7′), 7.98 (d, 2H, H16,16′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 15.4 (C20), 21.7 (C19), 98.3 (C4), 121.3 (C11), 125.0 (C17,17′), 127.2 (C9), 128.6 (C7,7′), 128.7 (C16,16′), 129.4 (C12,12′), 129.9 (C8,8′), 130.2 (C13,13′), 132.1 (C6), 133.5 (C14), 139.0 (C15), 145.0 (C3), 149.0 (C18), 162.8 (C10), 167.6 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 151.1 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 192.9 (N1), 276.4 (N2), N3 not observed. Stability Test: compound 2 was dissolved in DMSO-d6 and its 1H NMR spectrum immediately recorded. After 5 days the spectrum was unchanged. ESI-MS (+) CH3CN (m/z, relative intensity %): 957 [30] [Zn(HL2)(H2L2)]+; 979 [30] [Zn(HL2)2 + Na+]; 1467 [30] [Zn2(HL2)3].
N), 1481vs νas(SO2), 1475 ν(C–N), 1164vs νs(SO2), 1079 m ν(N–N), 551s ν(Zn–N), 430 m ν(Zn–O). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 1.22 (s, 3H, H20), 2.45 (s, 3H, H19), 6.61 (sbr, 1H, N4–H), 6.78 (d, 2H, H17,17′), 7.19 (d, 2H, H12,12′), 7.24 (t, 1H, H9), 7.42 (t, 2H, H8,8′), 7.45 (m, 3H, H13,13′, H14), 7.83 (d, 2H, H7,7′), 7.98 (d, 2H, H16,16′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 15.4 (C20), 21.7 (C19), 98.3 (C4), 121.3 (C11), 125.0 (C17,17′), 127.2 (C9), 128.6 (C7,7′), 128.7 (C16,16′), 129.4 (C12,12′), 129.9 (C8,8′), 130.2 (C13,13′), 132.1 (C6), 133.5 (C14), 139.0 (C15), 145.0 (C3), 149.0 (C18), 162.8 (C10), 167.6 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 151.1 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 192.9 (N1), 276.4 (N2), N3 not observed. Stability Test: compound 2 was dissolved in DMSO-d6 and its 1H NMR spectrum immediately recorded. After 5 days the spectrum was unchanged. ESI-MS (+) CH3CN (m/z, relative intensity %): 957 [30] [Zn(HL2)(H2L2)]+; 979 [30] [Zn(HL2)2 + Na+]; 1467 [30] [Zn2(HL2)3].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1575 m ν(C
N), 1575 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1558 m ν(C
O), 1558 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1526 m ν(C
C), 1526 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1480vs νas(NO2), 1319s, 1300vs νs(NO2), 1110 m ν(C–N), 961 m ν(N–N), 511 m ν(Zn–N), 475s ν(Zn–O). 1H NMR (CDCl3): δ, 1.57s (4H, H2O); 1.27s (6H, H19); 6.31 (sbr, 1H, N4–H), 6.68 (d, 2H, H17,17′), 7.14 (d, 2H, H12,12′), 7.25 (t, 1H, H9), 7.41 (t, 2H, H8,8′), 7.45 (m, 3H, H13,13′, H14), 7.77 (d, 2H, H7,7′), 7.97 (d, 2H, H16,16′). 13C NMR (CDCl3): δ, 15.2 (C19), 98.5 (C4), 112.6 (C17,17′), 125.7 (C7,7′), 126.2 (C9), 126.7 (C16,16′), 127.7 (C14), 128.8 (C12,12′), 129.2 (C8,8′), 130.6 (C13,13′), 133.0 (C6), 140.9 (C15), 150.8 (C18), 162.2 (C11), C3, C5 and C10 n.o. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 125.2 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 197.4 (N1), N2 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 478 [25] [Zn(HL3)]+; 891 [50] [Zn(HL3)(H2L3)]+; 913 [20] [Zn(HL3)2 + Na+].
C), 1480vs νas(NO2), 1319s, 1300vs νs(NO2), 1110 m ν(C–N), 961 m ν(N–N), 511 m ν(Zn–N), 475s ν(Zn–O). 1H NMR (CDCl3): δ, 1.57s (4H, H2O); 1.27s (6H, H19); 6.31 (sbr, 1H, N4–H), 6.68 (d, 2H, H17,17′), 7.14 (d, 2H, H12,12′), 7.25 (t, 1H, H9), 7.41 (t, 2H, H8,8′), 7.45 (m, 3H, H13,13′, H14), 7.77 (d, 2H, H7,7′), 7.97 (d, 2H, H16,16′). 13C NMR (CDCl3): δ, 15.2 (C19), 98.5 (C4), 112.6 (C17,17′), 125.7 (C7,7′), 126.2 (C9), 126.7 (C16,16′), 127.7 (C14), 128.8 (C12,12′), 129.2 (C8,8′), 130.6 (C13,13′), 133.0 (C6), 140.9 (C15), 150.8 (C18), 162.2 (C11), C3, C5 and C10 n.o. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 125.2 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 197.4 (N1), N2 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 478 [25] [Zn(HL3)]+; 891 [50] [Zn(HL3)(H2L3)]+; 913 [20] [Zn(HL3)2 + Na+].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1566s ν(C
N), 1566s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1520 m ν(C
O), 1520 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1478vs νas(NO2), 1321vs, 1303s νs(NO2), 1100 m ν(N–N), 548 m ν(Zn–N), 430 m ν(Zn–O). 1H NMR (CDCl3): δ, 2.29s (s, 3H, H20), 4.15 (s, 2H, H11) 6.14 (sbr, 1H, N4–H), 6.44 (d, 2H, H17,17′), 7.02 (d, 2H, H13,13′), 7.25 (m, 3H, H9, H8,8′), 7.41 (m, 3H, H14,14′, H15), 7.77 (d, 2H, H7,7′), 7.93 (d, 2H, H16,16′). 13C NMR (CDCl3): δ, 17.3 (C20), 35.9 (C11), 98.6 (C4), 112.4 (C17,17′), 121.2 (C7,7′), 125.6 (C13,13′), 125.9 (C16), 127.4 (C9), 127.8 (C8,8′), 127.9 (C12), 128.8 (C14,14′), 128.9 (C15), 129.3 (C18,18′), 134.8 (C6), 141.0 (C3), 151.2 (C19), 162.5 (C10), 178.3 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N4 not observed. {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N1, N2 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 492 [20] [Zn(HL4)]+; 919 [40] [Zn(HL4)(H2L4)]+; 941 [30] [Zn(HL4)2 + Na+].
C), 1478vs νas(NO2), 1321vs, 1303s νs(NO2), 1100 m ν(N–N), 548 m ν(Zn–N), 430 m ν(Zn–O). 1H NMR (CDCl3): δ, 2.29s (s, 3H, H20), 4.15 (s, 2H, H11) 6.14 (sbr, 1H, N4–H), 6.44 (d, 2H, H17,17′), 7.02 (d, 2H, H13,13′), 7.25 (m, 3H, H9, H8,8′), 7.41 (m, 3H, H14,14′, H15), 7.77 (d, 2H, H7,7′), 7.93 (d, 2H, H16,16′). 13C NMR (CDCl3): δ, 17.3 (C20), 35.9 (C11), 98.6 (C4), 112.4 (C17,17′), 121.2 (C7,7′), 125.6 (C13,13′), 125.9 (C16), 127.4 (C9), 127.8 (C8,8′), 127.9 (C12), 128.8 (C14,14′), 128.9 (C15), 129.3 (C18,18′), 134.8 (C6), 141.0 (C3), 151.2 (C19), 162.5 (C10), 178.3 (C5). {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N4 not observed. {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN N1, N2 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 492 [20] [Zn(HL4)]+; 919 [40] [Zn(HL4)(H2L4)]+; 941 [30] [Zn(HL4)2 + Na+].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1565s ν(C
N), 1565s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1525 m ν(C–N), 1321vs νs(CF3), 1100s νas(CF3), 1064s νs(CF3), 981 ν(N–N), 549 m ν(Zn–N), 425 m ν(Zn–O). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 0.97 (s, 9H, H13,13′,13′′), 1.60s (s, 2H, H2O), 2.33 (s, 3H, H19), 2.82 (s br, 2H, H11), 5.98 (s br, 1H, N4–H), 6.43 (d, 2H, H15,15′), 7.24 (t, 1H, H9), 7.35 (d, 2H, H16,16′), 7.46 (t, 2H, H8,8′), 8.01 (d, 2H, H7,7′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 18.0 (C19), 30.4 (C13,13′,13′′), 34.7 (C12), 40.6 (C11), 99.2 (C4), 114.3 (C15,15′), 120.4 (C7,7′), 123.5q (2JC–F = 33.0 Hz, C17), 124.2q (1JC–F = 271.3 Hz, C18), 125.2 (C9), 126.4q (3JC–F = 3.9 Hz, C16,16′), 128.7 (C8,8′), 139.0 (C6), 147.8 (C3), 149.1 (C14), 161.9 (C10), 180.1 (C5). 19F NMR (CDCl3): δ, −61.45. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 120.0 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 274.1 (N2), N1 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 925 [20] [Zn(HL5)(H2L5)]+.
O), 1525 m ν(C–N), 1321vs νs(CF3), 1100s νas(CF3), 1064s νs(CF3), 981 ν(N–N), 549 m ν(Zn–N), 425 m ν(Zn–O). 1H NMR (CDCl3 with 0.05% v/v TMS, 500 MHz, 298 K): δH 0.97 (s, 9H, H13,13′,13′′), 1.60s (s, 2H, H2O), 2.33 (s, 3H, H19), 2.82 (s br, 2H, H11), 5.98 (s br, 1H, N4–H), 6.43 (d, 2H, H15,15′), 7.24 (t, 1H, H9), 7.35 (d, 2H, H16,16′), 7.46 (t, 2H, H8,8′), 8.01 (d, 2H, H7,7′). 13C NMR (CDCl3, 125 MHz, 298 K): δC 18.0 (C19), 30.4 (C13,13′,13′′), 34.7 (C12), 40.6 (C11), 99.2 (C4), 114.3 (C15,15′), 120.4 (C7,7′), 123.5q (2JC–F = 33.0 Hz, C17), 124.2q (1JC–F = 271.3 Hz, C18), 125.2 (C9), 126.4q (3JC–F = 3.9 Hz, C16,16′), 128.7 (C8,8′), 139.0 (C6), 147.8 (C3), 149.1 (C14), 161.9 (C10), 180.1 (C5). 19F NMR (CDCl3): δ, −61.45. {1H,15N}gs-HSQC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 120.0 (N4). {1H,15N} gs-HMBC NMR (CDCl3, 51 MHz, 3J(N–H) = 3 Hz, 298 K): δN 274.1 (N2), N1 and N3 not observed. ESI-MS (+) CH3CN (m/z, relative intensity %): 925 [20] [Zn(HL5)(H2L5)]+.
Crystallographic refinement
X-ray data collection of suitable single crystals was done on a Bruker D8 VENTURE area detector equipped with graphite monochromated Mo-Kα radiation (λ = 0.71073 Å) by applying the ω-scan method. The data reduction was performed with the APEX3 suite64 and corrected for absorption using SADABS.65 The final structures were solved using SHELXT66 and refined on F2 by a full-matrix least-square technique using anisotropic displacement parameters67 by means of the Olex2 v1.3 crystallographic package68 and the SHELXL-2018 program.66 All hydrogen atoms were included as fixed contributions riding on attached atoms with isotropic thermal displacement parameters 1.2 or 1.5 times those of the respective atom. In complex 2, due to the disorder within the voids, the SQUEEZE procedure implemented in PLATON program69 was used during the refinement and two crystallization methanol molecules were removed from the structure. In addition, in this structure, one phenyl ring is disordered over two alternate positions so it was refined with occupancies of 40 and 60% for all atoms and anisotropic displacement parameters of the ring. CCDC numbers 2158176–2158180† contain the supplementary crystallographic data for compounds.Computational details
The electronic structure and geometries of the proligands H2L2, H2L4 and H2L5, their tautomers and anions, [HLn]−, and their zinc complexes were investigated by using density functional theory at the B3LYP level.62,63 For the proligands and their corresponding anions the 6-311G** basis set was used for the optimization, while for the Zn complexes the optimization was carried out using 6-311G*. Molecular geometries were optimized without symmetry restrictions. Frequency calculations were carried out at the same level of theory to identify all stationary points as minima (zero imaginary frequencies) and to provide the thermal correction to free energies at 298.15 K and 1 atm. Solution-phase SCF energies were calculated by a single-point calculation on the in vacuum optimized structure using the CPCM solvation model in chloroform.64 Gibbs free energies in chloroform solution were estimated from the equation Gsolv = Esolv + (Ggas − Egas). The GIAO method was used for the NMR calculations (1H, 13C and 15N NMR isotropic shielding tensors), which were carried out at the 6-311 + G(2d,p) level of theory. The computed IR spectra were scaled by a factor of 0.96.65,66 The DFT calculations were executed using the Gaussian 09 program package.67 Coordinates of all optimized compounds are collected in Table S6 (ESI†).Antibacterial activity
The antibacterial activity of the ligands H2L1–H2L5 and of the Zn(II) complexes 1–5 was investigated against Gram-positive bacteria, Staphylococcus aureus (S. aureus) ATCC 25923 and Gram-negative, Escherichia coli (E. coli) ATCC 25922. Bacteria were grown aerobically at 37 °C overnight using Tryptone Soya Broth (OXOID) as the growth medium. Bacteria proliferation was monitored by measuring the increase of optical density in the culture suspension at 600 nm (OD600). The enriched culture (log phase) obtained was further diluted to 106 CFU mL−1 concentration. A preliminary bacterial growth inhibition (BGI) analysis was carried out by using the Agar diffusion method (Bauer et al., 1966) with a fixed concentration of 2 mg mL−1 of the ligands and 4 mg mL−1 of the Zn(II) complexes (both corresponding to ∼ 5 μmol mL−1). A loop full of the given test strain (E. coli and S. aureus) was inoculated in 10 mL of N-broth (nutrient broth) and incubated for 24 h in an incubator at 37 °C in order to activate the bacterial strain. The various compounds were weighed into Eppendorf content and suspended in autoclaved physiological solution, to which 1 mL of the enriched culture was then added (reaching the concentration of 106 CFU ml−1) and kept on an IKA KS 130 BASIC platform shaker for 24 hours at slow speed. The final solution was included uniformly into Petri dishes containing Plate Count Agar (OXOID). Adopting the same procedure a strain without treatment were used as negative control. The tests were carried out at different time intervals (0, 2, 4, 8, 12, 24 hours) for viability measurements over time. The bacterial viability was reported as Growth Rate (%) using the following formula: Percentage of cell viability (%) = (CFU tx/CFU t0) 100 (where t0 is the time zero at the beginning of the experiment and tx is a specific time in hours of the experiment). The growth curves of E. coli and S. aureus without antibacterial agents were also measured as blanks.68 All tests were done in triplicate.69Reactive oxygen species (ROS) detection assay
To better evaluate the antibacterial activity of such compounds, the test for ROS measurement was carried out against the compounds 4 and 5, that showed better bacterial inhibition. To quantify the ROS generated in our system, the 2′,7′-dichlorofluorescein diacetate (DCFDA) assay was performed. The test protocol is based on the spread of 20 mL of DCFDA in the bacterial cultures in the range of 1–3 × 106 CFU mL−1 for 30 minutes at 37 °C. Then 10 mL are added to the Eppendorf containing 4 mg of 4 and 5 respectively, suspended in 1 mL of physiological solution. 100 μL of each Eppendorf were transferred to a 96-well plate in triplicate in the dark and ROS generation was assessed for the first two and four hours. Maintaining the same procedure, an untreated sample strain was used as a control (blank). Fluorescence from each sample well was read using the FLUOstar Omega fluorescence cytometer from BMG LABTECH at 485/20 nm and 528/20 nm wavelengths for excitation and emission respectively. The average of the triplicate was calculated and reported as the intensity of fluorescence in arbitrary unit (a.u.).Propidium iodide (PI) – viability assay
To assess the viability of bacteria treated with compounds 4 and 5, the fluorescent PI staining assay was carried out, since it allows to evaluate the level of damage related to membrane permeability. A suspension (106 CFU mL−1) of E. coli and S. aureus, treated with 4 mg of compounds 4 and 5, were incubated for a period of 24 h at 37 °C. After 4 h, 8 h, 12 h, and 24 h, 100 μL of the content was transferred in triplicate into 96-well plate in the dark and 1,5 mL of Propidium Iodide (PI) was added in each well. Maintaining the same procedure, an untreated sample strain was used as a control (blank). Fluorescence emitted by the cells was read using the FLUOstar Omega fluorescence cytometer from BMG LABTECH at 485/20 nm and 528/20 nm wavelengths for excitation and emission respectively. The average of the triplicate was calculated and reported as the intensity of fluorescence in arbitrary unit (a.u.).Confocal laser scanning microscopy (CLSM) study
To determine the damaging effect of 4 and 5 on the cell membranes of S. aureus and E. coli, CM analysis was carried out with LIVE/DEAD® BacLight™ Bacterial Viability Kits (Invitrogen), which utilize mixtures of SYTO®9 green-fluorescent nucleic acid stain and propidium iodide (PI) red-fluorescent nucleic acid stain. Briefly, a bacterial suspension in the range of 1 × 108 CFU mL−1 (∼0.03 OD670) for E. coli and 1 × 107 CFU mL−1 (∼0.15 OD670) for S. aureus were treated with 4 mg of 4 and 5 during the logarithmic growth phase for 4 h. After the incubation period, suspensions were concentrated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10–15 minutes, then the supernatant was removed and the pellet resuspended in 0.85% NaCl solution. Afterwards, equal volumes of Syto-9 and Propidium iodide were added to all samples and incubated for 15 min in the absence of light. Samples were then fixed between a slide and an 18 mm square coverslip to observe the fluorescence under confocal microscope (Nikon ECLIPSE Ti). The control assay was conducted without treatment. The excitation/emission maxima for these dyes are about 480/500 nm for SYTO 9 stain and 490/635 nm for PI.
000g for 10–15 minutes, then the supernatant was removed and the pellet resuspended in 0.85% NaCl solution. Afterwards, equal volumes of Syto-9 and Propidium iodide were added to all samples and incubated for 15 min in the absence of light. Samples were then fixed between a slide and an 18 mm square coverslip to observe the fluorescence under confocal microscope (Nikon ECLIPSE Ti). The control assay was conducted without treatment. The excitation/emission maxima for these dyes are about 480/500 nm for SYTO 9 stain and 490/635 nm for PI.
Scanning electron microscopy (SEM) study
The morphology of the bacterial cells after treating with 4 and 5 was determined by SEM (Sigma 300, Zeiss) operating at 15.0 kV, Bruker. For SEM sample preparation, log phase cells of E. coli and S. aureus (106 CFUs) were incubated with 4 mg of complexes 4 and 5 for 4 hours at 37 °C. After incubation period, bacterial strains were centrifugated (2000g × 5 min), washed thrice with physiological solution and fixed with 200 μL of 2.5% (v/v) glutaraldehyde solution for 2 hours on glass slide. The fixed pellets were washed twice with physiological solution, then were immersed in a solution with increasing concentrations of ethanol (10, 30, 50, 70, 90 and 100%), 5 min for each concentration. After drying, samples were placed on aluminium stubs using self-adhesive carbon conductive tab and chrome-coated. The untreated bacterial cells were taken as control.Conclusions
We have reported new hydrazone-pyrazolone ligands and investigated their tautomerism in the solid state and solution. The X-ray studies on free ligands H2L2 and H2L4 show they exist in the NH,NH tautomeric form. Then, five new zinc(II) complexes have been synthesized and characterized in the solid state and solution. Complexes 1 and 2 are mononuclear anhydrous compounds, while 3 and 5 are mononuclear diaqua compounds. Remarkable is the result of the X-ray studies on compound 4, showing a polynuclear species assembled through involvement of nitro groups of the ligand (HL4)− in the interaction with zinc atoms of vicinal units. These interactions are however weak and long, with a Zn–O distance of 2.342(3) Å, as compared to the Zn–O distances of the carbonyl oxygen of pyrazolone moiety, which is equal to 2.004 (3) Å. This evidence allows us to rationalize how such weak interactions of nitro groups can be easily broken even in chlorinated solution, where 4 is soluble, or they can be replaced by molecules of water or alcohols during the synthesis in methanol, as for compound 3 which is instead isolated as a molecular diaqua compound. The theoretical DFT study have been used to rationalize the structures experimentally found (for example, polymeric nature of 4), to support the spectroscopical assignments (IR and NMR) and to have a better knowledge of the behaviour of pyrazolone-based hydrazones as ligands in coordination chemistry. Exploration of antibacterial potential of both free ligands and zinc(II) complexes evidenced that coordination to zinc(II) strongly improves the efficiency against both the Gram- and Gram + bacteria. Among them, the polynuclear Zn(II) complex 4 and the mononuclear dihydrate Zn(II) complex 5 with the ligand having hydrophobic substituents, display an exceptional activity against both bacterial cell lines.Author contributions
F. Marchetti – conceptualization, funding acquisition and writing – review & editing; R. Pettinari – resources and writing – original draft; F. Verdicchio – DFT calculations and visualization; A. Tombesi – synthesis and characterization of zinc complexes; S. Scuri – antimicrobial study validation and data curation; S. Xhafa – synthesis and characterization of ligands; L. Olivieri – antimicrobial and biological investigation; C. Pettinari – project administration and ligands and zinc complexes synthesis supervision; D. Choquesillo-Lazarte – X-ray crystallographic investigation; A. García-García – X-ray crystallographic study visualization; A. Rodríguez-Diéguez – X-ray crystallographic study supervision, validation and writing; A. Galindo – DFT study supervision, validation and writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
In memory of Stefania Scuri, who left us prematurely on 24th August 2022 after a brief and terrible disease.Thanks are due to the University of Camerino (Italy) for financial support. The research grant of Dr Laura Olivieri was funded under the frame of the project Nano4-Fresh – Nanomaterials for an environmentally friendly and sustainable handling of perishable products (PRIMA19_00246), which is part of the Partnership on Research and Innovation in the Mediterranean Area (PRIMA) Programme supported by the European Union and funded by the national funding bodies of Participating States (MUR in Italy is gratefully acknowledged). We thank the Marche Region for supporting this research by funding the two-year research grant of Dr Sonila Xhafa as part of the POR Marche FSE 2014–2020 P.I. 8.1 R.A. 8.5. Financial support from the Spanish Ministerio de Ciencia e Innovación (PGC2018-093443-B-I00) is gratefully acknowledged.
References
- Z. Zhao, X. Dai, C. Li, X. Wang, J. Tian, Y. Feng, J. Xie, C. Ma, Z. Nie, P. Fan, M. Qian, X. He, S. Wu, Y. Zhang and X. Zheng, Pyrazolone Structural Motif in Medicinal Chemistry: Retrospect and Prospect, Eur. J. Med. Chem., 2020, 186, 111893 CrossRef CAS PubMed.
- C. Cusan, G. Altinier, S. Sosa, F. Sibilla, F. Bucar, A. Tubaro, M. Prato, G. Spalluto and T. D. Ros, Anti-Inflammatory and Anti-Oxidant Activity of a New Class of Phenyl-Pyrazolone Derivatives, Curr. Drug Discovery Technol., 2006, 3(1), 67–73 CrossRef CAS PubMed.
- X. Fan, X. Zhang, L. Zhou, K. A. Keith, E. R. Kern and P. F. Torrence, A Pyrimidine–Pyrazolone Nucleoside Chimera with Potent in Vitro Anti-Orthopoxvirus Activity, Bioorg. Med. Chem. Lett., 2006, 16(12), 3224–3228 CrossRef CAS PubMed.
- F. Caruso, C. Pettinari, F. Marchetti, M. Rossi, C. Opazo, S. Kumar, S. Balwani and B. Ghosh, Inhibitory Effect of β-Diketones and Their Metal Complexes on TNF-α Induced Expression of ICAM-1 on Human Endothelial Cells, Bioorg. Med. Chem., 2009, 17(17), 6166–6172 CrossRef CAS PubMed.
- M. F. Brana, A. Gradillas, A. G. Ovalles, B. López, N. Acero, F. Llinares and D. M. Mingarro, Synthesis and Biological Activity of N, N-Dialkylaminoalkyl-Substituted Bisindolyl and Diphenyl Pyrazolone Derivatives, Bioorg. Med. Chem., 2006, 14(1), 9–16 CrossRef CAS PubMed.
- T. Watanabe, S. Yuki, M. Egawa and H. Nishi, Protective Effects of MCI-186 on Cerebral Ischemia: Possible Involvement of Free Radical Scavenging and Antioxidant Actions, J. Pharmacol. Exp. Ther., 1994, 268(3), 1597–1604 CAS.
- H. Kawai, H. Nakai, M. Suga, S. Yuki, T. Watanabe and K.-I. Saito, Effects of a Novel Free Radical Scavenger, MCI-186, on Ischemic Brain Damage in the Rat Distal Middle Cerebral Artery Occlusion Model, J. Pharmacol. Exp. Ther., 1997, 281(2), 921–927 CAS.
- T.-W. Wu, L.-H. Zeng, J. Wu and K.-P. Fung, Myocardial Protection of MCI-186 in Rabbit Ischemia–Reperfusion, Life Sci., 2002, 71(19), 2249–2255 CrossRef CAS PubMed.
- M. Himly, B. Jahn-Schmid, K. Pittertschatscher, B. Bohle, K. Grubmayr, F. Ferreira, H. Ebner and C. Ebner, IgE-Mediated Immediate-Type Hypersensitivity to the Pyrazolone Drug Propyphenazone, J. Allergy Clin. Immunol., 2003, 111(4), 882–888 CrossRef CAS PubMed.
- D. Costa, A. P. Marques, R. L. Reis, J. L. F. C. Lima and E. Fernandes, Inhibition of Human Neutrophil Oxidative Burst by Pyrazolone Derivatives, Free Radicals Biol. Med., 2006, 40(4), 632–640 CrossRef CAS PubMed.
- J. B. Field, E. C. Dolendo, A. Mireles and B. H. Ershoff, Evaluation of the Effect of a Pyrazolone Derivative KB-95 on the Toxicity and Activity of Some Anticancer Drugs, Cancer Res., 1966, 26(7 Part 1), 1371–1375 CAS.
- C. Pettinari, F. Marchetti and A. Drozdov, β-Diketones and Related Ligands, in Comprehensive Coordination Chemistry II, ed. J. A. McCleverty and T. J. Meyer, Elsevier Ltd., 2004, vol. 1, pp. 97–115 Search PubMed.
- J. S. Casas, M. S. García-Tasende, A. Sánchez, J. Sordo and Á. Touceda, Coordination Modes of 5-Pyrazolones: A Solid-State Overview, Coord. Chem. Rev., 2007, 251(11–12), 1561–1589 CrossRef CAS.
- F. Marchetti, C. Pettinari and R. Pettinari, Acylpyrazolone Ligands: Synthesis, Structures, Metal Coordination Chemistry and Applications, Coord. Chem. Rev., 2005, 249(24), 2909–2945 CrossRef CAS.
- F. Marchetti, R. Pettinari and C. Pettinari, Recent Advances in Acylpyrazolone Metal Complexes and Their Potential Applications, Coord. Chem. Rev., 2015, 303, 1–31 CrossRef CAS.
- F. Caruso, M. Rossi, J. Tanski, R. Sartori, R. Sariego, S. Moya, S. Diez, E. Navarrete, A. Cingolani, F. Marchetti and C. Pettinari, Synthesis, Structure, and Antitumor Activity of a Novel Tetranuclear Titanium Complex, J. Med. Chem., 2000, 43(20), 3665–3670 CrossRef CAS PubMed.
- F. Caruso, L. Massa, A. Gindulyte, C. Pettinari, F. Marchetti, R. Pettinari, M. Ricciutelli, J. Costamagna, J. C. Canales, J. Tanski and M. Rossi, (4-Acyl-5-Pyrazolonato)Titanium Derivatives: Oligomerization, Hydrolysis, Voltammetry, and DFT Study, Eur. J. Inorg. Chem., 2003, 3221–3232 CrossRef CAS.
- F. Caruso, C. Pettinari, F. Marchetti, P. Natanti, C. Phillips, J. Tanski and M. Rossi, Synthesis, Molecular Structure (X-Ray and DFT), and Solution Behavior of Titanium 4-Acyl-5-Pyrazolonates. Correlations with Related Antitumor β-Diketonato Derivatives, Inorg. Chem., 2007, 46(18), 7553–7560 CrossRef CAS PubMed.
- R. Pettinari, C. Pettinari, F. Marchetti, C. M. Clavel, R. Scopelliti and P. J. Dyson, Cytotoxicity of Ruthenium-Arene Complexes Containing β-Ketoamine Ligands, Organometallics, 2013, 32(1), 309–316 CrossRef CAS.
- F. Caruso, E. Monti, J. Matthews, M. Rossi, M. B. Gariboldi, C. Pettinari, R. Pettinari and F. Marchetti, Synthesis, Characterization, and Antitumor Activity of Water-Soluble (Arene)Ruthenium(II) Derivatives of 1,3-Dimethyl-4-Acylpyrazolon-5- Ato Ligands. First Example of Ru(Arene)(Ligand) Antitumor Species Involving Simultaneous Ru−N7(Guanine) Bonding and Li, Inorg. Chem., 2014, 53, 3668–3677 CrossRef CAS PubMed.
- R. Pettinari, C. Pettinari, F. Marchetti, B. W. Skelton, A. H. White, L. Bonfili, M. Cuccioloni, M. Mozzicafreddo, V. Cecarini, M. Angeletti, M. Nabissi and A. M. Eleuteri, Arene-Ruthenium(II) Acylpyrazolonato Complexes: Apoptosis-Promoting Effects on Human Cancer Cells, J. Med. Chem., 2014, 57(11), 4532–4542 CrossRef CAS PubMed.
- R. Pettinari, F. Marchetti, C. Pettinari, A. Petrini, R. Scopelliti, C. M. Clavel and P. J. Dyson, Synthesis, Structure, and Antiproliferative Activity of Ruthenium(II) Arene Complexes with N,O-Chelating Pyrazolone-Based β-Ketoamine Ligands, Inorg. Chem., 2014, 53(24), 13105–13111 CrossRef CAS PubMed.
- R. Pettinari, F. Marchetti, C. Pettinari, A. Petrini, B. W. Skelton, A. H. White, L. Bonfili, M. Cuccioloni and A. M. Eleuteri, Dinuclear (N6-Arene) Ruthenium(II) Acylpyrazolone Complexes: Synthesis, Characterization and Cytotoxicity, J. Organomet. Chem., 2015, 791, 1–5 CrossRef CAS.
- J. Palmucci, F. Marchetti, R. Pettinari, C. Pettinari, R. Scopelliti, T. Riedel, B. Therrien, A. Galindo and P. J. Dyson, Synthesis, Structure, and Anticancer Activity of Arene-Ruthenium(II) Complexes with Acylpyrazolones Bearing Aliphatic Groups in the Acyl Moiety, Inorg. Chem., 2016, 55(22), 11770–11781 CrossRef CAS PubMed.
- F. Marchetti, R. Pettinari, C. Di Nicola, C. Pettinari, J. Palmucci, R. Scopelliti, T. Riedel, B. Therrien, A. Galindo and P. J. Dyson, Synthesis, Characterization and Cytotoxicity of Arene-Ruthenium(II) Complexes with Acylpyrazolones Functionalized with Aromatic Groups in the Acyl Moiety, Dalton Trans., 2018, 47(3), 868–878 RSC.
- S. A. De Pascali, D. Migoni, M. Monari, C. Pettinari, F. Marchetti, A. Muscella and F. P. Fanizzi, Synthesis, Crystal Structure, and Biological Study of PtII Complexes with 4-Acyl-5-Pyrazolones, Eur. J. Inorg. Chem., 2014, 1249–1259 CrossRef CAS.
- C. Pettinari, F. Caruso, N. Zaffaroni, R. Villa, F. Marchetti, R. Pettinari, C. Phillips, J. Tanski and M. Rossi, Synthesis, Spectroscopy (IR, Multinuclear NMR, ESI-MS), Diffraction, Density Functional Study and in Vitro Antiproliferative Activity of Pyrazole-Beta-Diketone Dihalotin(IV) Compounds on 5 Melanoma Cell Lines, J. Inorg. Biochem., 2006, 100(1), 58–69 CrossRef CAS PubMed.
- A. Jain, S. Saxena, A. K. Rai and P. N. Saxena, Assessment of Toxicity of Some Penta-and Hexacoordinated Organotin (IV) and Tetracoordinated Tin(II) Complexes of Heterocyclic β-Diketones, Bioinorg. Chem. Appl., 2006, 2006, 60140 Search PubMed.
- B. Zhao, X. Shang, L. Xu, W. Zhang and G. Xiang, Novel Mixed Ligand Di-n-Butyltin (IV) Complexes Derived from Acylpyrazolones and Fluorinated Benzoic Acids: Synthesis, Characterization, Cytotoxicity and the Induction of Apoptosis in Hela Cancer Cells, Eur. J. Med. Chem., 2014, 76, 87–97 CrossRef CAS PubMed.
- F. Marchetti, J. Palmucci, C. Pettinari, R. Pettinari, F. Condello, S. Ferraro, M. Marangoni, A. Crispini, S. Scuri, I. Grappasonni, M. Cocchioni, M. Nabissi, M. R. Chierotti and R. Gobetto, Novel Composite Plastics Containing Silver(I) Acylpyrazolonato Additives Display Potent Antimicrobial Activity by Contact, Chem. – Eur. J., 2015, 21(2), 836–850 CrossRef CAS PubMed.
- F. Marchetti, J. Palmucci, C. Pettinari, R. Pettinari, S. Scuri, I. Grappasonni, M. Cocchioni, M. Amati, F. Lelj and A. Crispini, Linkage Isomerism in Silver Acylpyrazolonato Complexes and Correlation with Their Antibacterial Activity, Inorg. Chem., 2016, 55(11), 5453–5466 CrossRef CAS PubMed.
- F. Marchetti, J. Palmucci, C. Pettinari, R. Pettinari, M. Marangoni, S. Ferraro, R. Giovannetti, S. Scuri, I. Grappasonni, M. Cocchioni, F. J. Maldonado Hodar and R. Gunnella, Preparation of Polyethylene Composites Containing Silver(I) Acylpyrazolonato Additives and SAR Investigation of Their Antibacterial Activity, ACS Appl. Mater. Interfaces, 2016, 8(43), 29676–29687 CrossRef CAS PubMed.
- L. Liu, D. Jia, Y. Qiao, Y. Ji and K. Yu, Synthesis and Crystal Structure of N-(1-Phenyl-3-Methyl-4-Benzal-Pyrazolone-5)-N′-Salicylidene Hydrazine, J. Chem. Crystallogr., 2002, 32(8), 255–259 CrossRef CAS.
- Y. Zhang, L. Zhang, L. Liu, J. Guo, D. Wu, G. Xu, X. Wang and D. Jia, Anticancer Activity, Structure, and Theoretical Calculation of N-(1-Phenyl-3-Methyl-4-Propyl-Pyrazolone-5)-Salicylidene Hydrazone and Its Copper(II) Complex, Inorg. Chim. Acta, 2010, 363(2), 289–293 CrossRef CAS.
- L. Zhang, L. Liu, G. C. Xu and D. Z. Jia, Syntheses, Crystal Structures and Fluorescence Properties of Zn(II) Complexes with Pyrazolone Derivatives, J. Chem. Crystallogr., 2008, 38(11), 837–843 CrossRef CAS.
- J. Li, L. Zhang, L. Liu, G. Liu, J. Guo and D. Jia, Syntheses, Structures and Fluorescence of Lanthanide Complexes of 4-Acyl Pyrazolone Derivatives, Inorg. Chim. Acta, 2007, 360(11), 3504–3510 CrossRef CAS.
- Y. Yang, L. Zhang, L. Liu, G. Liu, J. Guo and D. Jia, Syntheses, Crystal Structures and Thermal Behaviors of Three Complexes with 4-Acyl Pyrazolone Derivatives, Inorg. Chim. Acta, 2007, 360(8), 2638–2646 CrossRef CAS.
- Kirk-Othmer Encyclopedia of Chemical Technology, ed. M. Grayson and D. Eckroth, Wiley, New York, 3rd edn, 1978, p. 819 Search PubMed.
- F. Marchetti, C. Pettinari, R. Pettinari, A. Cingolani, D. Leonesi and A. Lorenzotti, Group 12 Metal Complexes of Tetradentate N2O2-Schiff-Base Ligands Incorporating Pyrazole: Synthesis, Characterisation and Reactivity toward S-Donors, N-Donors, Copper and Tin Acceptors, Polyhedron, 1999, 18(23), 3041–3050 CrossRef CAS.
- F. Marchetti, C. Pettinari, R. Pettinari, D. Arriva, S. Troyanov and A. Drozdov, On the Interaction of Acylpyrazolonates with Zinc(II) Acceptors: The Role of Ancillary Ligands, Inorg. Chim. Acta, 2000, 307(1–2), 97–105 CrossRef CAS.
- F. Marchetti, Zinc and Cadmium Derivatives Containing Several 4-Acyl-5-Pyrazolonate Donors and Additional Ancillary Ligands, Main Group Met. Chem., 2001, 24(5), 53–59 Search PubMed.
- P. F. Liguori, A. Valentini, M. Palma, A. Bellusci, S. Bernardini, M. Ghedini, M. L. Panno, C. Pettinari, F. Marchetti, A. Crispini and D. Pucci, Non-Classical Anticancer Agents: Synthesis and Biological Evaluation of Zinc(II) Heteroleptic Complexes, Dalton Trans., 2010, 39(17), 4205–4212 RSC.
- D. Pucci, A. Crispini, M. Ghedini, M. La Deda, P. F. Liguori, C. Pettinari and E. I. Szerb, “Green Light” for Zn(Ii) Mesogens, RSC Adv., 2012, 2(24), 9071–9078 RSC.
- R. Pettinari, F. Marchetti, C. Di Nicola, C. Pettinari, A. Galindo, R. Petrelli, L. Cappellacci, M. Cuccioloni, L. Bonfili, A. M. Eleuteri, M. F. C. Guedes Da Silva and A. J. L. Pombeiro, Ligand Design for N,O-or N,N-Pyrazolone-Based Hydrazones Ruthenium(II)-Arene Complexes and Investigation of Their Anticancer Activity, Inorg. Chem., 2018, 57, 14123–14133 CrossRef CAS PubMed.
- F. Marchetti, C. Pettinari, C. Di Nicola, A. Tombesi and R. Pettinari, Coordination Chemistry of Pyrazolone-Based Ligands and Applications of Their Metal Complexes, Coord. Chem. Rev., 2019, 401, 213069 CrossRef CAS.
- K. Parimala and V. Balachandran, Vibrational Spectroscopic (FTIR and FT Raman) Studies, First Order Hyperpolarizabilities and HOMO, LUMO Analysis of p-Toluenesulfonyl Isocyanate Using Ab Initio HF and DFT Methods, Spectrochim. Acta, Part A, 2011, 81(1), 711–723 CrossRef CAS PubMed.
- G. Shanmugam and S. Brahadeeswaran, Spectroscopic, Thermal and Mechanical Studies on 4-Methylanilinium p-Toluenesulfonate - A New Organic NLO Single Crystal, Spectrochim. Acta, Part A, 2012, 95, 177–183 CrossRef CAS PubMed.
- S. Vijayalakshmi and S. Kalyanaraman, Non-Linear Optical Analyses of Nitro Aniline Derived Schiff Bases of 9-Anthraldehyde, Opt. Mater., 2013, 35(3), 440–443 CrossRef CAS.
- J. Clarkson and W. E. Smith, A DFT Analysis of the Vibrational Spectra of Nitrobenzene, J. Mol. Struct., 2003, 655(3), 413–422 CrossRef CAS.
- N. M. Kreienborg and C. Merten, How to Treat C-F Stretching Vibrations? A Vibrational CD Study on Chiral Fluorinated Molecules, Phys. Chem. Chem. Phys., 2019, 21(7), 3506–3511 RSC.
- I. Shaikh, R. N. Jadeja, R. Patel, V. Mevada and V. K. Gupta, 4-Acylhydrazone-5-Pyrazolones and Their Zinc(II) Metal Complexes: Synthesis, Characterization, Crystal Feature and Antimalarial Activity, J. Mol. Struct., 2021, 1232, 130051 CrossRef CAS.
- Z.-Y. Yang, R.-D. Yang, F.-S. Li and K.-B. Yu, Crystal Structure and Antitumor Activity of Some Rare Earth Metal Complexes with Schiff Base, Polyhedron, 2000, 19(26–27), 2599–2604 CrossRef CAS.
- V. G. Vlasenko, D. A. Garnovskii, G. G. Aleksandrov, N. I. Makarova, S. I. Levchenkov, A. L. Trigub, Y. V. Zubavichus, A. I. Uraev, Y. V. Koshchienko and A. S. Burlov, Mixed Ligand Metal-Complexes of Tridentate N, N, S Pyrazole Containing Schiff Base and 2-Amino-1-Ethylbenzimidazole: Synthesis, Structure, Spectroscopic Studies and Quantum-Chemical Calculations, Polyhedron, 2017, 133, 245–256 CrossRef CAS.
- N. M. Hosny, A. Belal, R. Motawea, M. A. Hussien and M. H. Abdel-Rhman, Spectral Characterization, DFT, Docking and Cytotoxicity of N-Benzyl-4,5-Dihydro-3-Methyl-5-Oxo-1H-Pyrazole-4-Carbothioamide and Its Metal Complexes, J. Mol. Struct., 2021, 1232, 130020 CrossRef CAS.
- P. J. Petersen, C. H. Jones and P. A. Bradford, In Vitro Antibacterial Activities of Tigecycline and Comparative Agents by Time-Kill Kinetic Studies in Fresh Mueller-Hinton Broth, Diagn. Microbiol. Infect. Dis., 2007, 59(3), 347–349 CrossRef CAS PubMed.
- G. A. Pankey and L. D. Sabath, Clinical Relevance of Bacteriostatic versus Bactericidal Mechanisms of Action in the Treatment of Gram-positive Bacterial Infections, Clin. Infect. Dis., 2004, 38(6), 864–870 CrossRef CAS PubMed.
- S. V. Kumar, S. O. Scottwell, E. Waugh, C. J. McAdam, L. R. Hanton, H. J. L. Brooks and J. D. Crowley, Antimicrobial Properties of Tris(Homoleptic) Ruthenium(II) 2-Pyridyl-1,2,3-Triazole “Click” Complexes against Pathogenic Bacteria, Including Methicillin-Resistant Staphylococcus Aureus (MRSA), Inorg. Chem., 2016, 55(19), 9767–9777 CrossRef CAS PubMed.
- H. Kargar, A. Adabi, M. Nawaz, M. Ashfaq and K. Shahzad, Synthesis, Spectral Characterization, Crystal Structure and Antibacterial Activity of Nickel(II), Copper(II) and Zinc(II) Complexes Containing ONNO Donor Schiff Base Ligands, J. Mol. Struct., 2021, 1233, 130112 CrossRef CAS.
- R. S. Joseyphus and M. S. Nair, Antibacterial and Antifungal Studies on Some Schiff Base Complexes of Zinc(II), Mycobiology, 2018, 36, 93–98 CrossRef PubMed.
- Y. Anjaneyulu and R. P. Rao, Preparation, Characterization and Antimicrobial Activity Studies on Some Ternary Complexes of Cu(II) with Acetylacetone and Various Salicylic Acids, Synth. React. Inorg. Met.-Org. Chem., 1986, 16(2), 257–272 CrossRef CAS.
- L. Mishra and V. Singh, Synthesis, Structural and Antifungal Studies of Co(II), Ni(II), Cu(II) and Zn(II) Complexes with New Schiff Bases Bearing Benzimidazoles, Indian J. Chem., Sect. A: Inorg., Bio-inorg., Phys., Theor. Anal. Chem., 1993, 32(05), 446–449 Search PubMed.
- A. D. Becke, Density–functional Thermochemistry. III. The Role of Exact Exchange, J. Chem. Phys., 1993, 98(7), 5648–5652 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron Density, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37(2), 785–789 CrossRef CAS PubMed.
- M. Cossi, N. Rega, G. Scalmani and V. Barone, Energies, Structures, and Electronic Properties of Molecules in Solution with the C–PCM Solvation Model, J. Comput. Chem., 2003, 24(6), 669–681 CrossRef CAS PubMed.
- M. W. Wong, Vibrational Frequency Prediction Using Density Functional Theory, Chem. Phys. Lett., 1996, 256(4–5), 391–399 CrossRef CAS.
- A. P. Scott and L. Radom, Harmonic Vibrational Frequencies: An Evaluation of Hartree− Fock, Møller− Plesset, Quadratic Configuration Interaction, Density Functional Theory, and Semiempirical Scale Factors, J. Phys. Chem., 1996, 100(41), 16502–16513 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. J. A. Throssell, Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision B.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- L. B. Reller, M. Weinstein, J. H. Jorgensen and M. J. Ferraro, Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices, Clin. Infect. Dis., 2009, 49(11), 1749–1755 CrossRef PubMed.
- A. W. Bauer, W. M. M. Kirby, J. C. Sherris and M. Turck, Antibiotic Susceptibility Testing by a Standardized Single Disk Method, Am. J. Clin. Pathol., 1966, 45, 493–496 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2158176–2158180. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2dt02430f |
| ‡ Deceased. |
| This journal is © The Royal Society of Chemistry 2022 |