Open Access Article
Open Access ArticleA doxorubicin–peptide–gold nanoparticle conjugate as a functionalized drug delivery system: exploring the limits†
Kai S.
Exner
 ab and
Anela
Ivanova
ab and
Anela
Ivanova
 *a
*a
aSofia University, Faculty of Chemistry and Pharmacy, Department of Physical Chemistry, 1 James Bourchier blvd., 1164 Sofia, Bulgaria. E-mail: aivanova@chem.uni-sofia.bg; Tel: +359-2-8161520
bCluster of Excellence RESOLV, Bochum, Germany
First published on 25th May 2022
Abstract
Efficient transport of pharmaceuticals to malignant cells in the human body often requires the application of drug-delivery systems (DDSs) consisting of several building blocks, each of them bearing a specific function. While nanoparticles are promising as potential carrier moieties, biomolecules may add to the efficient delivery by binding several drug molecules simultaneously. In this contribution, we apply a combination of atomistic molecular dynamics simulations and density functional theory calculations to characterize a multi-component DDS for the transport of the anthracycline antibiotic doxorubicin (DOX), comprising a gold nanoparticle (NP) and a drug-binding peptide (DBP) grafted on the NP surface. We have shown previously that the DDS can stabilize one DOX per DBP. However, by increasing the drug load to a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DOX
1 DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio the two drug molecules compete for the available adsorption sites, which may cause spontaneous dissociation of one DOX molecule. We identify the chain length of the DBP as a limiting factor for the drug-loading capacity and provide important guidelines for further optimization of multi-component functionalized DDSs.
DBP ratio the two drug molecules compete for the available adsorption sites, which may cause spontaneous dissociation of one DOX molecule. We identify the chain length of the DBP as a limiting factor for the drug-loading capacity and provide important guidelines for further optimization of multi-component functionalized DDSs.
1 Introduction
Cancer is the leading cause of death worldwide.1 The treatment of malignant cancer cells requires chemotherapy, in which, among others, anthracycline antibiotics such as doxorubicin (DOX) are used as therapeutic agents.2 The direct use of DOX or other pharmaceuticals is, however, limited by severe side effects occurring when the drug enters the human body.3 These side effects comprise cytotoxicity to undifferentiated cells, cardiotoxicity, myelotoxicity, nausea, and vomiting. As such, it is an ultimate goal of researchers to develop strategies that help overcome these inadequacies. A forthcoming solution is the application of drug-delivery systems (DDSs).4 A DDS consists of several building blocks, in which each unit of the DDS bears a specific function. Besides the drug, prospective components of a DDS are a biomolecule and a nanoparticle, which in conjunction may add to the transport efficiency of the drug. While a biomolecule may bind several drug cargos at the same time, a nanoparticle may carry several biomolecule-drug entities simultaneously. Another advantage corresponds to the fact that ligands for targeted delivery or additional residues, facilitating water solubility or enabling membrane translocation, can be readily attached to the surface of a nanoparticle.Gold nanoparticles (Au-NPs) are an auspicious carrier material for the delivery of DOX.5 This can be explained by the fact that chemically modified Au-NPs have been shown to overcome multidrug resistance in cancer cells.6 Gold nanoconjugates can provide a highly effective method for introducing pharmaceutically active substances into cells.7 They can either entrap non-covalently or bind through a covalent linker the active substance, where the site and rate of release often determine the therapeutic efficacy.8 Their function may be understood even better if these conjugates are investigated by molecular modelling techniques on different time and length scales.9
Computational studies on the anthracycline antibiotic doxorubicin10 and DDS components11 for its transport increased in recent years. In these contributions, the geometry of DOX in the gas phase and its structural and dynamic parameters in aqueous solution under ambient and physiological conditions were resolved,10 or the interactions of DOX with a carrying unit were quantified by molecular simulations.11 Further molecular-level details of the interaction of DOX with the different building blocks of a DDS constituent are ultimately required to fine-tune components within the construction of a complex functionalized DDS. In a recent theoretical study, we investigated a DDS component consisting of a gold nanoparticle (Au-NP) and a drug-binding peptide (DBP) with one adsorbed DOX molecule by a combination of atomistic molecular dynamics (MD) simulations and density functional theory (DFT) calculations.12 Therein, the associate NP–DBP–DOX was identified as a prospective building block of a more complex DDS, corroborated by the fact that the drug was preferably intercalated in-between its carrier moieties, NP and DBP. It was concluded that this intercalation of the drug is beneficial for its delivery, as DOX is efficiently shielded from the electrolyte solution and, hence, is not directly exposed to external influences during the transport.
In the present work, we extend our previous study on the associate NP–DBP–DOX by increasing the number of DOX molecules in the investigated model to identify the maximum drug load per peptide chain for efficient drug delivery. Applying a combination of classical MD and DFT, we identify the chain length of the DBP as a limiting factor for the theoretically observed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DOX
1 DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio, which is in reasonable agreement with experimental studies.13 We also confirm the predisposition of DOX to interact profoundly with its carrier material.
DBP ratio, which is in reasonable agreement with experimental studies.13 We also confirm the predisposition of DOX to interact profoundly with its carrier material.
2 Computational details
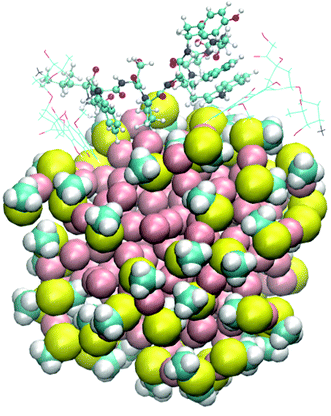
To describe the associate NP–DBP–DOX by computational methods, we adopt the atomic coordinates of a theoretically modeled Au-NP from the literature.14 It should be emphasized that the size of this Au-NP (2.46 nm in diameter; cf. ESI,† Section S1) is somewhat smaller than Au-NPs in therapeutic or other applications, typically ranging from about 5 nm to 10 nm.8 However, the reduction in size of the Au-NP is indispensable to apply a modeling approach that takes both dynamic (MD) and energetic (DFT) aspects of the multi-component DDS into account. Consequently, we restrict the theoretical model to one Au-NP covalently bound to one drug-binding peptide (DBP) molecule. DBP has the sequence NH3+-CGGLWSPWYGGSW-CONH2. It is attached to the NP by a covalent Au–S bond via the side chain of the N-terminal cysteine residue (C) of the peptide, which is coherent with experimental studies of the associate NP–DBP–DOX.13Two DOX molecules are present in the investigated model system. In the initial configuration, one DOX is already attached to the peptide and a second DOX molecule is randomly placed in the saline solution surrounding the NP–DBP–DOX complex obtained in our previous study.12 The last snapshot of the trajectory from the previous work12 is taken as the starting geometry. Fig. 1 provides a sketch of NP–DBP–2DOX after the system is fully relaxed. A complete illustration of the model system can be found in the ESI† (Fig. S1).
The associate NP–DBP–2DOX is solvated in a box with edge sizes of (6 × 6 × 7) nm, applying periodic boundary conditions. Sodium and chloride ions are added to mimic physiological conditions of the human body at T = 310 K, that is, the NaCl concentration amounts to 154 mmol L−1. The number of explicit water molecules in the simulations is 7767. The total number of atoms in the model is 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 111.
111.
Atomistic MD simulations are performed for the NP–DBP–2DOX model (Fig. 1), using the force field AMBER0315 for DOX, DBP, as well as for the sodium cations, chloride anions, and the caps of Au-NP. The solvent is modeled by TIP3P.16 Parameters used for the charged forms of the drug and the DBP are specified in previous publications.9,12 The parameters for the covalent Au–S junction between Au-NP and DBP as well as non-bonded parameters for Au are taken from the study of Giri and Spohr.17 RESP charges18 for the Au, C, S, and H atoms of the caps are given in our previous publication.12
For all Au atoms of the Au-NP, position restraints with a force constant of 1800 kJ mol−1 nm−2 are imposed during all MD simulations to prevent the Au-NP from dissociating, discussed in more detail in our previous work.9,12 Berendsen barostat19 and v-rescale thermostat20 are employed to maintain conditions close to those in vivo, i.e., constant pressure of 1 bar and temperature of 310 K. During the simulations, all hydrogen-containing bonds are restrained by LINCS21 or SETTLE22 for the organic molecules and water, respectively. A Lennard-Jones potential with a cutoff of 1.2 nm and a switch function activated at 1.0 nm describes the non-bonded interactions, while electrostatics is assessed in the monopole approximation with the PME method,23 applying a cutoff of 1.2 nm and a switch function turned on at 1.0 nm. The l-BFGS algorithm24 is used for energy minimization, and the integration of the equations of motion is performed with leap-frog,25 using a time step of 1 fs. Snapshots in the trajectory are saved every 2 ps.
All MD simulations, applying the software package Gromacs 2016.3,26 are carried out following a standard protocol, consisting of energy minimization with position restraints, energy minimization without position restraints (except for those on the Au atoms, which are maintained throughout the entire simulation), heating, equilibration, and data collection within a production phase of 200 ns. This procedure is described in detail in our previous work.9 The equilibration of the associate NP–DBP–2DOX is verified by the evolution of the total energy, temperature, pressure, and the root-mean-square deviation (RMSD) of the atomic coordinates of the highly flexible DBP with respect to the coordinates of the minimized structure (Fig. S2, ESI†).
Cluster analysis of the production part of the trajectory is performed with the method of Jarvis–Patrick,27 in which a cutoff distance of 0.13 nm is used. Clusters with a relative population of at least 3% are considered as representative of a given structural type, referring to previous studies on DOX.9,12 A given configuration is illustrated by its middle structure. VMD 1.8.628 is used for visualization.
For the middle structures of all representative clusters, a model consisting of 421 atoms (all 138 atoms of the two DOX residues, all 192 atoms of DBP, 91 atoms from the capped Au-NP) is subjected to DFT calculations. The parts of the Au-NP that do not interact directly with DBP–2DOX are cut out for computational feasibility. To obtain the binding energies (BEs) between the four parts of the DDS component, DFT with dispersion correction is employed, utilizing B3LYP-D3 as the functional29 and 6-31G* as the basis set for all atoms except Au, for which LANL2DZ30 is applied. The BEs are calculated for the dimers DOX1–DBP, DOX2–DBP, DOX1–NP, DOX2–NP, and DBP–NP in vacuo from the relation BE = E(X–Y) − E(X) − E(Y), in which E(X–Y), E(X), and E(Y) indicate the electronic energies of the respective dimer (X–Y) and the corresponding monomers (X and Y). Basis set superposition error (BSSE) counterpoise correction31 is applied to alleviate overbinding effects. All DFT calculations are performed with GAUSSIAN16.32
3 Results and discussion
In the initial configuration of the associate NP–DBP–2DOX, DOX1 is interacting with the caps of the Au-NP and the tryptophan residue W5, as observed in our previous study for the conjugate NP–DBP–DOX.12 DOX2 is placed randomly in the saline solution surrounding the NP–DBP–DOX complex. Already during the MD relaxation stage, DOX2 also adsorbs on the NP–DBP carrier, finding the tryptophan residue W8 in conjunction with the caps of the Au-NP as an adsorption site. In the subsequent production phase (200 ns), different adsorption structures of the two drug molecules are observed, in which DOX1 does not change its stacking partner W5, whereas DOX2 scans different adsorption sites on the surface of the conjugate NP–DBP. After 195 ns, DOX2 dissociates from the DDS component into the saline solution, indicating that the conjugate NP–DBP can stabilize two drug molecules in a limited time range only, as discussed below.In the ESI,† Section S1, we compare the physical properties of the Au-NP in the complex NP–DBP–2DOX to the associate NP–DBP–DOX modeled in our previous study.12 We observe that the presence of a second drug molecule results in an elongated, non-spherical NP, accompanied by reorientation of the Au atoms (Fig. S3 and S4, ESI†). We trace the more deformed-sphere shape of the NP to the fact that the peptide is more spread on the surface of the NP when two DOX entities are adsorbed, thereby aiming at sufficient stabilization of both drug molecules to prevent their dissociation. The configurational changes of the NP and DBP have direct implications on the surface chemistry of the two DOX residues within the DDS component, which is discussed next.
In the 200 ns production phase, we observe the following relatively long-living (with existence time ≥ 5 ns) adsorption structures, summarized in Fig. 2:
(a) DOX1 is intercalated between the caps of the NP and the tryptophan residue W5; DOX2 is intercalated between the caps of the NP and W8.
(b) DOX1 is intercalated between the caps of the NP and W5; DOX2 is non-specifically intercalated between the caps of the NP and the backbone of the peptide, that is, there is no specific interaction of the drug with any amino acid from the DBP.
(c) DOX1 is stacked by W5 in a perpendicular arrangement to the NP surface, in which part of the anthracycline fragment of DOX interacts with the caps of the NP; DOX2 is non-specifically intercalated between the caps of the NP and the peptide backbone.
(d) DOX1 is stacked by W5 in a perpendicular arrangement to the NP surface; DOX2 is intercalated between W8 and W13.
(e) DOX1 is stacked by W5 in a perpendicular arrangement to the NP surface; DOX2 is intercalated between the tyrosine residue Y9 and the caps of the NP.
Enlarged depictions of the adsorption configurations in Fig. 2 can be found in the ESI,† Section S2 (Fig. S5–S9). As evident from the representative structures, DOX has a pronounced affinity to interact both with the DBP and with the nanoparticle simultaneously. This was observed in the previous study with one DOX entity as well.12 In the associate NP–DBP–2DOX, both drug molecules attempt to intercalate their anthracycline part between a tryptophan residue from the DBP and the caps of the NP. Since DOX is positively charged, it happens as far apart as possible along the peptide chain (Fig. 2(a)). This causes significant stretching of the DBP structure, which tries to balance between the two drug molecules.
The observed adsorption configurations are similar to the structures obtained in our recent study of the conjugate NP–DBP–DOX.12 There, DOX was either intercalated between the caps of the NP and the tryptophan residue W5, intercalated between two tryptophan residues (W5 and W13), stacked by W5 in a perpendicular arrangement to the NP surface, or non-specifically intercalated between the caps of the NP and the backbone of the peptide.12 Due to the presence of a second drug molecule in the associate NP–DBP–2DOX, the tryptophan entity W8 also comes into play, which stabilizes DOX2 in the first part of the trajectory. Contrary to the conjugate NP–DBP–DOX, we observe interaction of the second drug molecule with tyrosine (Y9), indicating that DOX2 cannot find a stable adsorption site on the tryptophan. Indeed, the interaction of DOX2 with Y9 is short-term and, thereafter, DOX2 spontaneously dissociates from the DDS component at t = 195 ns and remains in the solution until the end of the trajectory. This is the reason why we did not evaluate further trajectories or did not extend the simulations beyond 200 ns. The analyzed trajectory already reveals that the conjugate NP–DBP–2DOX is not stable over an extended time range. Rather, the NP–DBP junction can stabilize two drug entities only for a limited period. Existence times at the timescale of the MD simulations for the detected adsorption configurations are given as a histogram in Fig. 3.
 | ||
| Fig. 3 Existence times, extracted from the MD simulations, for the five different adsorption states of DOX1 and DOX2 depicted in Fig. 2. The adsorption state on the right corresponds to the last 5 ns of the production phase, in which DOX2 has dissociated from the DDS component (Fig. S10, ESI†). | ||
Reading the histogram in Fig. 3 from left to right provides an overview of the time evolution of the analyzed trajectory, since the structures are ordered as they appear during the simulation. A more detailed discussion of the MD trajectory can be found in the ESI,† Section S3. Adsorption state (a) prevails in the first 70 ns of the trajectory, but at t = 70 ns DOX2 leaves its stacking partner W8 and intercalates non-specifically between the backbone of the peptide and the caps of the Au-NP within configuration (b). Hence, DOX2 is closer in space to DOX1, but dimer formation of the DOX residues, observed in previous studies of Gocheva et al. for the conjugate DBP–DOX without the NP,11 does not take place. As discussed in our previous contribution,12 the presence of the Au-NP increases the degrees of freedom and provides an additional option to interact with. It becomes evident from Fig. 2 and 3 that the Au-NP is of utmost importance to stabilize both drug entities, discussed in more detail below within the analysis of the DFT calculations.
After 85 ns, the adsorption configuration of DOX1 switches from the sandwich complex CAPS–DOX1–W5 to the perpendicular configuration W5–DOX1, in which part of the drug is still interacting with the caps of the NP. As such, both DOX residues are separated from each other in that the tryptophan residue W5 serves as a protection shield for DOX1 (Fig. S7, ESI†). The adsorption configuration (c) corresponds to the state with the longest existence time of about 100 ns (Fig. 3). We trace this finding to the fact that in this structure the peptide structure is less strained compared to adsorption configurations (a) or (d), in which several tryptophan residues of the DBP need to cope with both drug entities at the same time. After 185 ns, DOX2 intercalates between W8 and W13 within the adsorption state (d). It can be deduced from Fig. S8 that the anthracycline fragment of the drug is not ideally aligned between the two tryptophan residues, but rather the sandwich configuration W8–DOX2–W13 is somewhat twisted. This might explain the short existence time of this adsorption state, unlike the complex with one DOX, where it was much more stable.12 After that, DOX2 switches its position to intercalate between tyrosine (Y9) and the caps of the NP within adsorption configuration (e). However, this placement is energetically unstable (Table 1) so that DOX2 dissociates from the DDS component. At the end, only DOX1 is carried by the conjugate NP–DBP.
| Configuration | (a) | (b) | (c) | (d) | (e) |
|---|---|---|---|---|---|
| # Clusters | 5 | 1 | 6 | 1 | 1 |
| DOX1–NP | −14.11 ± 5.52 | −3.72 | 0.86 ± 6.82 | +6.26 | −3.00 |
| DOX1–DBP | +7.92 ± 7.07 | −8.96 | −37.74 ± 8.32 | −48.77 | −30.16 |
| DBP1–NP | −67.07 ± 11.66 | −56.20 | −46.77 ± 6.28 | −51.29 | −54.52 |
| DOX2–NP | −15.40 ± 5.51 | −30.91 | −22.18 ± 6.87 | −14.17 | −3.72 |
| DOX2–DBP | −5.34 ± 10.33 | −20.49 | −23.32 ± 20.15 | +2.73 | +8.72 |
| DBP2–NP | −64.11 ± 7.58 | −31.60 | −30.83 ± 11.87 | −26.88 | −40.97 |
To comprehend the energetics of the reported adsorption configurations, we apply DFT calculations. Cluster analysis, which brings together configurations of the same structural types, reveals 14 representative geometries for the NP–DBP–2DOX associate. For them, we calculate for both drug entities separately the two-body binding energies between DOX and DBP, DOX and NP, as well as DBP and NP (Table 1). When the energetics for DOX1 is assessed, DOX2 is treated as a non-interacting part together with the DBP and vice versa.
Table 1 indicates that the energetics of the two drug residues significantly changes within the observed adsorption states. While in adsorption configuration (a) both drug entities are mainly stabilized by the caps of the NP, the interaction with the peptide is only weakly attractive or even repulsive for DOX2 and DOX1, respectively. Since both pharmaceuticals are capped in a sandwich configuration between the peptide and the caps of the NP (Fig. S5, ESI†), the peptide needs to spread over the NP surface to provide each drug molecule a single tryptophan residue for intercalation. This may explain the strong interaction between the peptide and the nanoparticle, quantified within the dimers DBP1–NP and DBP2–NP.
As soon as DOX2 leaves its intercalation partner W8 (adsorption state (b)), the contribution of the peptide to the stabilization of both drug entities significantly increases. This is particularly visible in the adsorption state (c) with the longest existence time, where DOX1, stacked by W5 in perpendicular configuration to the NP, is entirely stabilized by the peptide, while the peptide and the caps of the NP equally contribute to the stabilization of DOX2. The enhanced attraction of both drug residues with the DBP, though, is accompanied by a deteriorated interaction of the peptide and the caps of the NP (dimer DBP–NP). Stabilization of both drug entities to a comparable extent by the DBP is only observed in configuration (c), indicating that the DBP lacks balance between the two DOX molecules in the other adsorption states.
In the last two adsorption configurations (d) and (e), the BE between the peptide and the caps of the NP is enhanced. In turn, this results in significant shrinking of the interaction energy between DOX2 and the DBP, while the stabilization of DOX1 by the peptide is almost unaffected. In other words, DOX1 keeps its attractive interaction with the DBP from configuration (c) in configurations (d) and (e) at the expense of DOX2, which is even repelled by the peptide in these adsorption states. We conclude that the BEs of the calculated dimers (Table 1) are intrinsically coupled, involving a redistribution of the interaction energies between the different building blocks of the DDS component when a change in adsorption configuration takes place.12
In our previous contribution referring to the associate NP–DBP–DOX,12 we identified the CAPS–DOX–W5 configuration (comparable to the stabilization of DOX1 in (a) and (b) without the presence of the second DOX molecule) as the longest-existing and most stabilized adsorption state. We use the energetics of the CAPS–DOX–W5 configuration obtained there as a benchmark to assess whether the drug entities DOX1 and DOX2 in the conjugate NP–DBP–2DOX are sufficiently stabilized. The BEs for the dimers DOX–NP and DOX–DBP from NP–DBP–DOX are (−16.83 ± 8.38) kcal mol−1 and (−21.46 ± 10.31) kcal mol−1, respectively.12 Thus, the overall stabilization of DOX in the associate NP–DBP–DOX is given by the sum of these two contributions, that is, (−38.29 ± 18.69) kcal mol−1. Table 2 summarizes the overall stabilization energy of DOX1 and DOX2 in the conjugate NP–DBP–2DOX, derived by summing up the BEs reported in Table 1.
| Configuration | (a) | (b) | (c) | (d) | (e) |
|---|---|---|---|---|---|
| Overall DOX1 stabilization | −6.19 ± 12.59 | −12.68 | −36.88 ± 15.14 | −42.51 | −33.16 |
| Overall DOX2 stabilization | −20.74 ± 15.84 | −51.40 | −45.50 ± 27.02 | −11.44 | +5.00 |
Table 2 shows that the presence of DOX2 significantly affects the stabilization of DOX1 in its sandwich configuration between the caps of the NP and W5 (adsorption states (a) and (b)), resulting in insufficient stabilization of DOX1 when compared to the benchmark value (−38.29 ± 18.69) kcal mol−1. In contrast, DOX2 is adequately stabilized in its intercalation complex between the caps of the NP and W8 (adsorption state (a)) and even improves its interaction with the building blocks of the DDS component when intercalating non-specifically between the NP and the DBP (adsorption state (b)). This finding indicates that the short peptide chain is not capable of stabilizing two drug residues in a balanced way by a sandwich arrangement between the caps of the NP and a tryptophan residue at the same time, albeit this type of sandwich arrangement is the energetically most favorable configuration for one DOX molecule in the associate NP–DBP–DOX.12 A potential solution to guarantee sufficient stabilization of both drug molecules refers to the π-stacking of DOX1 by W5 perpendicular to the NP surface in conjunction with the non-specific intercalation of DOX2, as encountered with the longest-existing adsorption state (c). In this case, both drug entities are stabilized similarly to DOX in the associate NP–DBP–DOX. However, as discussed above, this adsorption configuration suffers from a deteriorated interaction between the peptide and the NP, which may explain why, despite stabilizing both drugs sufficiently, a change in this adsorption configuration takes place. In the short-living adsorption states (d) and (e), the increased interaction between the DBP and NP (Table 1) is at the cost of the residue DOX2, which, compared to the benchmark value (−38.29 ± 18.69) kcal mol−1, is not sufficiently stabilized anymore. Even worse, in adsorption state (e) the interaction energy of DOX2 with its partners is repulsive, explaining the spontaneous release of the drug from the DDS component shortly afterwards.
In conclusion, the DFT calculations reveal that the conjugate NP–DBP–2DOX is able to stabilize two drug residues sufficiently in a limited time range only within the adsorption state (c). The existence time of this configuration amounts to 100 ns (Fig. 3). This may indicate that the associate NP–DBP is not stable on the long term when carrying several drug cargos, coinciding with the spontaneous dissociation of DOX2 at the end of the analyzed trajectory. The obtained results corroborate the experimentally observed DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio of about 0.7 at pH = 9,13 providing a molecular-level explanation of why the ratio DOX
DBP ratio of about 0.7 at pH = 9,13 providing a molecular-level explanation of why the ratio DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP within the DDS component does not exceed 1
DBP within the DDS component does not exceed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Even if the caps of the NP significantly contribute to stabilize both DOX entities within the DDS component, the short peptide chain is not capable of handling more than one drug residue in the presence of the NP.
1. Even if the caps of the NP significantly contribute to stabilize both DOX entities within the DDS component, the short peptide chain is not capable of handling more than one drug residue in the presence of the NP.
It should be noted, though, that a different behavior of the peptide is witnessed in the absence of the Au-NP, in which several drug cargos can be stacked on the tryptophan residues within the DBP.11d A potential strategy to overcome this issue in the presence of the gold nanoparticle could be to replace the short-sequenced DBP by a more extended peptide chain. This may guarantee that the peptide can maintain its attractive interaction with the caps of the NP, at the same time being able to provide multiple stable adsorption sites for the drug residues. Another option corresponds to a surface modification of the Au-NP, such as by introducing functionalized groups that add to the stabilization of DOX. Peptide ligands containing tryptophan residues appear promising since tryptophan entities are present in the adsorption configurations (a) and (c) with longest existence times (Fig. 3). Last, the pH of the medium is a parameter to be tuned for efficient drug delivery. In the ESI,† Section S4, we report MD simulations for the conjugate NP–DBP–2DOX, in which we mimic higher pH values with deprotonated amine groups of the two DOX entities and the cysteine residue of the DBP. In this higher-pH medium, the adsorption state of DOX1 is no longer linked to the location of the tryptophan residue W8, which has a beneficial effect for the entire DDS component, expressed in that DOX2 can enter long-living adsorption states with W13 or W13 and W8 that were previously not accessible (Fig. S14 and S15, ESI†). Therefore, variation of the medium acidity, or alternatively the way of administration of the pharmaceutical, may be used as an efficient tool for stabilization of the DOX–DBP–Au-NP drug delivery component.
4 Conclusions
In the present manuscript, we explore theoretically a drug-delivery system (DDS) component for the cytostatic doxorubicin (DOX), considering as a carrier a gold nanoparticle (NP) with a short chain drug-binding peptide (DBP) grafted on its surface. While experimentally it was found that a maximum of about one drug molecule can be attached to the carrier peptide, we investigate the limiting 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DOX
1 DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio by molecular modeling, applying a combination of atomistic molecular dynamics simulations and density functional theory calculations for a well-defined NP–DBP–2DOX model system. We demonstrate that by increasing the DOX
DBP ratio by molecular modeling, applying a combination of atomistic molecular dynamics simulations and density functional theory calculations for a well-defined NP–DBP–2DOX model system. We demonstrate that by increasing the DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio from 1
DBP ratio from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, a different surface chemistry of the drug entities in the DDS component is witnessed. This includes the presence of unstable adsorption sites for at least one DOX molecule, corroborated by the fact that one drug entity is spontaneously released at the end of the MD trajectory. This result can be understood by the application of DFT calculations, indicating that the interaction of the released drug molecule with the associate NP–DBP is repulsive before the pharmaceutical leaves its carrier.
1, a different surface chemistry of the drug entities in the DDS component is witnessed. This includes the presence of unstable adsorption sites for at least one DOX molecule, corroborated by the fact that one drug entity is spontaneously released at the end of the MD trajectory. This result can be understood by the application of DFT calculations, indicating that the interaction of the released drug molecule with the associate NP–DBP is repulsive before the pharmaceutical leaves its carrier.
The DFT calculations reveal that the conjugate NP–DBP can stabilize two drug molecules in a limited time range only. Otherwise, the interaction of at least one DOX molecule with its carriers is merely slightly attractive or even repulsive. We identify that the DBP is the critical factor for the observed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DOX
1 DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio. Due to the short chain of the peptide, the DBP is distinctly strained when interacting with two drug entities in the presence of the Au-NP. This is because the peptide strives to keep its additional attractive interaction with the NP surface as well. Therefore, it is not possible for the peptide to maintain balance between two drug molecules and the NP, and thus the DBP cannot handle more than one drug entity in the long term. To overcome this, we suggest different strategies, including the usage of an extended peptide chain, modification of the NP surface by adding functional groups, or variation of the medium acidity, to increase the drug load beyond the observed 1
DBP ratio. Due to the short chain of the peptide, the DBP is distinctly strained when interacting with two drug entities in the presence of the Au-NP. This is because the peptide strives to keep its additional attractive interaction with the NP surface as well. Therefore, it is not possible for the peptide to maintain balance between two drug molecules and the NP, and thus the DBP cannot handle more than one drug entity in the long term. To overcome this, we suggest different strategies, including the usage of an extended peptide chain, modification of the NP surface by adding functional groups, or variation of the medium acidity, to increase the drug load beyond the observed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DOX
1 DOX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio. The obtained results show that DOX is a very active cargo, which perturbs its carriers by interacting strongly with them. This is in line with previous reports33 and implies that DDS components for the transport of DOX should be selected carefully, preferably after pre-screening of their coupling to the drug.
DBP ratio. The obtained results show that DOX is a very active cargo, which perturbs its carriers by interacting strongly with them. This is in line with previous reports33 and implies that DDS components for the transport of DOX should be selected carefully, preferably after pre-screening of their coupling to the drug.
In conclusion, the present study underlines that the grafting of DBP–DOX complexes on an Au-NP could be regarded as an auspicious strategy to enhance the load of DOX per carrier unit after directed tuning. It may contribute to the efficient transport of anthracycline antibiotics to malignant cells within the construction of a more complex DDS for the delivery of doxorubicin in the future.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
K. S. E. gratefully acknowledges funding from the Alexander von Humboldt Foundation. K. S. E. is associated with the RESOLV Cluster of Excellence, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy-EXC 2033-390677874-RESOLV. Computational time was provided in part through Project No. 80-10-21/2020 of the Sofia University Research Fund.References
- R. L. Siegel, K. D. Miller and A. Jemal, Cancer statistics, CA-Cancer J. Clin., 2019, 69, 7–34 CrossRef PubMed.
- (a) G. Minotti, P. Menna, E. Salvatorelli, G. Cairo and L. Gianni, Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity, Pharmacol. Rev., 2004, 56, 185–229 CrossRef CAS PubMed; (b) J. Nadas and D. Sun, Anthracyclines as effective anticancer drugs, Expert Opin. Drug Discovery, 2006, 1, 549–568 CrossRef CAS PubMed; (c) P. Agrawal, S. K. Barthwal and R. Barthwal, Studies on self-aggregation of anthracycline drugs by restrained molecular dynamics approach using nuclear magnetic resonance spectroscopy supported by absorption, fluorescence, diffusion ordered spectroscopy and mass spectrometry, Eur. J. Med. Chem., 2009, 44, 1437–1451 CrossRef CAS PubMed.
- N. Ivanova, Y. Tsoneva, N. Ilkova and A. Ivanova, Complex systems for drug transport across cell membranes, Chem. Bulg. J. Sci. Edu., 2015, 24, 825–848 CAS.
- (a) O. Tacara, P. Sriamornsak and C. R. Dass, Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems, J. Pharm. Pharmacol., 2013, 65, 157–170 CrossRef PubMed; (b) A. Bunker and T. Róg, Mechanistic Understanding From Molecular Dynamics Simulation in Pharmaceutical Research 1: Drug Delivery, Front. Mol. Biosci., 2020, 7, 604770 CrossRef CAS PubMed.
- D. Curry, et al., Adsorption of doxorubicin on citrate-capped gold nanoparticles: Insights into engineering potent chemotherapeutic delivery system, Nanoscale, 2015, 7, 19611–19619 RSC.
- (a) F. Wang, Y.-C. Wang, S. Dou, M.-H. Xiong, T.-M. Sun and J. Wang, Doxorubicin-tethered responsive gold nanoparticles facilitate intracellular drug delivery for overcoming multidrug resistance in cancer cells, ACS Nano, 2011, 5, 3679–3692 CrossRef CAS PubMed; (b) Z. Wang, Z. Wang, D. Liu, X. Yan, F. Wang, G. Niu, M. Yang and X. Chen, Biomimetic RNA-silencing nanocomplexes: Overcoming multidrug resistance in cancer cells, Angew. Chem., Int. Ed., 2014, 53, 1997–2001 CrossRef CAS PubMed.
- (a) S. Aryal, J. J. Grailer, S. Pilla, D. A. Steeber and S. Gong, Doxorubicin conjugated gold nanoparticles as water-soluble and pH-responsive anticancer drug nanocarriers, J. Mater. Chem., 2009, 19, 7879–7884 RSC; (b) P. Manivasagan, S. Bharathiraja, N. Q. Bui, B. Jang, Y.-O. Oh, I. G. Lim and J. Oh, Doxorubicin-loaded fucoidan capped gold nanoparticles for drug delivery and photoacoustic imaging, Int. J. Biol. Macromol., 2016, 91, 578–588 CrossRef CAS PubMed.
- K. Kumar, P. Moitra, M. Bashir, P. Kondaiah and S. Bhattacharya, Natural tripeptide capped pH-sensitive gold nanoparticles for efficacious doxorubicin delivery both in vitro and in vivo, Nanoscale, 2020, 12, 1067–1074 RSC.
- K. S. Exner and A. Ivanova, Method to construct volcano relations by multiscale modeling: Building Bridges between the catalysis and biosimulation communities, J. Phys. Chem. B, 2020, 125, 2098–2104 CrossRef PubMed.
- (a) S. Zhu, L. Yan, X. Ji and W. Lu, Conformational diversity of anthracycline anticancer antibiotics: A density functional theory calculation, J. Molec. Struct., 2010, 951, 60–68 CrossRef CAS; (b) Y. Tsoneva, A. Tadjer, M. Lelle, K. Peneva and A. Ivanova, Molecular structure and pronounced conformational flexibility of doxorubicin in free and conjugated state within a drug-peptide compound, J. Phys. Chem. B, 2015, 119, 3001–3013 CrossRef CAS PubMed.
- (a) G. Gocheva, N. Ilieva, K. Peneva and A. Ivanova, Characterization of the interaction forces in a drug carrier complex of doxorubicin with a drug-binding peptide, Chem. Biol. Drug Des., 2018, 91, 874–884 CrossRef CAS PubMed; (b) L. Zhang, G. Peng, J. Li, L. Liang, Z. Kong, H. Wang, L. Jia, X. Wang, W. Zhang and J.-W. Shen, Molecular dynamics study on the configuration and arrangement of doxorubicin in carbon nanotubes, J. Mol. Liq., 2018, 262, 295–301 CrossRef CAS; (c) P. Wolski, K. Nieszporek and T. Panczyk, Multimodal, pH sensitive, and magnetically assisted carrier of doxorubicin designed and analyzed by means of computer simulations, Langmuir, 2018, 34, 2543–2550 CrossRef CAS PubMed; (d) G. Gocheva, K. Peneva and A. Ivanova, Self-assembly of doxorubicin and a drug-binding peptide studied by molecular dynamics, Chem. Phys., 2019, 525, 110380 CrossRef CAS.
- K. S. Exner and A. Ivanova, Identifying a gold nanoparticle as a proactive carrier for transport of a doxorubicin-peptide complex, Colloids Surf., B, 2020, 194, 111155 CrossRef CAS PubMed.
-
K. Peneva, personal communication: For Au-NPs with a mean size of 6 nm, the experimentally observed DOX
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBP ratio is about 0.7 at pH = 9. Unpublished results.
DBP ratio is about 0.7 at pH = 9. Unpublished results. - O. Lopez-Acevedo, J. Akola, R. L. Whetten, H. Grönbeck and H. Häkkinen, Structure and bonding in the ubiquitous icosahedral metallic gold cluster Au144(SR)60, J. Phys. Chem. Lett., 2009, 113, 5035–5038 CrossRef CAS.
- Y. Duan, C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang and P. Kollman, A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations, J. Comput. Chem., 2003, 24, 1999–2012 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- A. K. Giri and E. Spohr, Conformational equilibria of organic adsorbates on nanostructures in aqueous solution: MD simulations, J. Phys. Chem. C, 2015, 119, 25566–25575 CrossRef CAS.
- C. I. Bayly, P. Cieplak, W. D. Cornell and P. A. Kollman, A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- J. P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- S. Miyamoto and P. A. A. Kollman, Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N⊕log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- (a) R. H. Byrd, P. Lu and J. Nocedal, A limited memory algorithm for bound constrained optimization, SIAM J. Sci. Statistic. Comput., 1995, 16, 1190–1208 CrossRef; (b) C. Zhu, R. H. Byrd and J. Nocedal, Algorithm 778: L-BFGS-B: Fortran subroutines for large-scale bound-constrained optimization, ACM Trans. Math. Softw., 1997, 23, 550–560 CrossRef.
- M. P. Allen and D. J. Tildesley, Computer Simulation of Liquids, Clarendon Press, Oxford, 1987 Search PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Pálla, J. C. Smith, B. Hess and E. Lindahl, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1-2, 19–25 CrossRef.
- R. A. Jarvis and E. A. Patrick, Clustering Using a Similarity Measure Based on Shared Near Neighbors, IEEE Trans. Comput., 1973, C22, 1025–1034 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- (a) S. Grimme, J. Antony, S. Ehrlich and S. Krieg, A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H–Pu, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed; (b) A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS; (c) C. Lee, W. Yang and R. G. Parr, Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals, J. Chem. Phys., 1985, 82, 299–310 CrossRef CAS.
- S. F. Boys and F. Bernardi, The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors, Mol. Phys., 1970, 19, 553–566 CrossRef CAS.
- M. J. Frisch; G. W. Trucks; H. B. Schlegel; G. E. Scuseria; M. A. Robb; J. R. Cheeseman; G. Scalmani; V. Barone; G. A. Petersson; H. Nakatsuji; X. Li; M. Caricato; A. V. Marenich; J. Bloino; B. G. Janesko; R. Gomperts; B. Mennucci; H. P. Hratchian; J. V. Ortiz; A. F. Izmaylov; J. L. Sonnenberg; D. Williams-Young; F. Ding; F. Lipparini; F. Egidi; J. Goings; B. Peng; A. Petrone; T. Henderson; D. Ranasinghe; V. G. Zakrzewski; J. Gao; N. Rega; G. Zheng; W. Liang; M. Hada; M. Ehara; K. Toyota; R. Fukuda; J. Hasegawa; M. Ishida; T. Nakajima; Y. Honda; O. Kitao; T. Nakai; K. Vreven; J. A. Throssell; J. E. Montgomery, Jr.; F. Peralta; M. J. Ogliaro; H. Bearpark; J. J. Heyd; E. N. Brothers; K. N. Kudin; V. N. Staroverov; T. A. Keith; R. Kobayashi; J. Normand; K. Raghavachari; A. P. Rendell; J. C. Burant; S. S. Iyengar; J. Tomasi; M. Cossi; J. M. Millam; M. Klene; C. Adamo; R. Cammi; J. W. Ochterski; R. L. Martin; K. Morokuma; O. Farkas; J. B. Foresman and D. J. Fox, Gaussian 16, Revision B.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- (a) J. H. Poupaert and P. Couvreur, A computationally derived structural model of doxorubicin interacting with oligomeric polyalkylcyanoacrylate in nanoparticles, J. Controlled Release, 2003, 92, 19–26 CrossRef CAS; (b) B. Jawad, L. Poudel, R. Podgornik, N. F. Steinmetze and W.-Y. Ching, Molecular mechanism and binding free energy of doxorubicin intercalation in DNA, Phys. Chem. Chem. Phys., 2019, 211, 3877–3893 RSC; (c) K. R. Karnati and Y. Wang, Understanding the co-loading and releasing of doxorubicin and paclitaxel using chitosan functionalized single-walled carbon nanotubes by molecular dynamics simulations, Phys. Chem. Chem. Phys., 2018, 20, 9389–9400 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Section 1: investigated model NP-DBP-2DOX; Section 2: adsorption configurations in the associate NP-DBP-2DOX; Section 3: analysis of the MD trajectories; Section 4: analysis of MD trajectories at higher pH. See DOI: https://doi.org/10.1039/d2cp00707j |
| This journal is © the Owner Societies 2022 |